Abstract
The indolocarbazole (ICZ) alkaloids have attracted much attention due to their unique structures and potential therapeutic applications. A series of ICZs were recently isolated and identified from a marine-derived actinomycete strain, Streptomyces sanyensis FMA. To elucidate the biosynthetic machinery associated with ICZs production in S. sanyensis FMA, PCR using degenerate primers was carried out to clone the FAD-dependent monooxygenase gene fragment for ICZ ring formation, which was used as a probe to isolate the 34.6-kb DNA region containing the spc gene cluster. Sequence analysis revealed genes for ICZ ring formation (spcO, D, P, C), sugar unit formation (spcA, B, E, K, J, I), glycosylation (spcN, G), methylation (spcMA, MB), as well as regulation (spcR). Their involvement in ICZ biosynthesis was confirmed by gene inactivation and heterologous expression in Streptomyces coelicolor M1152. This work represents the first cloning and characterization of an ICZ gene cluster isolated from a marine-derived actinomycete strain and would be helpful for thoroughly understanding the biosynthetic mechanism of ICZ glycosides.
1. Introduction
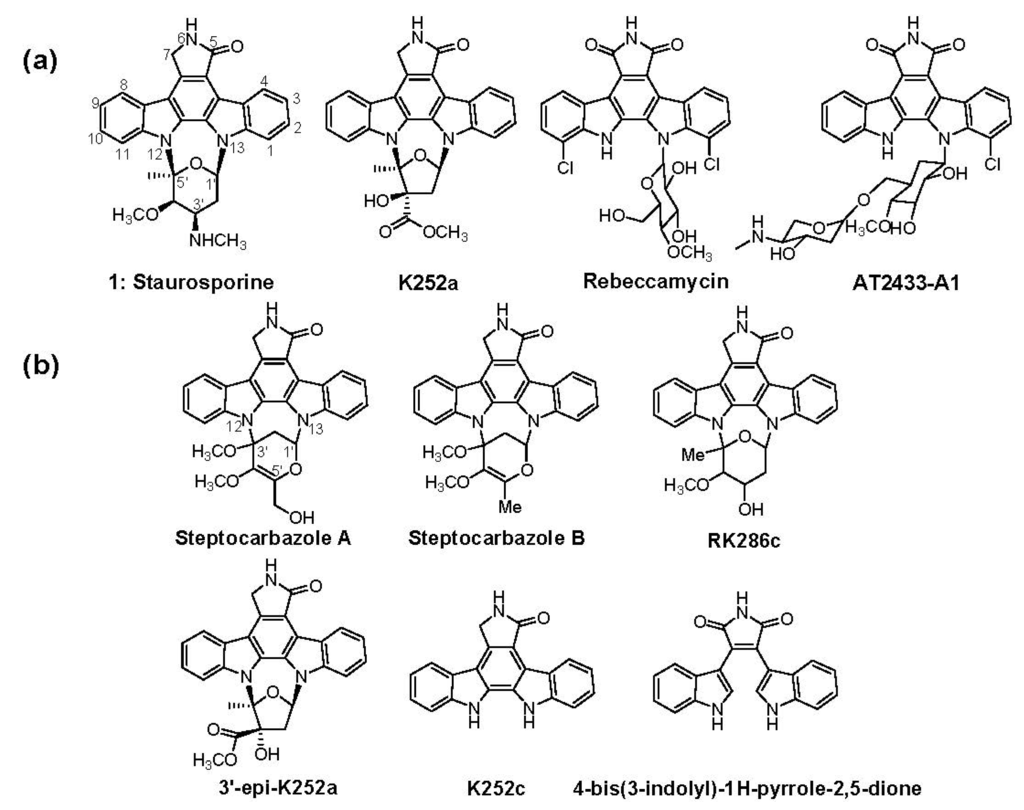
The firstly reported indolocarbazole (ICZ) was staurosporine (STA, 1), which was isolated from the fermentation culture of Streptomyces staurosporeus AM-2282 (ATCC 55006) in 1977 (Scheme 1) [1]. Since then, more than 130 ICZs have been isolated from various organisms, including bacteria, fungi and invertebrates, during the last 35 years [2,3,4]. Due to their unique structures and important biological bioactivities, this family of compounds has attracted much attention ever since its discovery. ICZs have been found to inhibit protein kinase, topoisomerase and ATP-binding cassette transporter [2,5,6,7,8,9]. Several ICZs, such as UCN-01 (7-hydroxy-STA), lestaurtinib (CEP-701), midostaurin (PKC412), edotecarin, becatecarin and NSC655649, are presently undergoing clinical trials for novel antitumor therapies [10].
