Triphlorethol-A from Ecklonia cava Up-Regulates the Oxidant Sensitive 8-Oxoguanine DNA Glycosylase 1
Abstract
:1. Introduction
2. Results
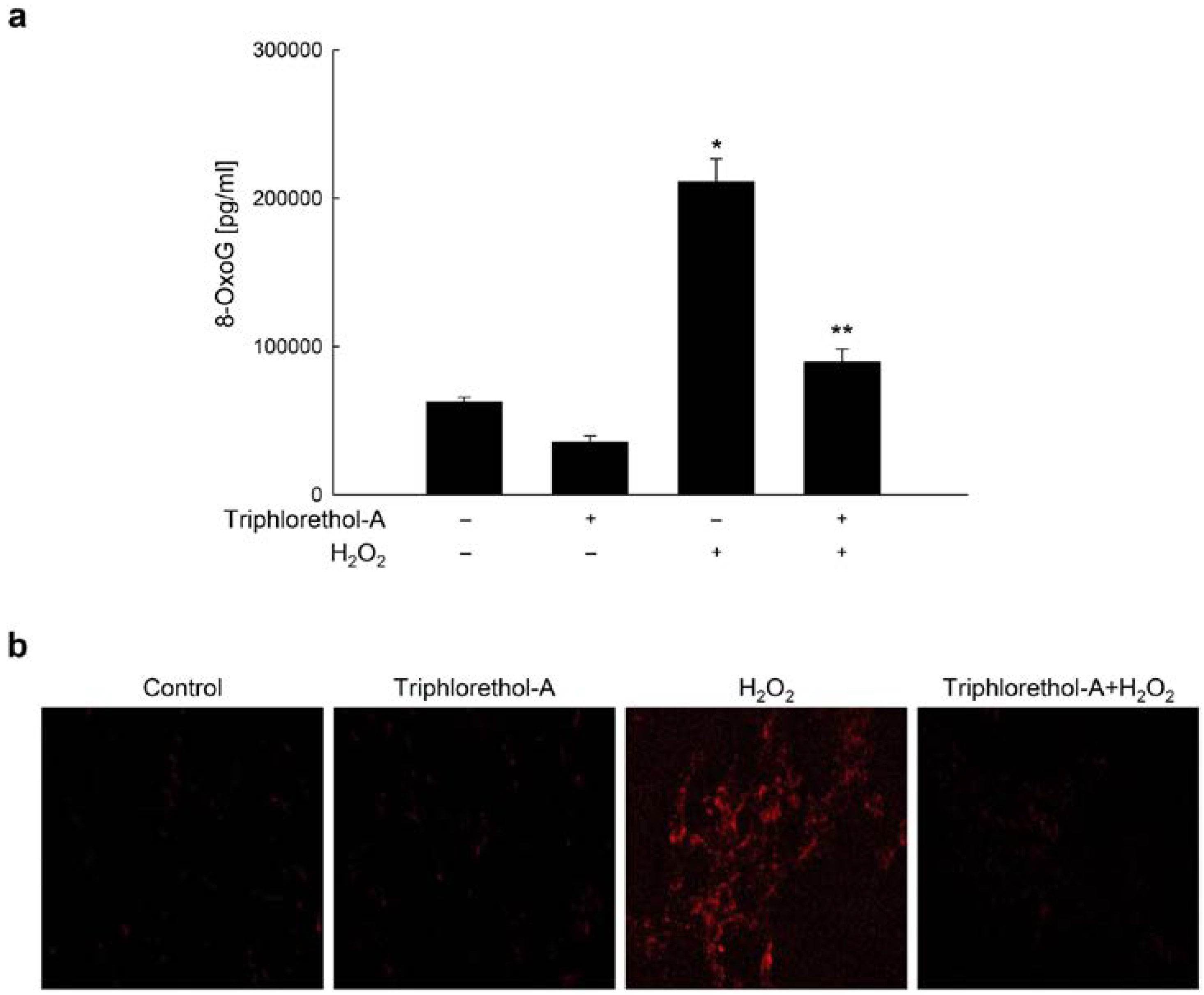
2.1. Triphlorethol-A Suppresses H2O2-induced 8-oxoG Formation
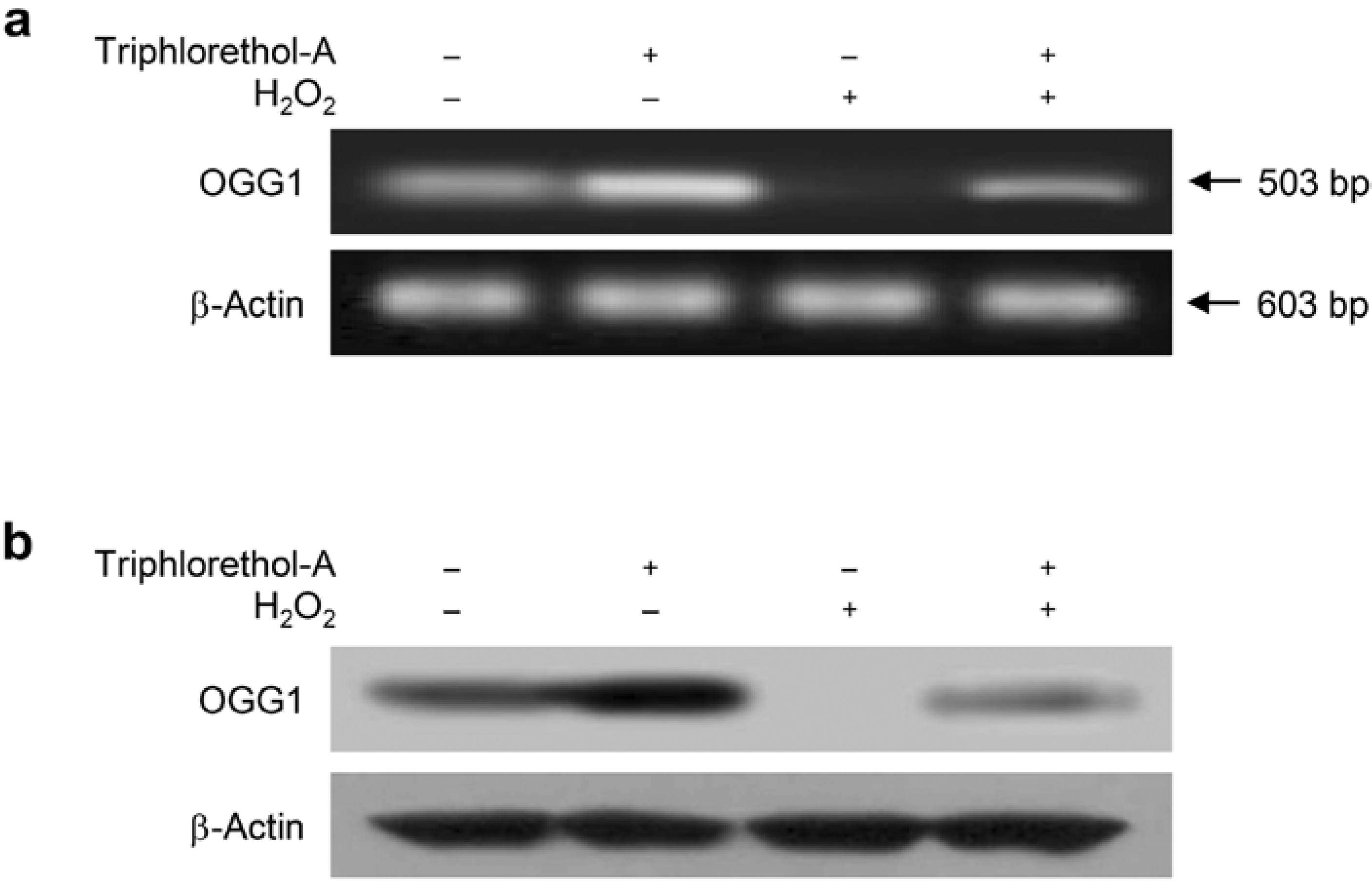
2.2. Triphlorethol-A Reverses the Suppression of OGG1 mRNA and Protein Expression by H2O2 Treatment
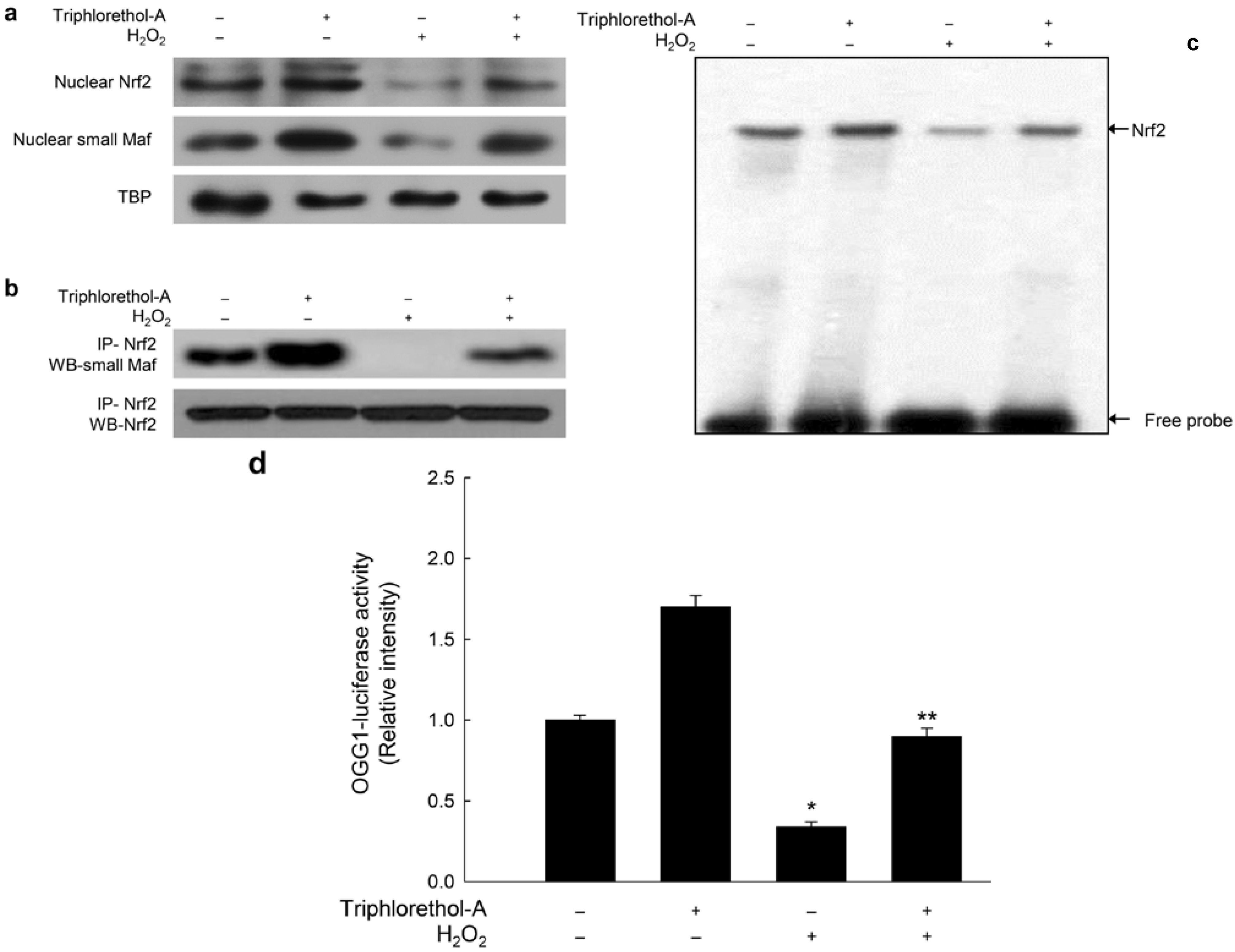
2.3. Triphlorethol-A Blocks Inhibition of OGG1 Transcription by H2O2 Treatment
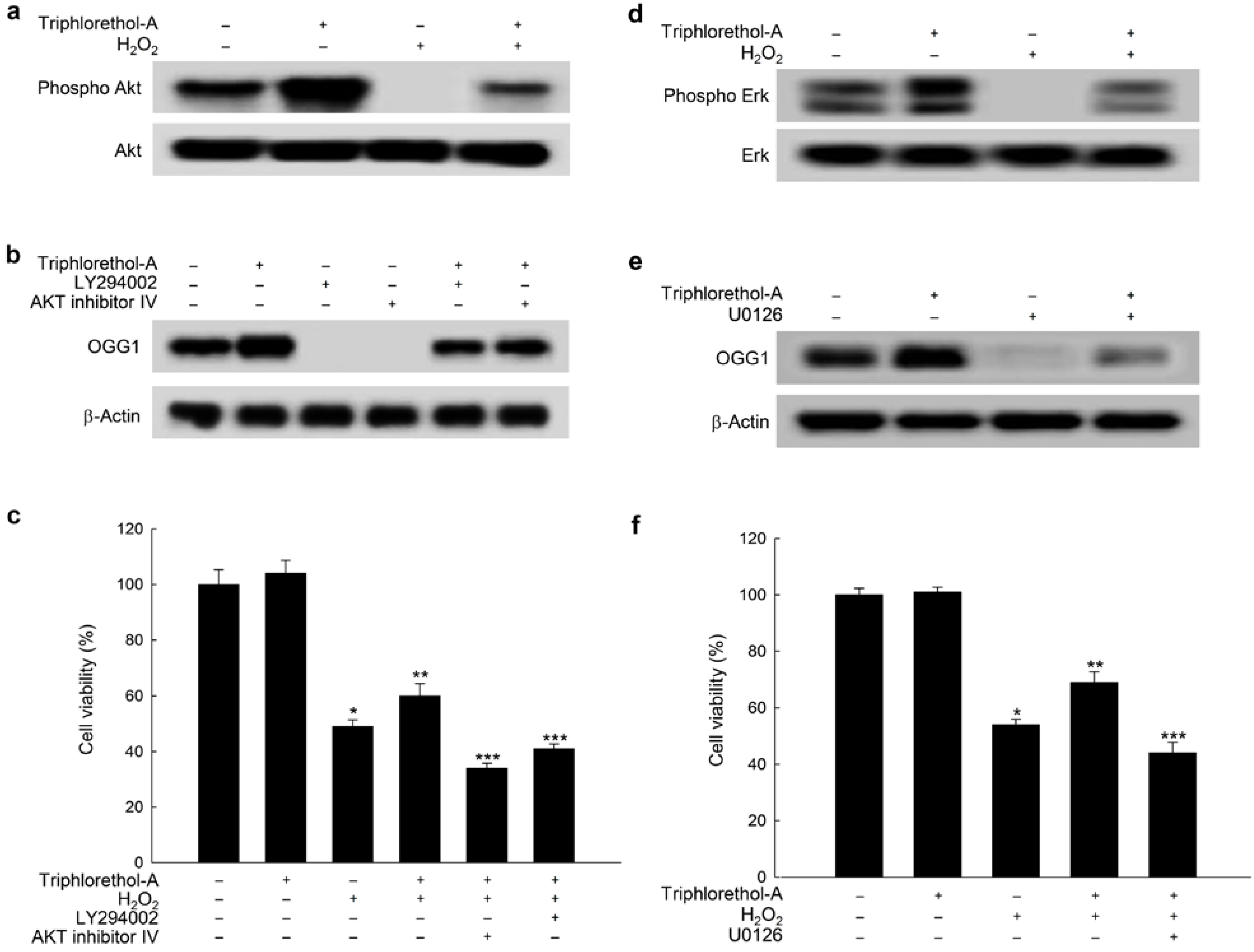
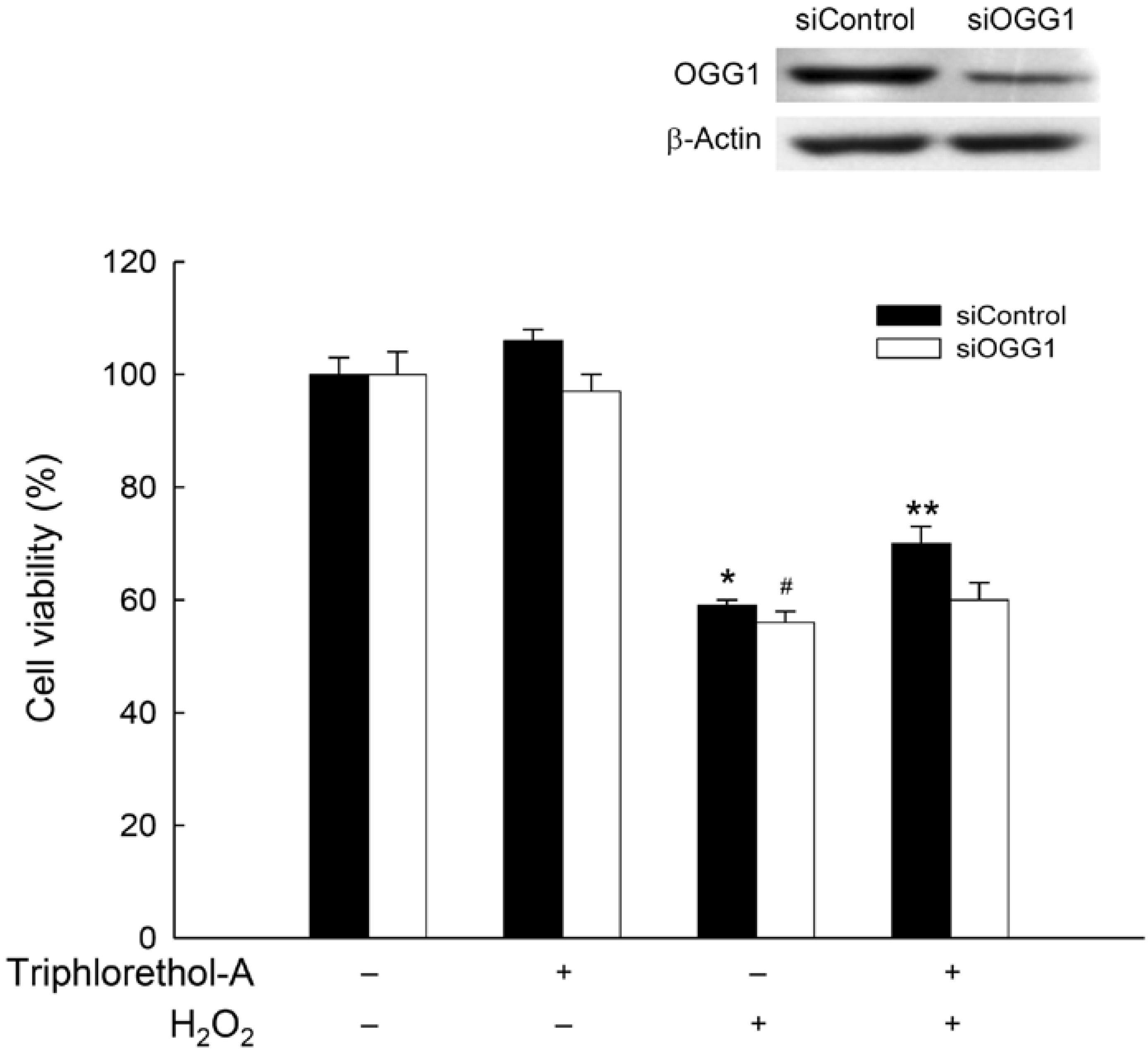
2.4. Triphlorethol-A Induces OGG1 Expression via the PI3K/Akt and Erk Pathways
3. Experimental Section
3.1. Reagents
3.2. Isolation of Triphlorethol-A
3.3. Cell Culture
3.4. Detection of 8-oxoG
3.5. Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
3.6. Extraction of Total Cellular Proteins and a Nucleus Protein Fraction
3.7. Western Blotting
3.8. Immunoprecipitation
3.9. Electrophoretic Mobility Shift Assay (EMSA)
3.10. Transient Transfection and OGG1-Promoter Luciferase Assay
3.11. Transient Transfection of Small Interfering RNA (siRNA)
3.12. Cell Viability
3.13. Statistical Analysis
4. Discussion and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Saul, R.L.; Ames, B.N. Background levels of DNA damage in the population. Basic Life Sci. 1986, 38, 529–535. [Google Scholar]
