Oxygenated Polyketides from Plakinastrella mamillaris as a New Chemotype of PXR Agonists
Abstract
:1. Introduction
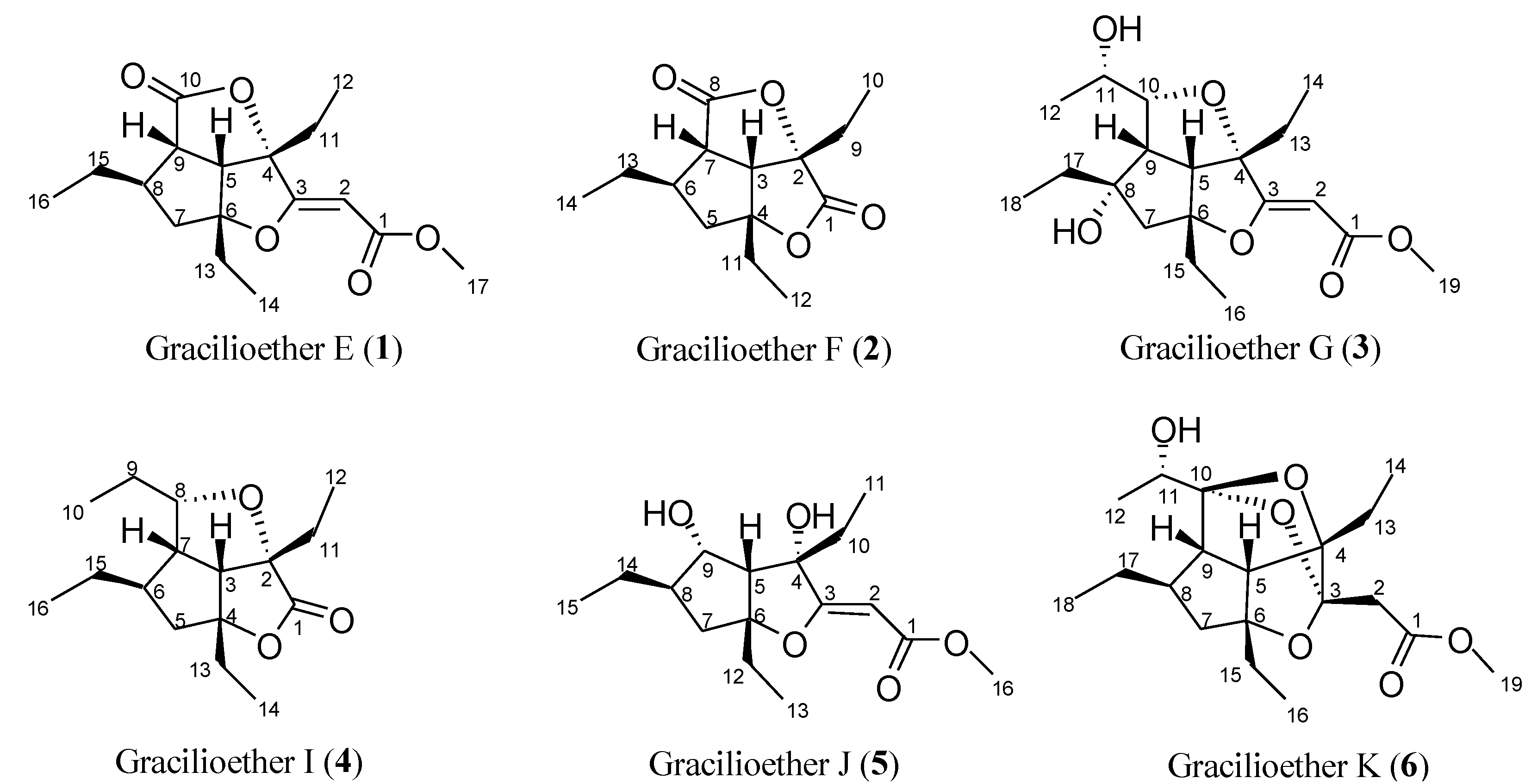
2. Results and Discussion
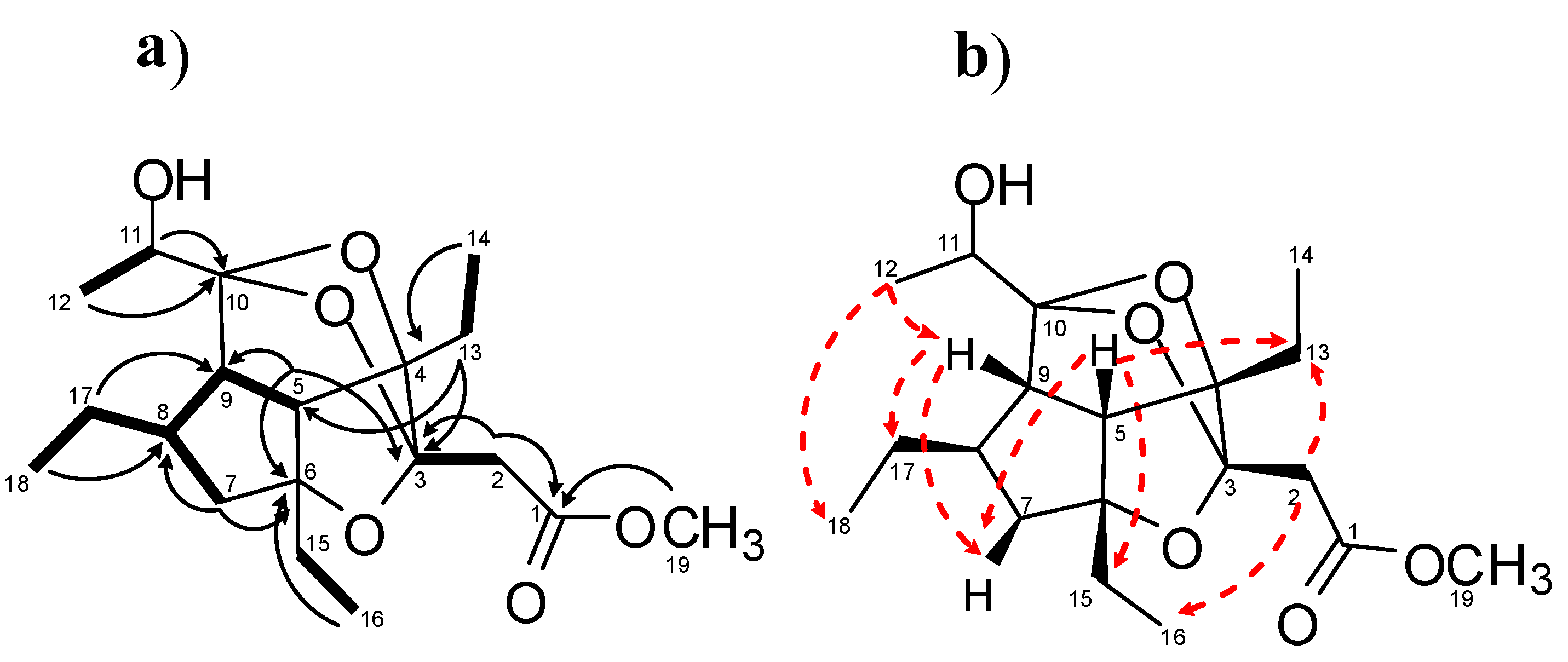
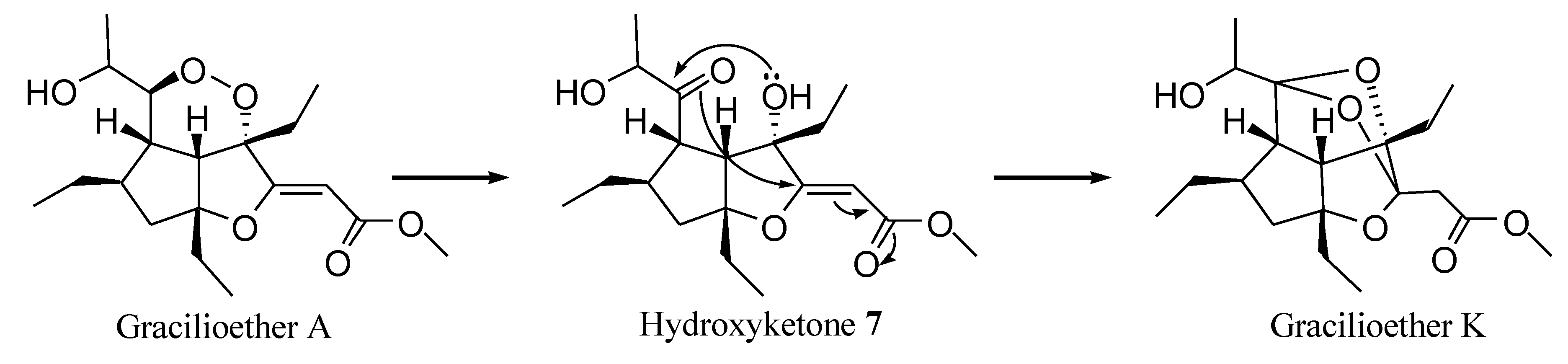
2.1. Isolation of Gracilioethers and Structural Characterization of Gracilioether K (6)
| Position | δH a | δC | HMBC | ROESY b |
|---|---|---|---|---|
| 1 | - | 171.1 | ||
| 2 | 2.63 d (14.2) 2.71 d (14.2) | 40.2 | C1, C3 | H2-13, H3-16 |
| 3 | - | 110.49 | ||
| 4 | - | 96.7 | ||
| 5 | 2.80 d (10.5) | 54.6 | C3, C6, C9, C15 | H-7b, H2-13, H3-14, H2-15, H3-16 |
| 6 | - | 93.8 | ||
| 7 | 1.51 m 2.28 dd (7.6, 13.8) | 47.9 | C5, C6, C8, C9 | H-5, H-9, H3-16 |
| 8 | 2.06 ovl | 39.4 | ||
| 9 | 2.74 dd (3.7, 10.5) | 59.0 | H-7b, H3-12, H2-17, H3-18 | |
| 10 | - | 110.51 | ||
| 11 | 3.88 q (6.5) | 66.0 | C10 | |
| 12 | 1.22 d (6.5) | 17.7 | C10, C11 | H-9, H3-18 |
| 13 | 1.74 quint (7.5) 2.07 ovl | 21.9 | C4, C14 C3, C4, C5 | H2-2, H-5 H2-2 |
| 14 | 1.07 t (7.4) | 9.1 | C4 | H-5 |
| 15 | 1.55 m 1.65 quint (7.3) | 32.6 | C5, C7 | H-5 H-5 |
| 16 | 0.93 t (7.5) | 9.8 | C6 | H2-2, H-5, H-7b |
| 17 | 1.32 m 1.53 m | 30.5 | C7, C8, C9, C18 | H-9 H-9 |
| 18 | 0.91 t (7.4) | 12.9 | C8 | H-9, H3-12 |
