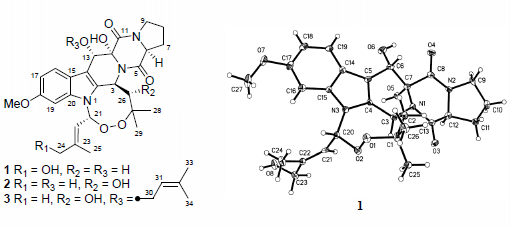
Prenylated Indolediketopiperazine Peroxides and Related Homologues from the Marine Sediment-Derived Fungus Penicillium brefeldianum SD-273
Abstract
:1. Introduction
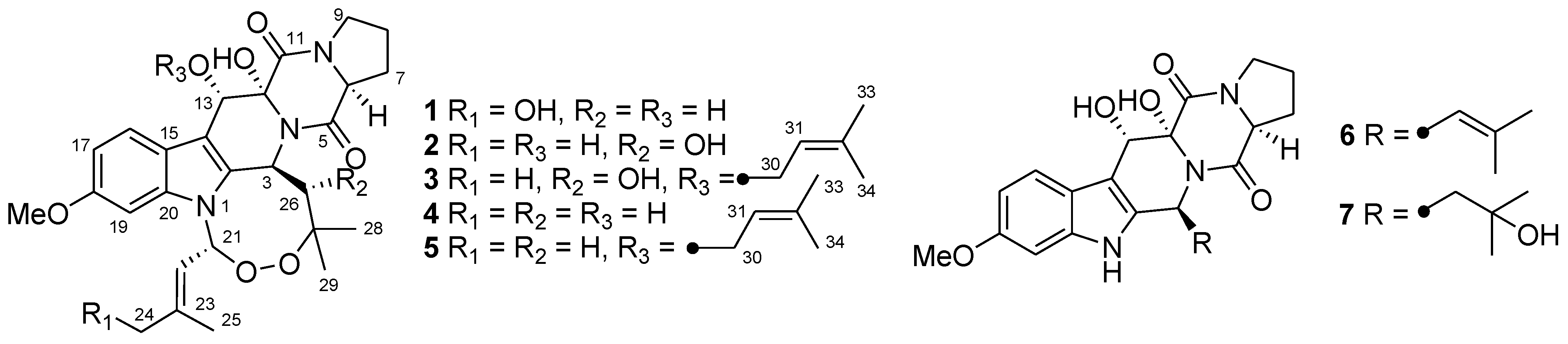
2. Results and Discussion
2.1. Structure Elucidation of the New Compounds 1–3
| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 2 | 131.4, C | 129.6, C | 129.8, C | |||
| 3 | 5.89, d (10.0) | 48.3, CH | 6.13, d (8.8) | 49.1, CH | 6.20, d (9.0) | 48.4, CH |
| 5 | 171.1, C | 170.7, C | 171.4, C | |||
| 6 | 4.45, t (8.0) | 58.8, CH | 4.35, t (7.9) | 58.7, CH | 4.45, t (8.2) | 58.9, CH |
| 7 | α 2.30, m β 1.88, m | 29.3, CH2 | α 2.33, m β 1.90, m | 29.5, CH2 | α 2.30, m β 1.87, m | 29.2, CH2 |
| 8 | 1.94, m | 22.6, CH2 | 1.90, m | 22.1, CH2 | 1.87, m | 22.5, CH2 |
| 9 | α 3.44, m β 3.50, t, 8.2 | 45.5, CH2 | α 3.44, m β 3.52, m | 44.8, CH2 | α 3.40, m β 3.52, m | 45.2, CH2 |
| 11 | 166.5, C | 166.0, C | 165.5, C | |||
| 12 | 82.1, C | 82.6, C | 84.1, C | |||
| 13 | 5.40, s | 68.4, CH | 5.43, s | 68.1, CH | 5.12, s | 73.5, CH |
| 14 | 107.9, C | 107.8, C | 107.5, C | |||
| 15 | 121.3, C | 120.6, C | 120.4, C | |||
| 16 | 7.72, d (8.7) | 121.8, CH | 7.72, d (9.3) | 121.1, CH | 7.57, d (8.7) | 120.3, CH |
| 17 | 6.69, dd (8.7, 1.3) | 109.3, CH | 6.69, dd (9.3, 2.1) | 108.7, CH | 6.74, dd (8.7, 2.2) | 108.8, CH |