- Fortini, P.; Pascucci, B.; Parlanti, E.; D’Errico, M.; Simonelli, V.; Dogliotti, E. 8-Oxoguanine DNA damage: At the crossroad of alternative repair pathways. Mutat. Res. 2003, 531, 127–139. [Google Scholar] [CrossRef]
- Loft, S.; Fischer-Nielsen, A.; Jeding, I.B.; Vistisen, K.; Poulsen, H.E. 8-Hydroxydeoxyguanosine as a urinary biomarker of oxidative DNA damage. J. Toxicol. Environ. Health 1993, 40, 391–404. [Google Scholar]
- Honda, M.; Yamada, Y.; Tomonaga, M.; Ichinose, H.; Kamihira, S. Correlation of urinary 8-hydroxy-2'-deoxyguanosine (8-OHdG), a biomarker of oxidative DNA damage, and clinical features of hematological disorders: A pilot study. Leuk. Res. 2000, 24, 461–468. [Google Scholar] [CrossRef]
- Kikuchi, A.; Takeda, A.; Onodera, H.; Kimpara, T.; Hisanaga, K.; Sato, N.; Nunomura, A.; Castellani, R.J.; Perry, G.; Smith, M.A.; et al. Systemic increase of oxidative nucleic acid damage in Parkinson’s Disease and multiple system atrophy. Neurobiol. Dis. 2002, 9, 244–248. [Google Scholar] [CrossRef]
- Matsui, M.; Nishigori, C.; Toyokuni, S.; Takada, J.; Akaboshi, M.; Ishikawa, M.; Imamura, S.; Miyachi, Y. The role of oxidative DNA damage in human arsenic carcinogenesis: Detection of 8-hydroxy-2'-deoxyguanosine in arsenic-related Bowen’s Disease. J. Invest. Dermatol. 1999, 113, 26–31. [Google Scholar] [CrossRef]
- Sato, S.; Mizuno, Y.; Hattori, N. Urinary 8-hydroxydeoxyguanosine levels as a biomarker for progression of Parkinson Disease. Neurology 2005, 64, 1081–1083. [Google Scholar] [CrossRef]