| 19 | 3.62 s | 52.2 | C1 |
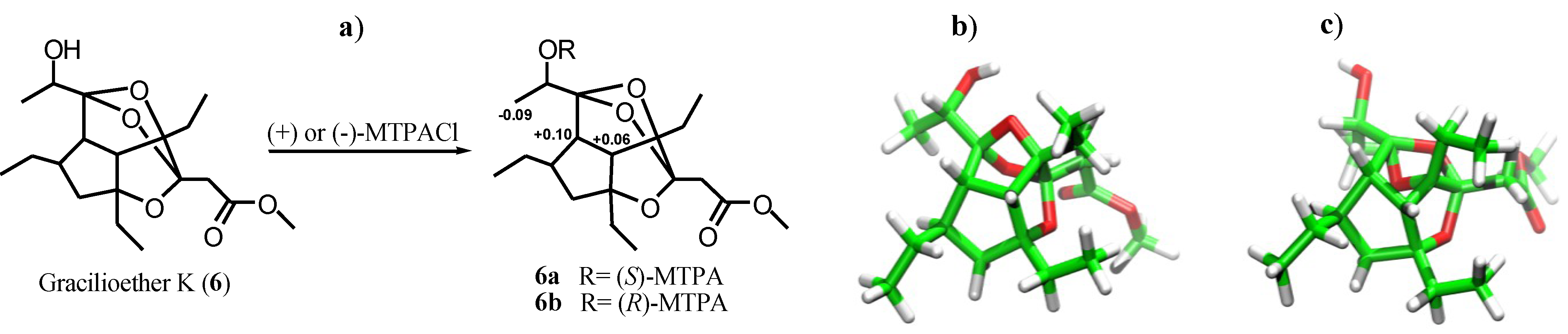
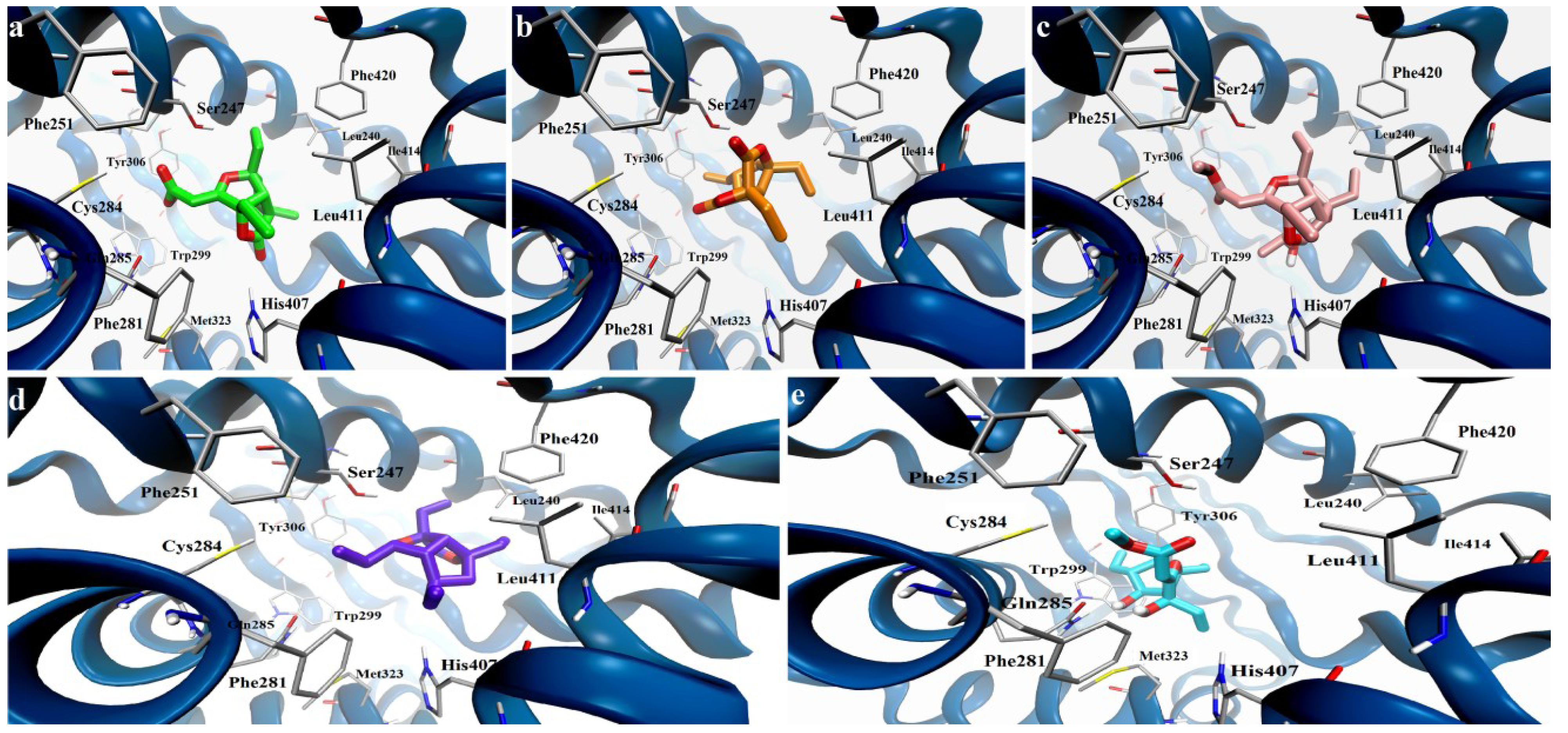
2.2. Pharmacological Evaluation
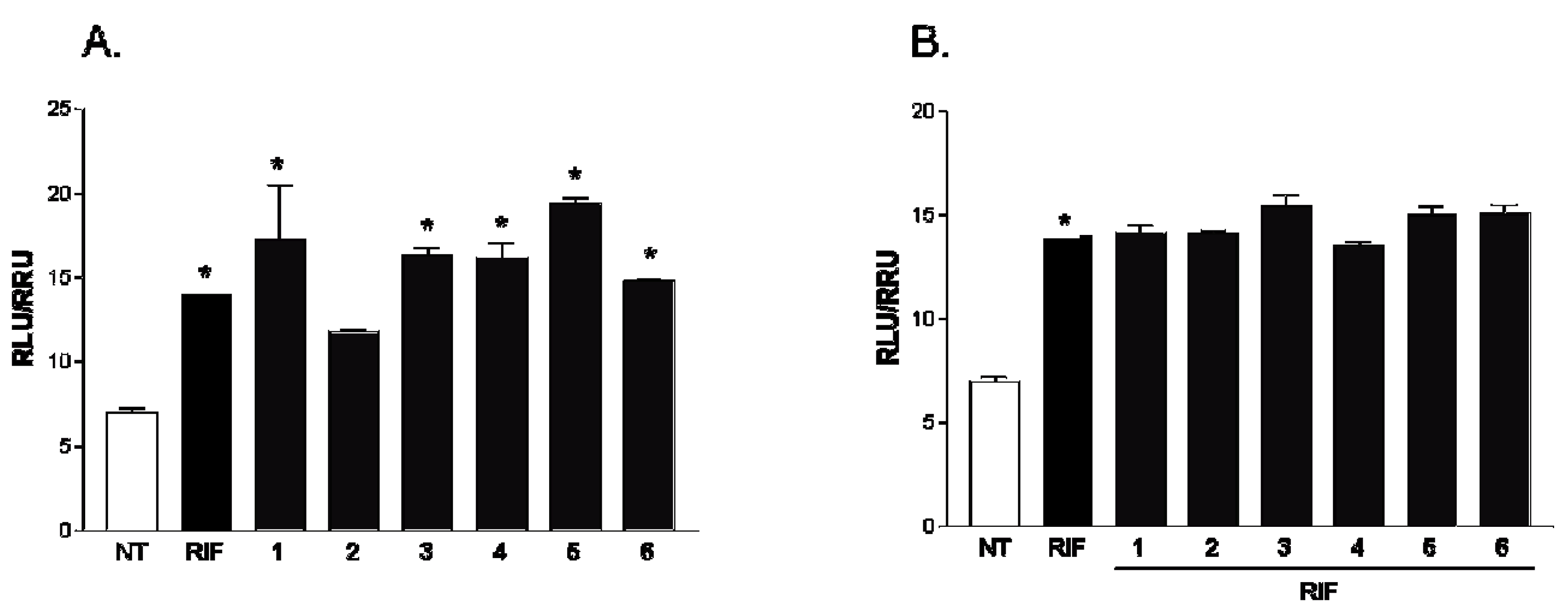
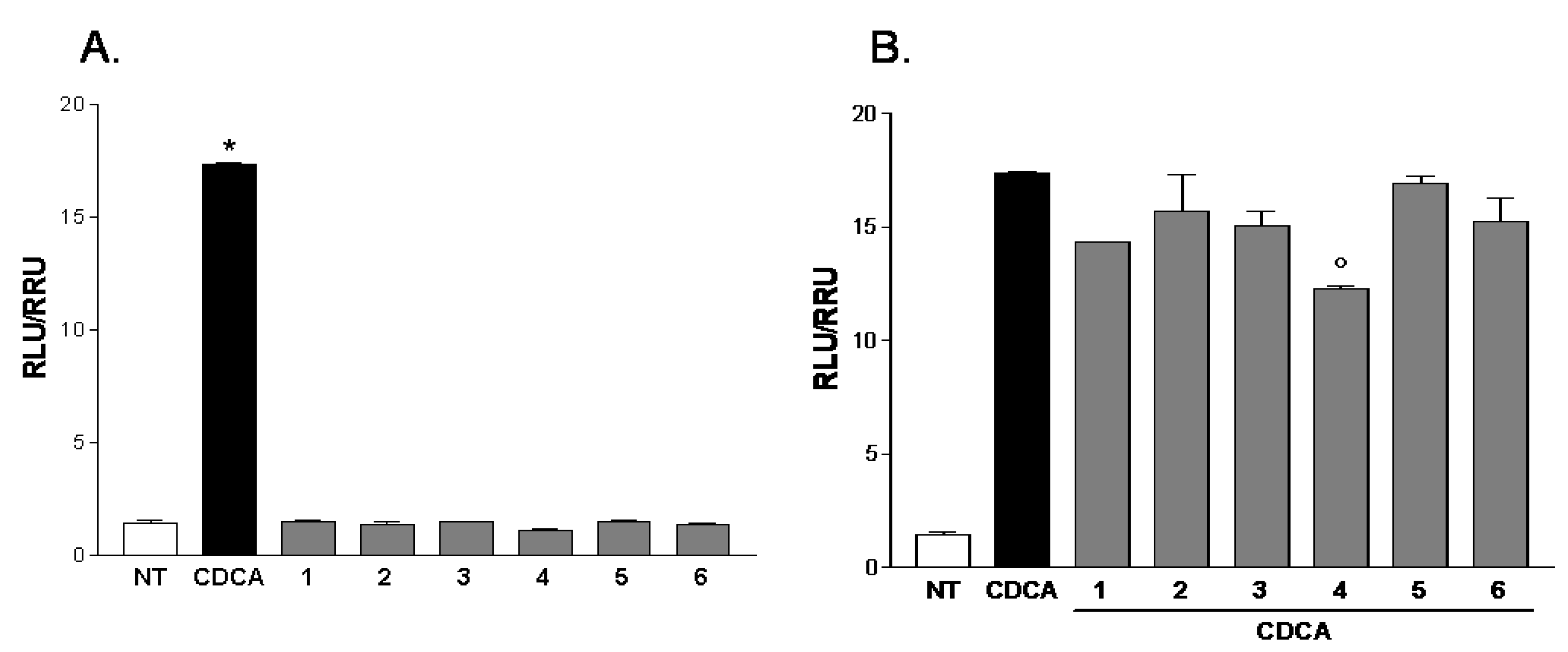
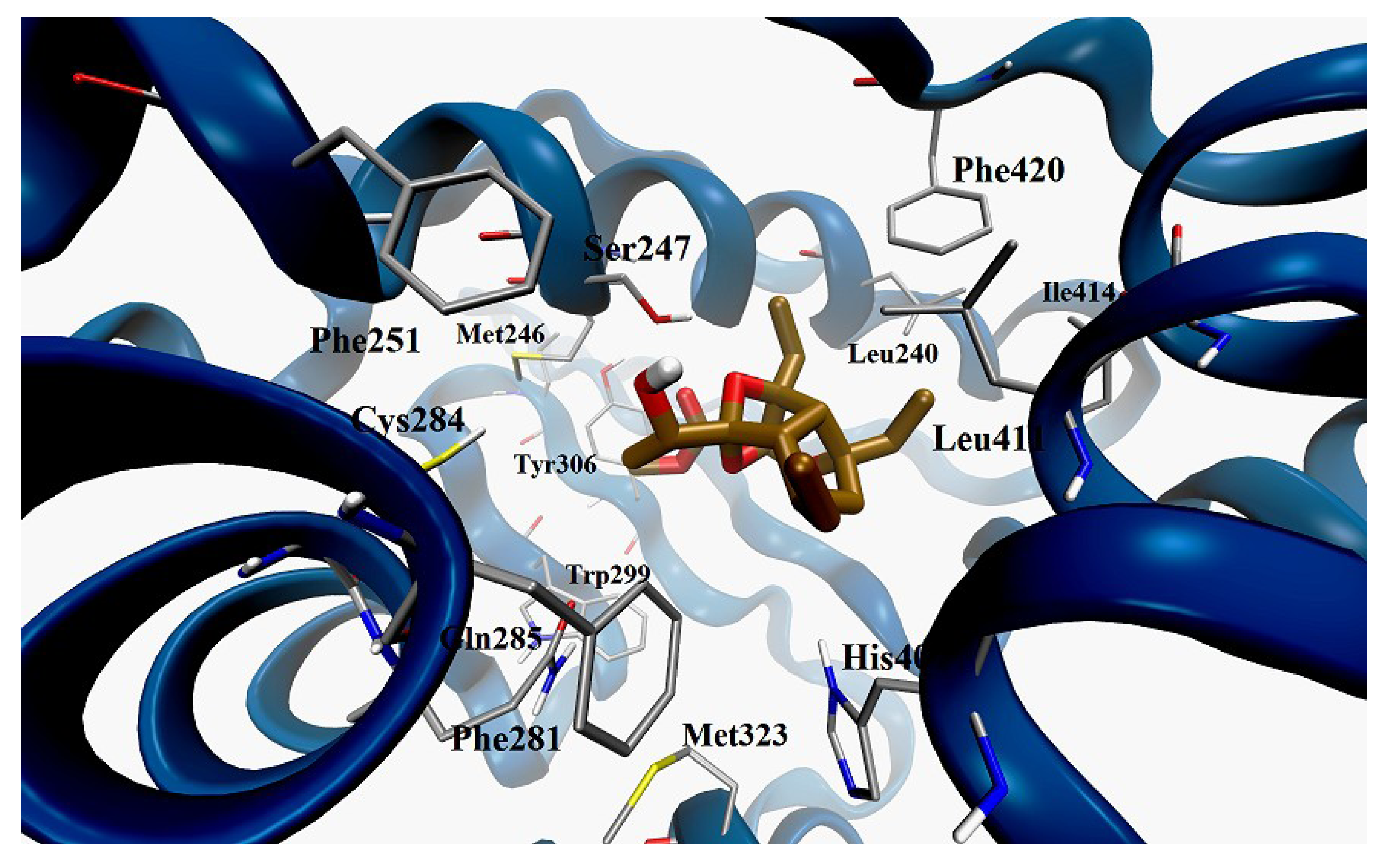
3. Experimental Section
3.1. General Experimental Procedures
3.2. Sponge Material and Separation of Gracilioethers
3.3. Characteristic Data for Gracilioether K (6)
3.4. General Procedure for the Preparation of MTPA Esters (6a) and (6b)
3.5. Computation Details
Docking Calculation
3.6. In Vitro Transactivation
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Lehmann, J.M.; McKee, D.D.; Watson, M.A.; Willson, T.M.; Moore, J.T.; Kliewer, S.A. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J. Clin. Invest. 1998, 102, 1016–1023. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Goodwin, B.; Willson, T.M. The nuclear pregnane X receptor: A key regulator of xenobiotic metabolism. Endocr. Rev. 2002, 23, 687–702. [Google Scholar] [CrossRef]
- Reschly, E.J.; Krasowski, M.D. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr. Drug Metab. 2006, 7, 349–365. [Google Scholar] [CrossRef]