Scheme 1.
(a) Structures of staurosporine (STA, 1), K252a, rebeccamycin and AT2433-A1; (b) Structures of indolocarbazoles (ICZs) isolated from S. sanyensis FMA besides STA and K252a.
The therapeutic diversity and medicinal potential of ICZs have inspired interest in their biosynthesis research. The biosynthetic gene clusters for rebeccamycin (REB), STA, AT2433-A1 and K252a (Scheme 1) have been reported [11,12,13,14,15], and their assembling mechanisms have been widely studied [2,16,17,18,19,20,21]. REB and AT2433-A1 are halogenated and contain a fully oxidized C-7 carbon and a β-glycosidic bond to only one indole nitrogen; conversely, STA and K252a are not halogenated and bear a fully reduced C-7 carbon and a sugar attached to both indole nitrogens (Figure 1). The ICZ rings are synthesized from two molecules of tryptophan involving a series of oxidation steps. These reactions include amino oxidation (catalyzed by amino oxidases RebO/StaO/AtmO/InkO/NokA), chromopyrrolic acid formation (catalyzed by heme-dependent oxidases RebD/StaD/AtmD/InkD/NokB), ring-closing reaction (catalyzed by cytochrome P450s RebP/StaP/AtmP/InkP/NokC) and oxidative decarboxylation (catalyzed by FAD-dependent monooxygenases RebC/StaC/AtmC/InkE/NokD) [16,19,20,22,23,24]. As a result, two classes of aglycone scaffolds—arcyriaflavin A (for REB and AT2433-A1) and K252C (for STA and K252a), are generated, respectively.
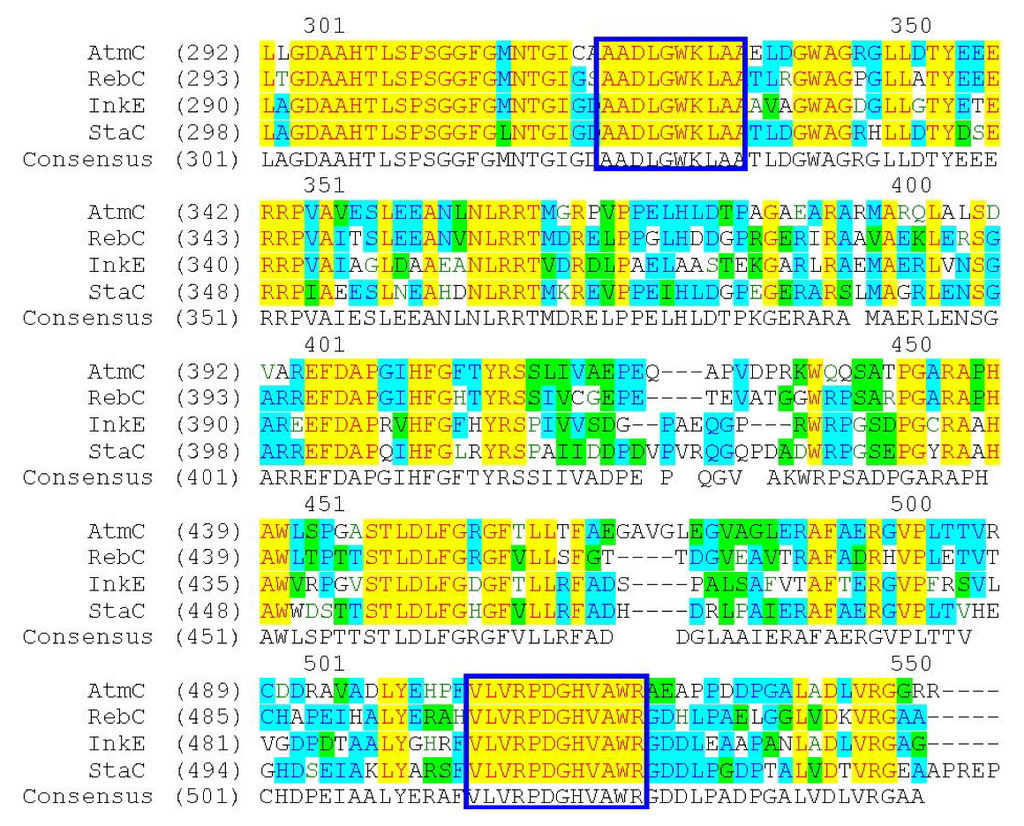
Figure 1.