- Wu, L.L.; Chiou, C.C.; Chang, P.Y.; Wu, J.T. Urinary 8-OHdG: A marker of oxidative stress to DNA and a risk factor for cancer, atherosclerosis and diabetics. Clin. Chim. Acta 2004, 339, 1–9. [Google Scholar] [CrossRef]
- Boiteux, S.; O’Connor, T.R.; Lederer, F.; Gouyette, A.; Laval, J. Homogeneous Escherichia Coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 1990, 265, 3916–3922. [Google Scholar] [PubMed]
- Krokan, H.E.; Nilsen, H.; Skorpen, F.; Otterlei, M.; Slupphaug, G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000, 476, 73–77. [Google Scholar] [CrossRef]
- Bogliolo, M.; Cappelli, E.; D'Osualdo, A.; Rossi, O.; Barbieri, O.; Kelley, M.R.; Frosina, G. Effect of S. Cerevisiae APN1 protein on mammalian DNA base excision repair. Anticancer Res. 2003, 23, 3727–3734. [Google Scholar] [PubMed]
- de Souza-Pinto, N.C.; Eide, L.; Hogue, B.A.; Thybo, T.; Stevnsner, T.; Seeberg, E.; Klungland, A.; Bohr, V.A. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 2001, 61, 5378–5381. [Google Scholar]
- Dhénaut, A.; Boiteux, S.; Radicella, J.P. Characterization of the hOGG1 promoter and its expression during the cell cycle. Mutat. Res. 2000, 461, 109–118. [Google Scholar] [CrossRef]
- Ueta, E.; Sasabe, E.; Yang, Z.; Osaki, T.; Yamamoto, T. Enhancement of apoptotic damage of squamous cell carcinoma cells by inhibition of the mitochondrial DNA repairing system. Cancer Sci. 2008, 99, 2230–2237. [Google Scholar] [CrossRef]