- Saini, S.P.; Mu, Y.; Gong, H.; Toma, D.; Uppal, H.; Ren, S.; Li, S.; Poloyac, S.M.; Xie, W. Dual role of orphan nuclear receptor pregnane X receptor in bilirubin detoxification in mice. Hepatology 2005, 41, 497–505. [Google Scholar] [CrossRef]
- Xie, W.; Radominska-Pandya, A.; Shi, Y.; Simon, C.M.; Nelson, M.C.; Ong, E.S.; Waxman, D.J.; Evans, R.M. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl. Acad. Sci. USA 2001, 98, 3375–3380. [Google Scholar] [CrossRef]
- Staudinger, J.L.; Goodwin, B.; Jones, S.A.; Hawkins-Brown, D.; MacKenzie, K.I.; LaTour, A.; Liu, Y.; Klaassen, C.D.; Brown, K.K.; Reinhard, J.; et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. USA 2001, 98, 3369–3374. [Google Scholar] [CrossRef]
- Fiorucci, S.; Cipriani, S.; Baldelli, F.; Mencarelli, A. Bile acid-activated receptors in the treatment of dyslipidemia and related disorders. Prog. Lipid Res. 2010, 49, 171–185. [Google Scholar] [CrossRef]
- Sonoda, J.; Chong, L.W.; Downes, M.; Barish, G.D.; Coulter, S.; Liddle, C.; Lee, C.H.; Evans, R.M. Pregnane X receptor prevents hepatorenal toxicity from cholesterol metabolites. Proc. Natl. Acad. Sci. USA 2005, 102, 2198–2203. [Google Scholar]
- Ihunnah, C.A.; Jiang, M.; Xie, W. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim. Biophys. Acta 2011, 1812, 956–963. [Google Scholar] [CrossRef]