| 18 | 156.0, C | 155.3, C | 155.3, C | |||
| 19 | 6.75, d (1.3) | 94.3, CH | 6.70, d (2.1) | 93.4, CH | 6.69, d (2.2) | 93.6, CH |
| 20 | 136.3, C | 135.3, C | 135.3, C | |||
| 21 | 6.88, d (8.4) | 85.5, CH | 6.78, d (8.2) | 85.0, CH | 6.77, d (8.2) | 85.1, CH |
| 22 | 5.26, d (8.4) | 116.0, CH | 5.06, d (8.2) | 118.3, CH | 5.02, d (8.2) | 118.1, CH |
| 23 | 146.9, C | 142.8, C | 143.0, C | |||
| 24 | 3.80, s | 65.2 CH2 | 1.73, s | 25.2, CH3 | 1.74, s | 25.4, CH3 |
| 25 | 1.87, s | 14.5, CH3 | 2.00, s | 18.3, CH3 | 2.00, s | 18.4, CH3 |
| 26 | α 1.91, m β 1.55, m | 51.3, CH2 | 2.92, d (8.8) | 80.0, CH | 2.92, d (9.0) | 79.3, CH |
| 27 | 83.2, C | 85.0, C | 85.2, C | |||
| 28 | 1.57, s | 24.6, CH3 | 1.44, s | 18.4, CH3 | 1.41, s | 18.3, CH3 |
| 29 | 0.95, s | 27.2, CH3 | 0.99, s | 25.3, CH3 | 0.98, s | 25.2, CH3 |
| 30 | 4.56, dd (7.0, 11.0) 4.88, dd (6.5, 11.0) | 68.5, CH2 | ||||
| 31 | 5.57, t (6.5) | 122.2, CH | ||||
| 32 | 134.5, C | |||||
| 33 | 1.75, s | 18.1, CH3 | ||||
| 34 | 1.77, s | 25.2, CH3 | ||||
| 18-OCH3 | 3.75, s | 55.8, CH3 | 3.76, s | 55.2, CH3 | 3.76, s | 55.2, CH3 |
| 12-OH | 5.10, br s b | 5.26, br s b | 6.16, br s | |||
| 13-OH | 6.68, br s b | 6.32, br s b | ||||
| 24-OH | 3.34, br s | |||||
| 26-OH | 5.52, br s | 5.37, br s | ||||