Alignment of AtmC (ABC02791), RebC (CAC93716), InkE (ABD59214) and StaC (BAF47693). The conserved regions of amino acids for degenerate primers design are indicated in the squares.
In recent years, an increasing amount of ICZs were isolated from marine-derived strains; in addition, the isolated ICZs are usually comprised of multiple analogs [4,25,26,27]. However, genetic information regarding these compounds has been seldom reported. Recently, a series of ICZs were isolated and identified from S . sanyensis FMA(=219808), which was isolated from mangrove soil samples collected in Sanya, Hainan Province of China [28]. The characterized ICZs include K252c, K252a, 3′-epi-K252a, RK286c, 4-bis(3-indolyl)-1H-pyrrole-2,5-dione and two novel ICZs, streptocarbazoles A and B (Scheme 1). The bioassay experiments revealed that streptocarbazole A was cytotoxic on HL-60 and A-549 cell lines and could arrest the cell cycle of Hela cells at the G2/M phase [4]. In contrast to all the other reported cyclic ICZ glycosides, which typically bear cyclic N-glycosidic linkages between the 1,5-carbons of the glycosyl moiety and two indole nitrogens of K252c, the aglycones of streptocarbazoles A and B are linked to the 1,3-carbons of the glycosyl moiety, indicating that a novel enzymatic mechanism might be involved in the C–N bond formation between the C-3′ of deoxysugar and the N-12 of aglycone (Scheme 1). This exceptional cyclic N-glycosidic linkage prompted us to investigate the biosynthetic mechanism of ICZs compounds in the marine-derived S. sanyensis FMA. Therefore, we did alignments of the ICZ ring biosynthetic genes and designed a pair of degenerate primers to amplify the corresponding DNA fragment encoding the FAD-dependent monooxygenase from strain FMA. A cosmid library of strain FMA was constructed, from which the spc biosynthetic gene cluster encoding ICZ production was isolated, which was surprisingly highly homologous to the STA gene cluster from Streptomyces sp. TP-A0274. Here, we report the cloning, characterization and heterologous expression of the spc gene cluster from S. sanyensis FMA.
2. Results and Discussion
2.1. Cloning and Sequencing of the spc Gene Cluster from S. sanyensis FMA
The formation of ICZ rings involves four conserved enzymes; therefore, a pair of degenerate primers were designed according to the alignment result of RebC (CAC93716), StaC (BAF47693), AtmC (ABC02791) and InkE (ABD59214), which revealed highly conserved regions of AADLGWKLAA and VLVRPDGHVAWR, as shown in Figure 1. A distinct product at the expected size of 0.6 kb was obtained by PCR from genomic DNA of the strain FMA using the degenerate primers and was cloned into pUM-T to yield pWLI601. Sequencing results showed that the PCR-amplified product was very similar to known FAD-dependent monooxygenases for ICZ ring biosynthesis, with 59% identity to RebC, 71% identity to StaC, 59% identity to AtmC and 55% identity to InkE, indicating that the amplified gene fragment is probably involved in ICZ biosynthesis in strain FMA. This cloning strategy would be applicable for probing the biosynthetic genes for other ICZ ring-containing natural products.