- Xie, W.; Tian, Y. Xenobiotic receptor meets NF-kappaB, a collision in the small bowel. Cell Metab. 2006, 4, 177–178. [Google Scholar] [CrossRef]
- Zhou, C.; Tabb, M.M.; Nelson, E.L.; Grun, F.; Verma, S.; Sadatrafiei, A.; Lin, M.; Mallick, S.; Forman, B.M.; Thummel, K.E.; et al. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J. Clin. Invest. 2006, 116, 2280–2289. [Google Scholar] [CrossRef]
- D’Auria, M.V.; Sepe, V.; Zampella, A. Natural ligands for nuclear receptors: Biology and potential therapeutic applications. Curr. Top. Med. Chem. 2012, 12, 637–669. [Google Scholar] [CrossRef]
- Gao, J.; Xie, W. Targeting xenobiotic receptors PXR and CAR for metabolic diseases. Trends Pharmacol. Sci. 2012, 33, 552–558. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E.; Bifulco, G.; D’Auria, M.V.; Zampella, A. Marine sponge steroids as nuclear receptor ligands. Trends Pharmacol. Sci. 2012, 33, 591–601. [Google Scholar] [CrossRef]
- De Marino, S.; Ummarino, R.; D’Auria, M.V.; Chini, M.G.; Bifulco, G.; Renga, B.; D’Amore, C.; Fiorucci, S.; Debitus, C.; Zampella, A. Theonellasterols and conicasterols from Theonella swinhoei. Novel marine natural ligands for human nuclear receptors. J. Med. Chem. 2011, 54, 3065–3075. [Google Scholar] [CrossRef]