- Keum, Y.S. Regulation of Nrf2-mediated phase II detoxification and anti-oxidant genes. Biomol. Ther. Seoul 2012, 20, 144–151. [Google Scholar] [CrossRef]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/Small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef]
- Motohashi, H.; Katsuoka, F.; Engel, J.D.; Yamamoto, M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 6379–6384. [Google Scholar] [CrossRef]
- Nakaso, K.; Yano, H.; Fukuhara, Y.; Takeshima, T.; Wada-Isoe, K.; Nakashima, K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003, 546, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, Y.; Sternberg, P.; Cai, J. Essential roles of the PI3 Kinase/Akt pathway in regulating Nrf2-dependent antioxidant functions in the RPE. Invest. Ophthalmol. Vis. Sci. 2008, 49, 1671–1678. [Google Scholar] [CrossRef]
- Zipper, L.M.; Mulcahy, R.T. Erk Activation is required for Nrf2 nuclear localization during pyrrolidine dithiocarbamate induction of glutamate cysteine ligase modulatory gene expression in HepG2 cells. Toxicol. Sci. 2003, 73, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Lee, K.H.; Park, J.W.; Lee, N.H.; Na, H.K.; Surh, Y.J.; You, H.J.; Chung, M.H.; Hyun, J.W. Triphlorethol-A induces heme oxygenase-1 via activation of ERK and NF-E2 related factor 2 transcription factor. FEBS Lett. 2007, 581, 2000–2008. [Google Scholar] [CrossRef]
