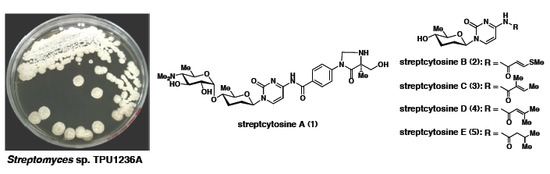
Anti-Mycobacterial Nucleoside Antibiotics from a Marine-Derived Streptomyces sp. TPU1236A
Abstract
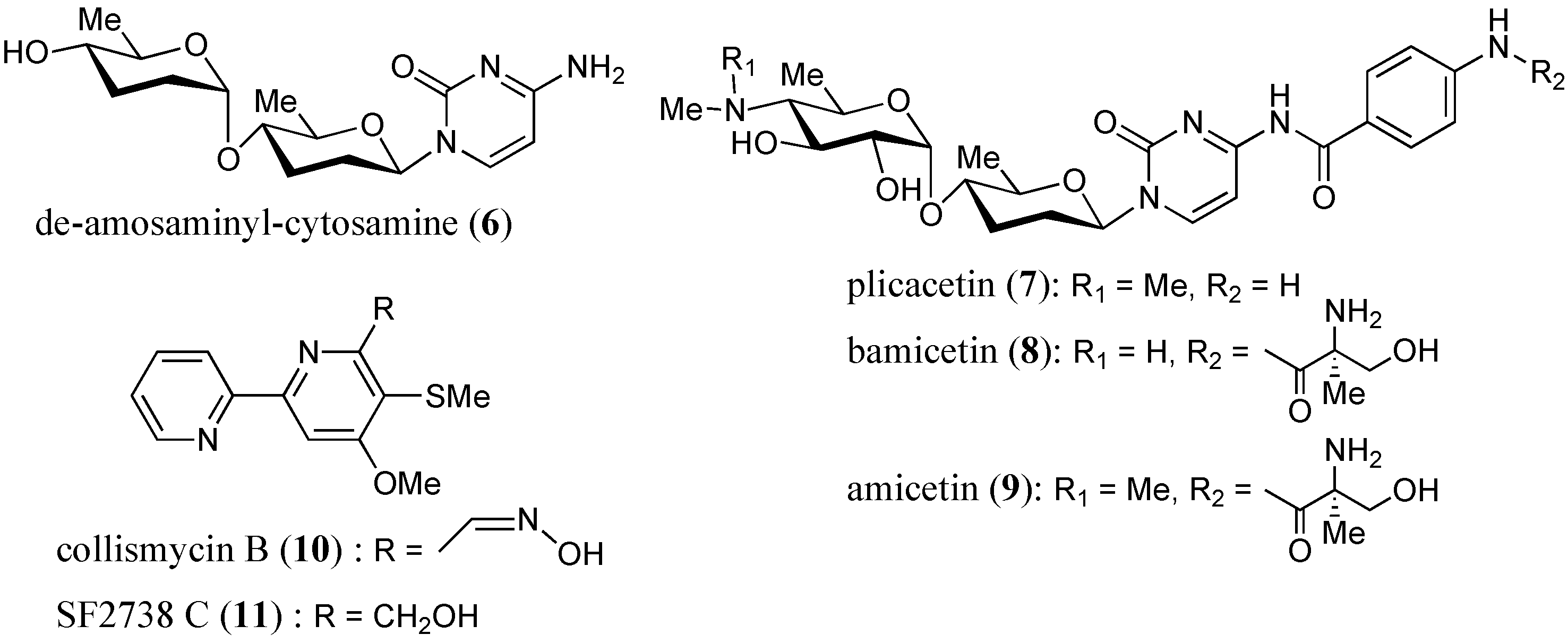
:1. Introduction

2. Results and Discussion
2.1. Structure of Streptcytosine A (1)

| C# | δC | δH, mult. (J in Hz) | HMBC |
|---|---|---|---|
| 2 | 157.4 | ||
| 4 | 165.0 | ||
| 5 | 99.1 | 7.58, d, (7.5) | 6 |
| 6 | 146.7 | 8.21, d, (7.5) | 2, 4, 5, 1′ |
| 8 | 168.4 | ||
| 9 | 131.6 | ||
| 10 | 130.8 | 8.10, d, (8.8) | 8, 11, 12, 14 |
| 11 | 120.6 | 7.87, d, (8.8) | 10, 13 |
| 12 | 142.0 | ||
| 13 | 120.6 | 7.87, d, (8.8) | 11, 14 |
| 14 | 130.8 | 8.10, d, (8.8) | 8, 10, 12, 13 |
| 16 | 172.2 | ||
| 17 | 68.3 | ||
| 19 | 66.1 | (a) 3.83, d, (11.6) | 16, 17 |
| (b) 4.03. d, (11.6) | 16 | ||
| 20 | 17.6 | 1.57, s | 16, 17, 19 |
| 21 | 61.1 | 5.28, br s | 16, 17 |
| 1′ | 84.9 | 5.79, d, (7.8) | |
| 2′ | 31.1 | (a) 2.19, br d, (9.0) | 4′ |
| (b) 1.71, m | 4′ | ||
| 3′ | 28.1 | (a) 1.71, m | 1′ |
| (b) 2.41, m | |||
| 4′ | 76.7 | 3.46, m | |
| 5′ | 78.4 | 3.77, dq, (9.0, 6.0) | 4′ |
| 6′ | 19.3 | 1.39, d, (6.0) | 4′, 5′ |
| 1″ | 96.8 | 5.03, d, (3.6) | 4′, 3″, 5″ |
| 2″ | 74.0 | 3.56, dd, (9.1, 3.6) | 3″ |
| 3″ | 68.1 | 3.98, dd, (11.0, 9.1) | 2″, 4″ |
| 4″ | 72.1 | 3.12, dd (11.0, 10.0) | 3″, 5″, 6″, 7″, 8″ |
| 5″ | 64.2 | 4.10, dq, (10.0, 6.2) | |
| 6″ | 19.1 | 1.47, d, (6.2) | 4″, 5″ |
| 7″ | 42.6 | 3.01, s | 4″, 8″ |
| 8″ | 42.6 | 3.01, s | 4″, 7″ |
2.2. Structures of Streptcytosines B–E (2–5)
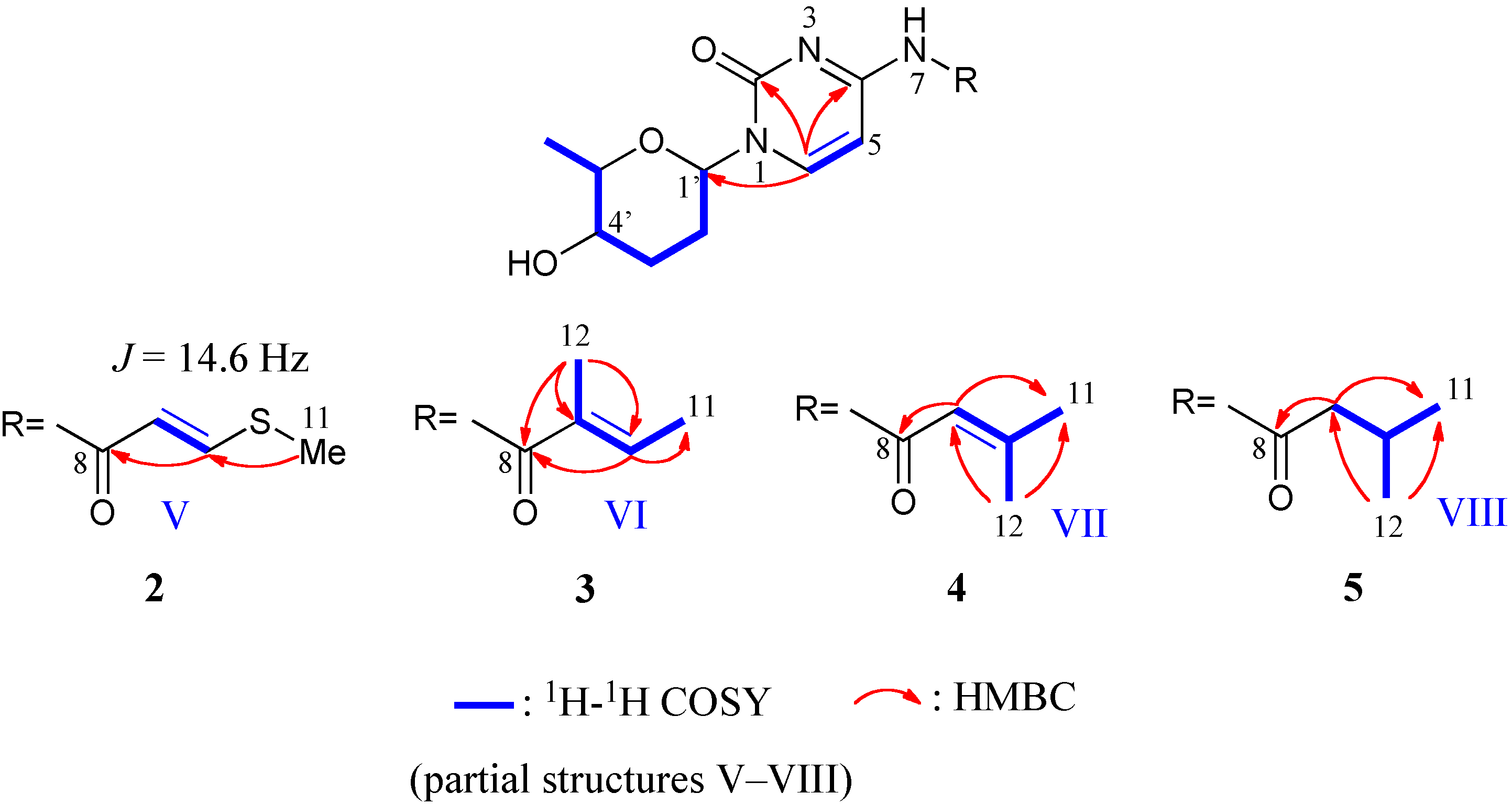
| 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|
| C# | δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) |
| 2 | 156.6 | 155.5 | 154.2 | 157.8 | ||||
| 4 | 164.6 | 164.2 | 164.3 | 164.8 | ||||