The cosmid library of strain FMA was constructed using SuperCos1 as the vector. A pair of specific primers was designed according to the internal sequence of the 0.6 kb fragment and was used for library screening. Five overlapped positive cosmids (pWLI611-615) were identified, and pWLI615 was chosen for further sequencing, ultimately giving a 34.6 kb continuous DNA region. The overall G + C content of the region was 75.5%. The sequence was deposited in the GenBank database under the accession number KC182794.
2.2. Organization and Characterization of the spc Gene Cluster
In total, 19 open reading frames (ORFs) were identified, among which 15 were designated as ICZ glycoside biosynthetic genes and the other four were predicted to be beyond the cluster (Figure 2). The composition and organization of the cluster are highly conserved with the STA gene cluster from Streptomyces sp. TP-A0274. The results are summarized in Table 1. spcODPC genes, which exhibit 65%–78% identity to the known homologous genes, encode the ICZ ring K252c. spcABEKJI genes, showing 64%–86% identity to their homologs, are responsible for the assembly of the sugar moiety, followed by C–N bond formation, catalyzed by SpcG and SpcN, sequentially. The two methylation-tailoring steps are performed by SpcMA and SpcMB, respectively. Expression of the gene cluster is probably regulated by SpcR, a LuxR family transcriptional activator harboring a typical Helix-Turn-Helix (HTH) motif for DNA binding at the C-terminus.
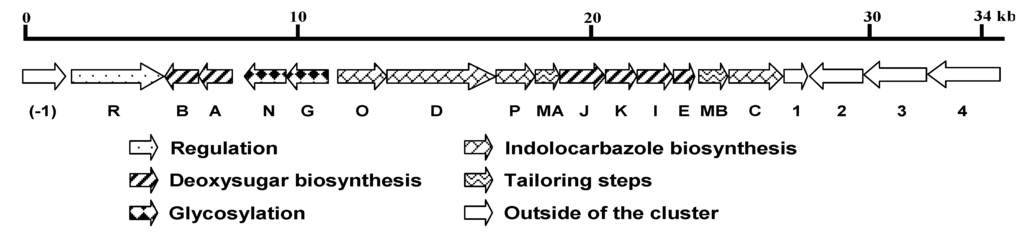
Figure 2.
Genetic organization of the spc biosynthetic gene cluster. Proposed functions of individual open reading frames are coded with various patterns and summarized in Table 1.

Table 1.
Proposed functions of proteins encoded by the spc biosynthetic gene cluster in S. sanyensis FMA andits comparison with the STA gene cluster in S treptomyces sp. TP-A0274.