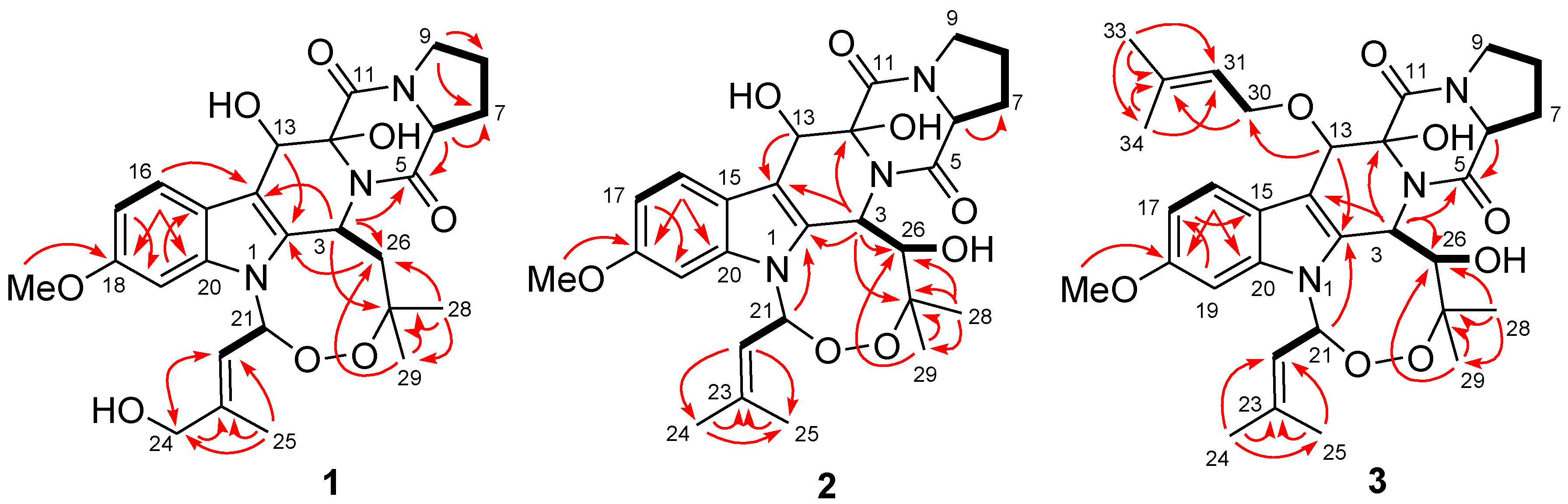


2.2. Biological Activities of the Isolated Compounds
3. Experimental Section
3.1. General
3.2. Fungal Material
3.3. Fermentation
3.4. Extraction and Isolation
 −12.5 (c 0.24, MeOH); UV (MeOH) λmax (log ε) 223 (4.51), 268 (3.70), 288 (3.64) nm; 1H and 13C NMR data, see Table 1; ESIMS m/z 550 [M + Na]+; HRESIMS m/z 550.2153 [M + Na]+ (calcd for C27H33N3O8Na+, 550.2165, Δ −1.2 mmu).
−12.5 (c 0.24, MeOH); UV (MeOH) λmax (log ε) 223 (4.51), 268 (3.70), 288 (3.64) nm; 1H and 13C NMR data, see Table 1; ESIMS m/z 550 [M + Na]+; HRESIMS m/z 550.2153 [M + Na]+ (calcd for C27H33N3O8Na+, 550.2165, Δ −1.2 mmu). −75.5 (c 0.23, MeOH); UV (MeOH) λmax (log ε) 223 (4.45), 271 (3.59), 287 (3.54) nm; 1H and 13C NMR data, see Table 1; ESIMS m/z 550 [M + Na]+; HRESIMS m/z 550.2156 [M + Na]+ (calcd for C27H33N3O8Na+, 550.2165, Δ −0.9 mmu).
−75.5 (c 0.23, MeOH); UV (MeOH) λmax (log ε) 223 (4.45), 271 (3.59), 287 (3.54) nm; 1H and 13C NMR data, see Table 1; ESIMS m/z 550 [M + Na]+; HRESIMS m/z 550.2156 [M + Na]+ (calcd for C27H33N3O8Na+, 550.2165, Δ −0.9 mmu). −10.1 (c 0.35, MeOH); UV (MeOH) λmax (log ε) 227 (4.60), 275 (3.88), 289 (3.80) nm; 1H and 13C NMR data, see Table 1; ESIMS m/z 618 [M + Na]+; HRESIMS m/z 618.2804 [M + Na]+ (calcd for C32H41N3O8Na+, 618.2791, Δ +1.3 mmu).