| 5 | 98.4 | 7.34, d (7.5) | 98.3 | 7.30, d (7.6) | 98.2 | 7.26, d (7.5) | 99.2 | 7.47, br d (7.6) |
| 6 | 146.8 | 8.19, d (7.5) | 147.5 | 8.23, d (7.6) | 147.0 | 8.21, d (7.5) | 146.7 | 8.12, d (7.6) |
| 8 | 166.2 | 171.3 | 168.2 | 176.5 | ||||
| 9 | 115.7 | 6.07, d (14.6) | 133.5 | 118.7 | 5.95, qq (1.3, 1.3) | 47.3 | 2.32, d (7.2) | |
| 10 | 150.7 | 8.01, d (14.6) | 137.4 | 6.71, q (7.0) | 161.3 | 27.1 | 2.10, m | |
| 11 | 14.7 | 2.42, s | 14.7 | 1.89, d (7.0) | 28.0 | 1.99, d (1.2) | 22.7 | 0.99, d (6.7) |
| 12 | –– | –– | 12.2 | 1.90, s | 20.8 | 2.24, d (1.2) | 22.7 | 0.99, d (6.7) |
| 1′ | 85.0 | 5.71, dd (9.9, 2.1) | 85.0 | 5.71, dd (10.1, 2.2) | 84.9 | 5.71, dd (10.0, 2.2) | 84.9 | 5.70, dd (9.9, 1.8) |
| 2′ | 32.4 | (a) 2.15, m (b) 1.67, m | 32.4 | (a) 2.16, m (b) 1.67, m | 32.4 | (a) 2.15, m (b) 1.67, m | 32.4 | (a) 2.15, m (b) 1.66, m |
| 3′ | 31.7 | (a) 1.67, m (b) 2.13, m | 31.7 | (a) 1.67, m (b) 2.13, m | 31.6 | (a) 1.67, m (b) 2.12, m | 31.7 | (a) 1.66, m (b) 2.12, m |
| 4′ | 71.8 | 3.28, m | 71.7 | 3.28, m | 71.8 | 3.27, m | 71.8 | 3.28, m |
| 5′ | 80.7 | 3.51, dq (9.1, 6.1) | 80.7 | 3.51, dq (9.2, 6.2) | 80.6 | 3.51, dq (9.1. 6.1) | 80.6 | 3.50, dq (9.2, 6.2) |
| 6′ | 18.6 | 1.34, d (6.1) | 18.6 | 1.34, d (6.2) | 18.6 | 1.33, d (6.1) | 18.6 | 1.33, d (6.2) |
2.3. Anti-Mycobacterial Activity of Compounds 1–11
| Compound | 5 μg/disc | 10 μg/disc | MIC (μg/mL) |
|---|---|---|---|
| 1 | 9 | 12 | 32 |
| 2 | –– a | –– | n.d.b |
| 3 | –– | –– | n.d. |
| 4 | –– | –– | n.d. |
| 5 | –– | –– | n.d. |
| 6 | –– | –– | n.d. |
| 7 | 9 | 13 | 32 |
| 8 | 18 | 24 | 16 |
| 9 | 21 | 26 | 8 |
| 10 | –– | 12 | >64 |
| 11 | 9 | 12 | 64 |
| streptomycin sulfate | 30 | 38 | 0.50 |
3. Experimental Section