- De Marino, S.; Ummarino, R.; D’Auria, M.V.; Chini, M.G.; Bifulco, G.; D’Amore, C.; Renga, B.; Mencarelli, A.; Petek, S.; Fiorucci, S.; et al. 4-Methylenesterols from Theonella swinhoei sponge are natural pregnane-X-receptor agonists and farnesoid-X-receptor antagonists that modulate innate immunity. Steroids 2012, 77, 484–495. [Google Scholar] [CrossRef]
- Festa, C.; de Marino, S.; D’Auria, M.V.; Bifulco, G.; Renga, B.; Fiorucci, S.; Petek, S.; Zampella, A. Solomonsterols A and B from Theonella swinhoei. The first example of C-24 and C-23 sulfated sterols from a marine source endowed with a PXR agonistic activity. J. Med. Chem. 2011, 54, 401–405. [Google Scholar] [CrossRef]
- De Marino, S.; Sepe, V.; D’Auria, M.V.; Bifulco, G.; Renga, B.; Petek, S.; Fiorucci, S.; Zampella, A. Towards new ligands of nuclear receptors. Discovery of malaitasterol A, an unique bis-secosterol from marine sponge Theonella swinhoei. Org. Biomol. Chem. 2011, 9, 4856–4862. [Google Scholar] [CrossRef]
- Sepe, V.; Ummarino, R.; D’Auria, M.V.; Mencarelli, A.; D’Amore, C.; Renga, B.; Zampella, A.; Fiorucci, S. Total synthesis and pharmacological characterization of solomonsterol A, a potent marine pregnane-X-receptor agonist endowed with anti-inflammatory activity. J. Med. Chem. 2011, 54, 4590–4599. [Google Scholar] [CrossRef]