- Zhang, R.; Kang, K.A.; Piao, M.J.; Kim, K.C.; Lee, N.H.; You, H.J.; Hyun, J.W. Triphlorethol-A improves the non-homologous end joining and base-excision repair capacity impaired by formaldehyde. J. Toxicol. Environ. Health A 2011, 74, 811–821. [Google Scholar] [CrossRef]
- Piao, M.J.; Zhang, R.; Lee, N.H.; Hyun, J.W. Protective effect of triphlorethol-A against ultraviolet B-mediated damage of human keratinocytes. J. Photochem. Photobiol. B. 2012, 106, 74–80. [Google Scholar] [CrossRef]
- Kang, K.A.; Lee, K.H.; Chae, S.; Koh, Y.S.; Yoo, B.S.; Kim, J.H.; Ham, Y.M.; Baik, J.S.; Lee, N.H.; Hyun, J.W. Triphlorethol-A from Ecklonia Cava protects V79-4 lung fibroblast against hydrogen peroxide induced cell damage. Free Radic. Res. 2005, 39, 883–892. [Google Scholar] [CrossRef]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, C. 8-Hydroxy-2'-deoxyguanosine (8-ohdg): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health. C. Environ. Carcinog. Ecotoxicol. Rev. 2009, 27, 120–139. [Google Scholar] [CrossRef] [PubMed]
- Struthers, L.; Patel, R.; Clark, J.; Thomas, S. Direct detection of 8-oxodeoxyguanosine and 8-oxoguanine by avidin and its analogues. Anal. Biochem. 1998, 255, 20–31. [Google Scholar] [CrossRef]