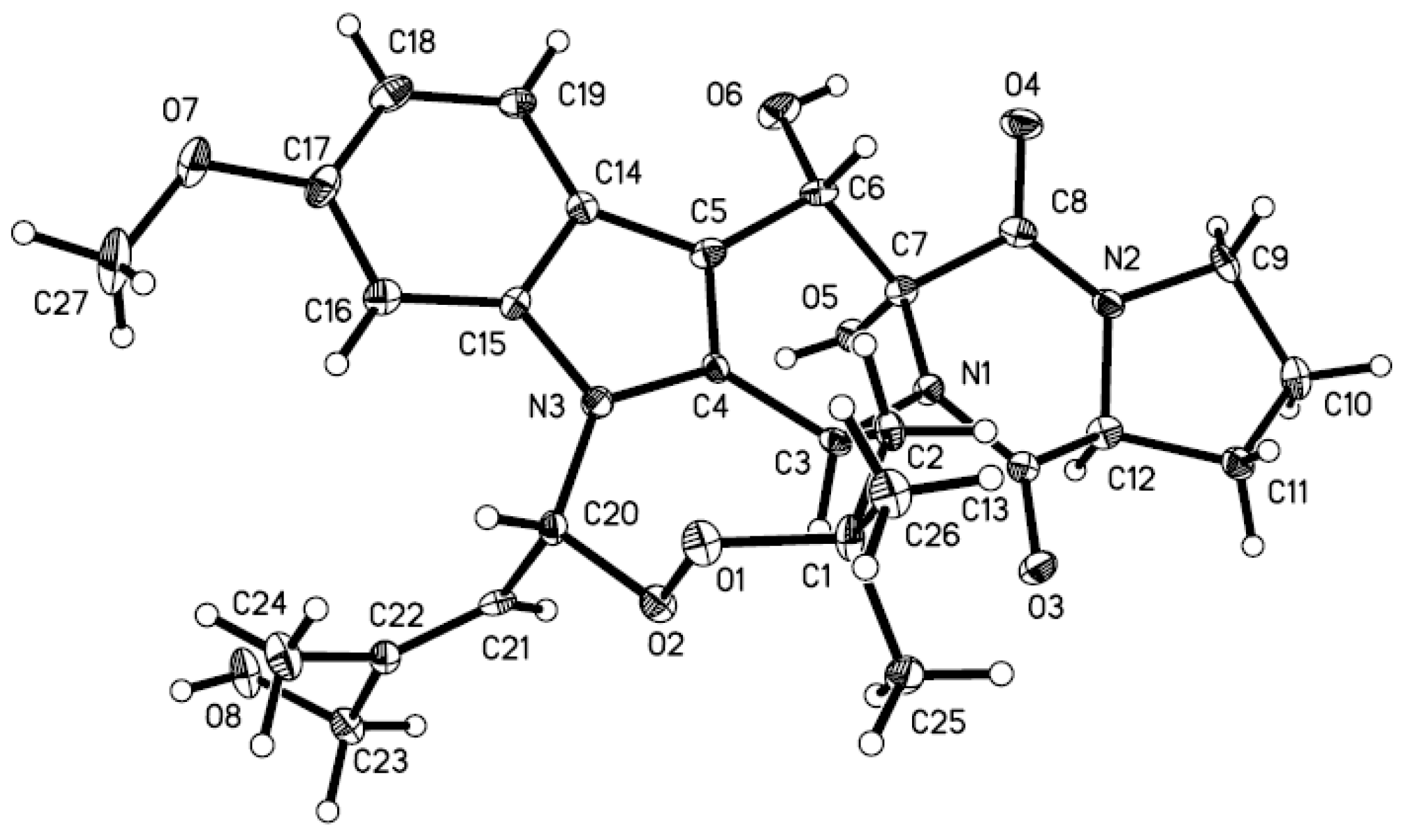
−10.1 (c 0.35, MeOH); UV (MeOH) λmax (log ε) 227 (4.60), 275 (3.88), 289 (3.80) nm; 1H and 13C NMR data, see Table 1; ESIMS m/z 618 [M + Na]+; HRESIMS m/z 618.2804 [M + Na]+ (calcd for C32H41N3O8Na+, 618.2791, Δ +1.3 mmu).3.5. X-ray Crystallographic Analysis of Compound 1
3.6. Amino Acid Analysis
3.7. Brine Shrimp Toxicity
3.8. Cytotoxicity Assay
3.9. Antibacterial Assay
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Wang, W.L.; Lu, Z.Y.; Tao, H.W.; Zhu, T.J.; Fang, Y.C.; Gu, Q.Q.; Zhu, W.M. Isoechinulin-type alkaloids, variecolorins A–L, from halotolerant Aspergillus variecolor. J. Nat. Prod. 2007, 70, 1558–1564. [Google Scholar] [CrossRef]
- Yamazaki, M.; Fujimoto, H.; Kawasaki, T. Chemistry of tremorogenic metabolites. I. Fumitremorgin A from Aspergillus fumigatus. Chem. Pharm. Bull. 1980, 28, 245–254. [Google Scholar] [CrossRef]
- Cui, C.B.; Kakeya, H.; Osada, H. Novel mammalian cell cycle inhibitors, tryprostatins A, B and other diketopiperazines produced by Aspergillus fumigatus II. Physico-chemical properties and structures. J. Antibiot. 1996, 49, 534–540. [Google Scholar] [CrossRef]
- Cole, R.J.; Kirksey, J.W.; Cox, R.H.; Clardy, J. Structure of the tremor-producing indole, TR-2. J. Agric. Food Chem. 1975, 23, 1015–1018. [Google Scholar] [CrossRef]
- Fayos, J.; Lokensgard, D.; Clardy, J.; Cole, R.J.; Kirksey, J.W. Structure of verruculogen, a tremor producing peroxide from Penicillium verruculosum. J. Am. Chem. Soc. 1974, 21, 6785–6787. [Google Scholar]
- Du, F.Y.; Li, X.M.; Li, C.S.; Shang, Z.; Wang, B.G. Cristatumins A–D, new indole alkaloids from the marine-derived endophytic fungus Eurotium cristatum EN-220. Bioorg. Med. Chem. Lett. 2012, 22, 4650–4653. [Google Scholar] [CrossRef]
- Wang, F.; Fang, Y.; Zhu, T.; Zhang, M.; Lin, A.; Gu, Q.; Zhu, W. Seven new prenylated indole diketopiperazine alkaloids from holothurian-derived fungus Aspergillus fumigatus. Tetrahedron 2008, 64, 7986–7991. [Google Scholar] [CrossRef]
- Willingale, J.; Perera, K.P.W.C.; Mantle, P.G. An intermediary role for the tremorgenic mycotoxin TR-2 in the biosynthesis of verruculogen. Biochem. J. 1983, 214, 991–993. [Google Scholar]