- Lau, A.J.; Yang, G.; Yap, C.W.; Chang, T.K.H. Selective agonism of human pregnane X receptor by individual ginkgolides. Drug Metab. Dispos. 2012, 40, 1113–1121. [Google Scholar] [CrossRef]
- Festa, C.; de Marino, S.; D’Auria, M.V.; Deharo, E.; Gonzalez, G.; Deyssard, C.; Petek, S.; Bifulco, G.; Zampella, A. Gracilioethers E–J, new oxygenated polyketides from the marine sponge Plakinastrella mamillaris. Tetrahedron 2012, 68, 10157–10163. [Google Scholar] [CrossRef]
- Ekins, S.; Kortagere, S.; Iyer, M.; Reschly, E.J.; Lill, M.A.; Redinbo, M.R.; Krasowski, M.D. Challenges predicting ligand-receptor interactions of promiscuous proteins: The nuclear receptor PXR. PLoS Comput. Biol. 2009, 5. [Google Scholar] [CrossRef]
- Watkins, R.E.; Maglich, J.M.; Moore, L.B.; Wisely, G.B.; Noble, S.M.; Davis-Searles, P.R.; Lambert, M.H.; Kliewer, S.A.; Redinbo, M.R. 2.1 Å crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry 2003, 42, 1430–1438. [Google Scholar]
- Xiao, L.; Nickbarg, E.; Wang, W.; Thomas, A.; Ziebell, M.; Prosise, W.W.; Lesburg, C.A.; Taremi, S.S.; Gerlach, V.L.; Le, H.V.; et al. Evaluation of in vitro PXR-based assays and in silico modeling approaches for understanding the binding of a structurally diverse set of drugs to PXR. Biochem. Pharmacol. 2011, 81, 669–679. [Google Scholar]