3.1. General Experimental Procedures
3.2. Isolation, Identification, and Fermentation of Producing Bacterium
3.3. Extraction and Isolation of Compounds 1–11
3.4. Anti-Mycobacterial Assay
4. Conclusions
Acknowledgments
Authors Contribution
Conflicts of Interest
References
- Zumla, A.; Raviglione, M.; Hafner, R.; von Reyn, C.F. Tuberculosis. N. Engl. J. Med. 2013, 368, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, G.; Pasca, M.R. Trends in discovery of new drugs for tuberculosis therapy. J. Antibiot. 2014, 67, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Ntie-Kang, F.; Yong, J.N.; Owono, L.C.; Sippl, W.; Megnassan, E. Perspectives on tuberculosis pathogenesis and discovery of anti-tubercular drugs. Curr. Med. Chem. 2014, 21, 3466–3477. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Global Tuberculosis Report 2013. Available online: http://www.who.int/tb/publications/global_report/en/ (accessed on 12 October 2014).
- Espinal, M.A. The global situation of MDR-TB. Tuberculosis 2003, 83, 44–51. [Google Scholar]
- Ryan, N.J.; Lo, J.H. Delamanid: first global approval. Drugs 2014, 74, 1041–1045. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.S.; Lakshmanan, M. Delamanid: A new armor in combating drug-resistant tuberculosis. J. Pharmacol. Pharmacother. 2014, 5, 222–224. [Google Scholar] [CrossRef] [PubMed]
- Reyrat, J.M.; Kahn, D. Mycobacterium smegmatis: An absurd model for tuberculosis? Trends Microbiol. 2001, 9, 472–473. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, H.; Zhang, G.; Li, S.; Zhang, G.; Zhu, Y.; Liu, J.; Zhang, C. Characterizing amosamine biosynthesis in amicetin reveals AmiG as a reversible retaining glocosyltransferase. J. Am. Chem. Soc. 2013, 135, 12152–12155. [Google Scholar] [CrossRef] [PubMed]