- An, C.Y.; Li, X.M.; Li, C.S.; Wang, M.H.; Xu, G.M.; Wang, B.G. Aniquinazolines A–D, four new quinazolinone alkaloids from marine-derived endophytic fungus Aspergillus nidulans. Mar. Drugs 2013, 11, 2682–2694. [Google Scholar] [CrossRef]
- Wang, M.H.; Li, X.M.; Li, C.S.; Ji, N.Y.; Wang, B.G. Secondary metabolites from Penicillium pinophilum SD-272, a marine sediment-derived fungus. Mar. Drugs 2013, 11, 2230–2238. [Google Scholar] [CrossRef]
- Li, C.S.; Li, X.M.; Gao, S.S.; Lu, Y.H.; Wang, B.G. Cytotoxic anthranilic acid derivatives from deep sea sediment-derived fungus Penicillium paneum SD-44. Mar. Drugs 2013, 11, 3068–3076. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, H.; Shang, Z.; Jiao, B.; Yuan, B.; Sun, W.; Wang, B.; Miao, M.; Huang, C. SD118-xanthocillin X (1), a novel marine agent extracted from Penicillium commune, induces autophagy through the inhibition of the MEK/ERK pathway. Mar. Drugs 2012, 10, 1345–1359. [Google Scholar] [CrossRef]
- An, C.Y.; Li, X.M.; Li, C.S.; Gao, S.S.; Shang, Z.; Wang, B.G. Triazoles and other N-containing metabolites from the marine-derived endophytic fungus Penicillium chrysogenum EN-118. Helv. Chim. Acta 2013, 96, 682–687. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.M.; Shang, Z.; Li, C.S.; Ji, N.Y.; Wang, B.G. Cytoglobosins A–G, cytochalasans from a marine-derived endophytic fungus, Chaetomium globosum QEN-14. J. Nat. Prod. 2010, 73, 729–733. [Google Scholar]
- Sun, H.F.; Li, X.M.; Meng, L.; Cui, C.M.; Gao, S.S.; Li, C.S.; Huang, C.G.; Wang, B.G. Asperolides A–C, tetranorlabdane diterpenoids from the marine alga-derived endophytic fungus Aspergillus wentii EN-48. J. Nat. Prod. 2012, 75, 148–152. [Google Scholar]
- Wang, S.; Li, X.M.; Teuscher, F.; Li, D.L.; Diesel, A.; Ebel, R.; Proksch, P.; Wang, B.G. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceola. J. Nat. Prod. 2006, 69, 1622–1625. [Google Scholar]
- Houbraken, J.; Samson, R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 2011, 70, 1–51. [Google Scholar] [CrossRef]
- Cambridge Crystallographic Data Centre (CCDC). Deposition No. CCDC923360. Available online: http://www.ccdc.cam.ac.uk/data_request/cif (accessed on 6 February 2013).