- Mo, J.; Xia, Y.; Wade, T.J.; Schmitt, M.; Le, X.C.; Dang, R.; Mumford, J.L. Chronic arsenic exposure and oxidative stress: OGG1 expression and arsenic exposure, nail selenium, and skin hyperkeratosis in inner Mongolia. Environ. Health Perspect. 2006, 114, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Sinha, D.; Biswas, J.; Bishayee, A. Nrf2-mediated redox signaling in arsenic carcinogenesis: A review. Arch. Toxicol. 2012, 87, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Kodama, M.; Miura, I.; Kinzyo, Z.; Kido, M.; Mori, H.; Nakayama, Y.; Takahashi, M. Structure of anti-plasmin inhibitor, eckol, isolated from the brown alga, Ecklonia kurome Okamura and inhibitory activities of its derivatives on plasma plasmin inhibitors. Chem. Pharm. Bull. 1989, 37, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, J.; DeGraff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: Assessment of chemosensitivity testing. Cancer Res. 1987, 47, 936–942. [Google Scholar] [PubMed]
- Mena, S.; Ortega, A.; Estrela, J.M. Oxidative stress in environmental-induced carcinogenesis. Mutat. Res. 2009, 674, 36–44. [Google Scholar] [CrossRef]
- Thannickal, V.J.; Fanburg, B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000, 279, L1005–L1028. [Google Scholar] [PubMed]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Hyun, J.W.; Choi, J.Y.; Zeng, H.H.; Lee, Y.S.; Kim, H.S.; Yoon, S.H.; Chung, M.H. Leukemic cell line, kg-1 has a functional loss of hOGG1 enzyme due to a point mutation and 8-hydroxydeoxyguanosine can kill KG-1. Oncogene 2000, 19, 4476–4479. [Google Scholar] [CrossRef]