- Ekins, S.; Chang, C.; Mani, S.; Krasowski, M.D.; Reschly, E.J.; Iyer, M.; Kholodovych, V.; Ai, N.; Welsh, W.J.; Sinz, M.; et al. Human pregnane X receptor antagonists and agonists define molecular requirements for different binding sites. Mol. Pharmacol. 2007, 72, 592–603. [Google Scholar] [CrossRef]
- Ueoka, R.; Nakao, Y.; Kawatsu, S.; Yaegashi, J.; Matsumoto, Y.; Matsunaga, S.; Furihata, K.; van Soest, R.W.M.; Fusetani, N. Gracilioethers A–C, antimalarial metabolites from the marine sponge Agelas gracilis. J. Org. Chem. 2009, 74, 4203–4207. [Google Scholar] [CrossRef]
- Ohtani, I.; Kusumi, T.; Kashman, Y.; Kakisawa, H. High-field FT NMR application of Mosher’s method. The absolute configurations of marine terpenoids. J. Am. Chem. Soc. 1991, 113, 4092–4096. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Festa, C.; Lauro, G.; de Marino, S.; D’Auria, M.V.; Monti, M.C.; Casapullo, A.; D’Amore, C.; Renga, B.; Mencarelli, A.; Petek, S.; et al. Plakilactones from the marine sponge Plakinastrella mamillaris. Discovery of a new class of marine ligands of peroxisome proliferator-activated receptor γ. J. Med. Chem. 2012, 55, 8303–8317. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Festa, C.; D'Amore, C.; Renga, B.; Lauro, G.; Marino, S.D.; D'Auria, M.V.; Bifulco, G.; Zampella, A.; Fiorucci, S. Oxygenated Polyketides from Plakinastrella mamillaris as a New Chemotype of PXR Agonists. Mar. Drugs 2013, 11, 2314-2327. https://doi.org/10.3390/md11072314
Festa C, D'Amore C, Renga B, Lauro G, Marino SD, D'Auria MV, Bifulco G, Zampella A, Fiorucci S. Oxygenated Polyketides from Plakinastrella mamillaris as a New Chemotype of PXR Agonists. Marine Drugs. 2013; 11(7):2314-2327. https://doi.org/10.3390/md11072314
Chicago/Turabian StyleFesta, Carmen, Claudio D'Amore, Barbara Renga, Gianluigi Lauro, Simona De Marino, Maria Valeria D'Auria, Giuseppe Bifulco, Angela Zampella, and Stefano Fiorucci. 2013. "Oxygenated Polyketides from Plakinastrella mamillaris as a New Chemotype of PXR Agonists" Marine Drugs 11, no. 7: 2314-2327. https://doi.org/10.3390/md11072314
APA StyleFesta, C., D'Amore, C., Renga, B., Lauro, G., Marino, S. D., D'Auria, M. V., Bifulco, G., Zampella, A., & Fiorucci, S. (2013). Oxygenated Polyketides from Plakinastrella mamillaris as a New Chemotype of PXR Agonists. Marine Drugs, 11(7), 2314-2327. https://doi.org/10.3390/md11072314