| Protein | Size (aa) | Proposed function | Homolog in strain TP-A0274 (Accession No.) | % Identity/Similarity |
|---|---|---|---|---|
| Orf(-1) | 341 | hypothetical protein | - | - |
| SpcR | 986 | transcriptional regulator | StaR (BAC55205.1) | 62/70% |
| SpcB | 357 | dTDP-glucose 4,6-dehydratase | StaB (BAC55206.1) | 82/87% |
| SpcA | 354 | glucose-1-phosphate thymidyltransferase | StaA (BAC55207.1) | 78/87% |
| SpcN | 390 | cytochrome P450 | StaN (BAC55208.1) | 76/81% |
| SpcG | 433 | N-glycosyltransferase | StaG (BAC55209.1) | 76/84% |
| SpcO | 503 | L-amino acid oxidase | StaO (BAC55210.1) | 77/85% |
| SpcD | 1123 | chromopyrrolic acid synthase | StaD (BAC55211.1) | 65/71% |
| SpcP | 427 | cytochrome P450 | StaP (BAC55212.1) | 71/79% |
| SpcMA | 277 | methyltransferase | StaMA (BAC55213.1) | 63/72% |
| SpcJ | 477 | 2,3-dehydratase | StaJ (BAC55214.1) | 64/70% |
| SpcK | 331 | 4-ketoreductase | StaK (BAC55215.1) | 75/83% |
| SpcI | 369 | aminotransferase | StaI (BAC55216.1) | 86/92% |
| SpcE | 208 | 3,5-epimerase | StaE (BAC55217.1) | 83/89% |
| SpcMB | 281 | methyltransferase | StaMB (BAC55218.1) | 75/83% |
| SpcC | 542 | monooxygenase | StaC (BAF47693.1) | 78/86% |
| Orf1 | 238 | hypothetical protein | - | - |
| Orf2 | 540 | integral membrane protein | - | - |
| Orf3 | 292 | outer membrane adhesion like protein | - | - |
| Orf4 | 660 | integral membrane protein | - | - |
| Orf5 | 884 | D-alanyl-D-alanine carboxypeptidase | - | - |
-: means not available.
Although 7 ICZs—K252c, K252a, 3′-epi-K252a, RK286c, 4-bis(3-indolyl)-1H-pyrrole-2,5-dione and streptocarbazoles A and B compounds have been originally isolated from S. sanyensis FMA, to our surprise, the isolated ICZ gene cluster exhibit high homology to that of the STA gene cluster from Streptomyces sp. TP-A0274. Further fermentation of strain FMA showed the major ICZ compound accumulated in strain FMA is indeed STA (Figure 3). We assume that the backbone formation of the previously isolated ICZs is directed by the spc gene cluster, and certain enzyme(s) beyond the cluster might be involved in the biosynthesis of some minor ICZ components as well. Culture conditions may influence gene expression, leading to metabolic changes and, consequently, resulting in the different metabolite profile of ICZs in this strain. Additional 20 kb DNA regions both upstream and downstream of the defined gene cluster were further analyzed for possible genes involving biosynthesis of the minor ICZs components (data not shown). No obvious hit was obtained. In addition, genome sequencing has been performed [29], revealing that the spc gene cluster is the only ICZ biosynthesis locus in the genome, and further analysis is currently going on.
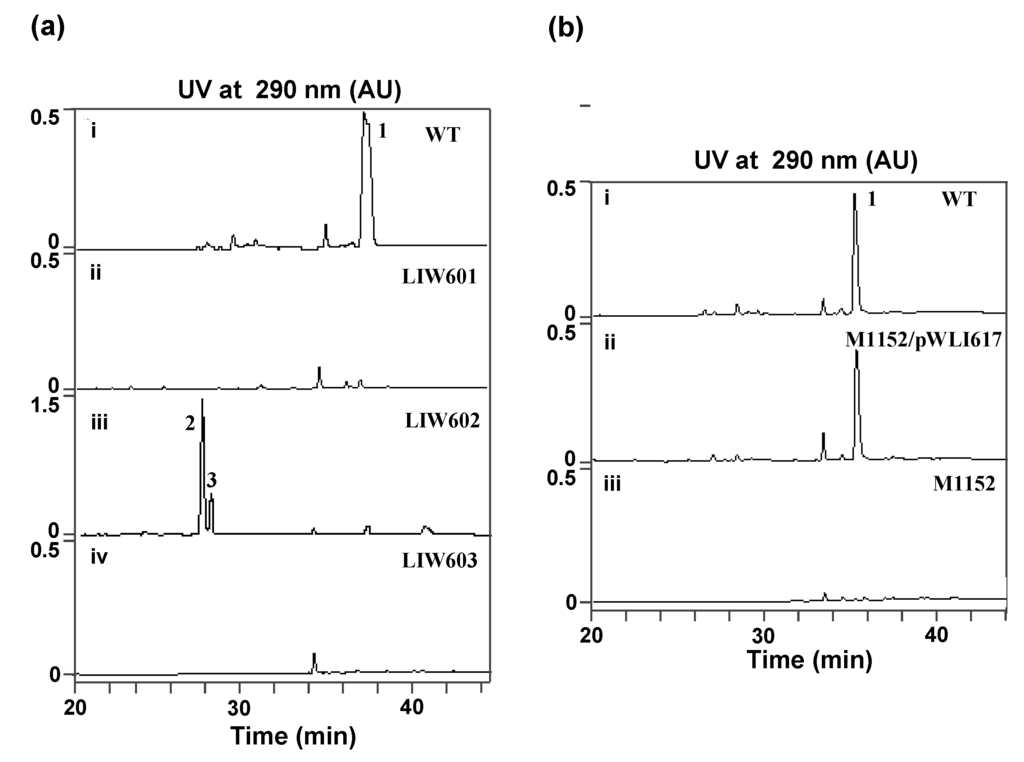
Figure 3.