- Hyun, J.W.; Jung, Y.C.; Kim, H.S.; Choi, E.Y.; Kim, J.E.; Yoon, B.H.; Yoon, S.H.; Lee, Y.S.; Choi, J.; You, H.J.; et al. 8-Hydroxydeoxyguanosine causes death of human leukemia cells deficient in 8-oxoguanine glycosylase 1 activity by inducing apoptosis. Mol. Cancer. Res. 2003, 1, 290–299. [Google Scholar]
- Barzilai, A.; Yamamoto, K. DNA damage responses to oxidative stress. DNA Repair Amst. 2004, 3, 1109–1115. [Google Scholar] [CrossRef]
- McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003, 278, 21592–21600. [Google Scholar]
- Davies, K.J. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life 2000, 50, 279–289. [Google Scholar] [CrossRef]
- Kang, K.A.; Zhang, R.; Lee, K.H.; Chae, S.; Kim, B.J.; Kwak, Y.S.; Park, J.W.; Lee, N.H.; Hyun, J.W. Protective effect of triphlorethol-a from Ecklonia Cava against ionizing radiation in vitro. J. Radiat. Res. Tokyo 2006, 47, 61–68. [Google Scholar] [CrossRef]
- Zhang, R.; Lee, I.K.; Kang, K.A.; Piao, M.J.; Kim, K.C.; Kim, B.J.; Lee, N.H.; Choi, J.Y.; Choi, J.; Hyun, J.W. Cytoprotective effects of triphlorethol-A against formaldehyde-induced oxidative damage and apoptosis: Role of mitochondria-mediated caspase-dependent pathway. J. Toxicol. Environ. Health A 2010, 73, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Sykiotis, G.P.; Bohmann, D. Stress-activated Cap'N'Collar transcription factors in aging and human disease. Sci. Signal. 2010, 3, re3. [Google Scholar]