- Sheldrick, G.M. SADABS, Software for Empirical Absorption Correction; University of Göttingen: Göttingen, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. SHELXTL, Structure Determination Software Programs; Bruker Analytical X-ray System Inc.: Madison, WI, USA, 1997. [Google Scholar]
- Sheldrick, G.M. SHELXL-97 and SHELXS-97,Program for X-ray Crystal Structure Solution and Refinement; University of Göttingen: Göttingen, Germany, 1997. [Google Scholar]
- Meyer, B.N.; Ferrigni, N.R.; Putnam, J.E.; Jacobsen, L.B.; Nichols, D.E.; McLaughlin, J.L. Brine shrimp: A convenient general bioassay for active plant constituents. Planta Med. 1982, 45, 31–34. [Google Scholar] [CrossRef]
- Bergeron, R.J.; Cavanaugh, P.F., Jr.; Kline, S.J.; Hughes, R.G., Jr.; Elliott, G.T.; Porter, C.W. Antineoplastic and antiherpetic activity of spermidine catecholamide iron chelators. Biochem. Biophys. Res. Commun. 1984, 121, 848–854. [Google Scholar] [CrossRef]
- Al-Burtamani, S.K.S.; Fatope, M.O.; Marwah, R.G.; Onifade, A.K.; Al-Saidi, S.H. Chemical composition, antibacterial and antifungal activities of the essential oil of Haplophyllum tuberculatum from Oman. J. Ethnopharmacol. 2005, 96, 107–112. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
An, C.-Y.; Li, X.-M.; Li, C.-S.; Xu, G.-M.; Wang, B.-G. Prenylated Indolediketopiperazine Peroxides and Related Homologues from the Marine Sediment-Derived Fungus Penicillium brefeldianum SD-273. Mar. Drugs 2014, 12, 746-756. https://doi.org/10.3390/md12020746
An C-Y, Li X-M, Li C-S, Xu G-M, Wang B-G. Prenylated Indolediketopiperazine Peroxides and Related Homologues from the Marine Sediment-Derived Fungus Penicillium brefeldianum SD-273. Marine Drugs. 2014; 12(2):746-756. https://doi.org/10.3390/md12020746
Chicago/Turabian StyleAn, Chun-Yan, Xiao-Ming Li, Chun-Shun Li, Gang-Ming Xu, and Bin-Gui Wang. 2014. "Prenylated Indolediketopiperazine Peroxides and Related Homologues from the Marine Sediment-Derived Fungus Penicillium brefeldianum SD-273" Marine Drugs 12, no. 2: 746-756. https://doi.org/10.3390/md12020746
APA StyleAn, C.-Y., Li, X.-M., Li, C.-S., Xu, G.-M., & Wang, B.-G. (2014). Prenylated Indolediketopiperazine Peroxides and Related Homologues from the Marine Sediment-Derived Fungus Penicillium brefeldianum SD-273. Marine Drugs, 12(2), 746-756. https://doi.org/10.3390/md12020746