- Haskell, T.H.; Ryder, A.; Frohardt, R.P.; Fusari, S.A.; Jakubowski, Z.L.; Bartz, Q.R. The isolation and characterization of three crystalline antibiotics from Streptomyces plicatus. J. Am. Chem. Soc. 1958, 80, 743–747. [Google Scholar] [CrossRef]
- DeBoer, C.; Caron, E.L.; Hinman, J.W. Amicetin, a new streptomyces antibiotic. J. Am. Chem. Soc. 1953, 75, 499–500. [Google Scholar] [CrossRef]
- Hinman, J.W.; Caron, E.L.; DeBoer, C. The isolation and purification of amicetin. J. Am. Chem. Soc. 1953, 75, 5864–5866. [Google Scholar] [CrossRef]
- Flynn, E.H.; Hinman, J.W.; Caron, E.L.; Woolf, D.O., Jr. The chemistry of Amicetin, a new antibiotic. J. Am. Chem. Soc. 1953, 75, 5867–5871. [Google Scholar] [CrossRef]
- Shindo, K.; Yamagishi, Y.; Okada, Y.; Kawai, H. Collismycins A and B, novel non-steroidal inhibitors of dexamethasone-glucocorticoid receptor binding. J. Antibiot. 1994, 47, 1072–1074. [Google Scholar] [CrossRef] [PubMed]
- Gomi, S.; Amano, S.; Sato, E.; Miyadoh, S.; Kodama, Y. Novel antibiotics SF2738A, B and C, and their analogs produced by Streptomyces sp. J. Antibiot. 1994, 47, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Shammas, C.; Donarski, J.A.; Ramesh, V. NMR structure of the peptidyl transferase RNA inhibitor antibiotic amicetin. Magn. Reson. Chem. 2007, 45, 133–141. [Google Scholar] [CrossRef]
- Haskell, T.H. Amicetin, bamicetin and plicacetin. Chemical studies. J. Am. Chem. Soc. 1958, 80, 747–751. [Google Scholar] [CrossRef]
- Shiomi, K.; Haneda, K.; Tomoda, H.; Iwai, Y.; Omura, S. Cytosaminomycins, new anticoccidial agents produced by Streptomyces sp. KO-8119. II. Structure elucidation of cytosaminomycins A, B, C and D. J. Antibiot. 1994, 47, 782–786. [Google Scholar]