- Boutten, A.; Goven, D.; Artaud-Macari, E.; Boczkowski, J.; Bonay, M. Nrf2 targeting: A promising therapeutic strategy in chronic obstructive pulmonary disease. Trends Mol. Med. 2011, 17, 363–371. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.C.; Lee, I.K.; Kang, K.A.; Piao, M.J.; Ryu, M.J.; Kim, J.M.; Lee, N.H.; Hyun, J.W. Triphlorethol-A from Ecklonia cava Up-Regulates the Oxidant Sensitive 8-Oxoguanine DNA Glycosylase 1. Mar. Drugs 2014, 12, 5357-5371. https://doi.org/10.3390/md12115357
Kim KC, Lee IK, Kang KA, Piao MJ, Ryu MJ, Kim JM, Lee NH, Hyun JW. Triphlorethol-A from Ecklonia cava Up-Regulates the Oxidant Sensitive 8-Oxoguanine DNA Glycosylase 1. Marine Drugs. 2014; 12(11):5357-5371. https://doi.org/10.3390/md12115357
Chicago/Turabian StyleKim, Ki Cheon, In Kyung Lee, Kyoung Ah Kang, Mei Jing Piao, Min Ju Ryu, Jeong Mi Kim, Nam Ho Lee, and Jin Won Hyun. 2014. "Triphlorethol-A from Ecklonia cava Up-Regulates the Oxidant Sensitive 8-Oxoguanine DNA Glycosylase 1" Marine Drugs 12, no. 11: 5357-5371. https://doi.org/10.3390/md12115357
APA StyleKim, K. C., Lee, I. K., Kang, K. A., Piao, M. J., Ryu, M. J., Kim, J. M., Lee, N. H., & Hyun, J. W. (2014). Triphlorethol-A from Ecklonia cava Up-Regulates the Oxidant Sensitive 8-Oxoguanine DNA Glycosylase 1. Marine Drugs, 12(11), 5357-5371. https://doi.org/10.3390/md12115357