- Ericsson, H. The paper disc method for determination of bacterial sensitivity to antibiotics. Studies on the accuracy of the technique. Scand. J. Clin. Lab. Invest. 1960, 12, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Amsterdam, D. Susceptibility testing of antimicrobials in liquid media. In Antibiotics in Laboratory Medicine, 4th ed.; Loman, V., Ed.; Williams & Wilkins: Baltimore, MD, USA, 1996; pp. 52–111. [Google Scholar]
- Mincer, T.J.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 2002, 68, 5005–5011. [Google Scholar] [CrossRef] [PubMed]
- Koyama, N.; Kojima, S.; Nonaka, K.; Masuma, R.; Matsumoto, M.; Omura, S.; Tomoda, H. Calpinactam, a new anti-mycobacterial agent, produced by Mortierella alpina FKI-4905. J. Antibiot. 2010, 63, 183–186. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bu, Y.-Y.; Yamazaki, H.; Ukai, K.; Namikoshi, M. Anti-Mycobacterial Nucleoside Antibiotics from a Marine-Derived Streptomyces sp. TPU1236A. Mar. Drugs 2014, 12, 6102-6112. https://doi.org/10.3390/md12126102
Bu Y-Y, Yamazaki H, Ukai K, Namikoshi M. Anti-Mycobacterial Nucleoside Antibiotics from a Marine-Derived Streptomyces sp. TPU1236A. Marine Drugs. 2014; 12(12):6102-6112. https://doi.org/10.3390/md12126102
Chicago/Turabian StyleBu, Ying-Yue, Hiroyuki Yamazaki, Kazuyo Ukai, and Michio Namikoshi. 2014. "Anti-Mycobacterial Nucleoside Antibiotics from a Marine-Derived Streptomyces sp. TPU1236A" Marine Drugs 12, no. 12: 6102-6112. https://doi.org/10.3390/md12126102
APA StyleBu, Y.-Y., Yamazaki, H., Ukai, K., & Namikoshi, M. (2014). Anti-Mycobacterial Nucleoside Antibiotics from a Marine-Derived Streptomyces sp. TPU1236A. Marine Drugs, 12(12), 6102-6112. https://doi.org/10.3390/md12126102