(a) HPLC traces of fermentation products of S. sanyensis FMA strains; (b) HPLC traces of fermentation products of S. coelicolor M1152 strains. (a): (i) Wild-type strain FMA; (ii) spcC mutant LIW601; (iii) spcI mutant LIW602; (iv) spcR mutant LIW603. (b): (i) Wild-type strain FMA; (ii) S. coelicolor M1152/pWLI617; (iii) S. coelicolor M1152. The ICZs compounds STA (1), K252d (2) and K252c (3) are indicated.
2.3. Involvement of the spc Gene Cluster in ICZs Biosynthesis in S. sanyensis FMA
To support the predicted involvement of this locus in the ICZs biosynthesis in S. sanyensis FMA, spcCIR genes were inactivated by using the PCR targeting strategy (Table S2, Supplementary Materials). The target genes were replaced with the aac(3)IV/oriT cassette, resulting in mutant cosmids pWLI621 (ΔspcC), pWLI622 (ΔspcI) and pWLI623 (ΔspcR), which were then transferred into the wild type S. sanyensis FMA. Apramycin-resistant (AprR) and kanamycin-sensitive (KanS) exconjugants were selected as double crossover mutants. LIW601 (ΔspcC), LIW602 (ΔspcI) and LIW603 (ΔspcR) and their genotypes were confirmed by PCR (Figure S1, Figure S2, Figure S3, Supplementary Materials). All the mutants were fermented and tested for ICZs formation, using the wild-type strain as a positive control. Inactivation of spcC almost completely abolished ICZs production in LIW601 (Figure 3A, panel ii), which proved its essential role for ICZs biosynthesis, consistent with previously reported results [19,23]. No ICZs production was observed in LIW603, indicating that spcR is a positive regulator (Figure 3a, panel iv). Conversely, spcI mutant accumulated two compounds (2 and 3) with very similar UV-vis spectra to that of STA (Figure 3a, panel iii).
The identity of the predominant ICZ compound, STA, produced by strain FMA was confirmed by MS and 1H NMR analysis, which were identical to those previously reported [30] (Figure S4 and Figure S5, Supplementary Materials). The two compounds produced by spcI mutant were isolated, and their structures were determined by MS, 1H NMR, 13C NMR, 1H–1H COSY, HSQC and HMBC (for two) data (Figure S6, Figure S7, Figure S8, Figure S9, Figure S10, Figure S11, Figure S12, Figure S13, Figure S14, Supplementary Materials). Compound 2 was identified as K252d, with a molecular weight of 457, while compound 3 is K252c, with a molecular weight of 311. Interestingly, only K252c was reported to be accumulated in the ΔstaI mutant of Streptomyces sp. TP-A0274 [31]. Therefore, we proposed that inactivation of spcI led to accumulation of both TDP-sugar and ICZ ring, and further, spcG may use TDP-L-rhamnose as a sugar donor to catalyze its attachment onto the N-13 atom of K252c to afford K252d (Scheme 2). The substrate promiscuity of SpcG would make it an alternative tool for glycodiversification.
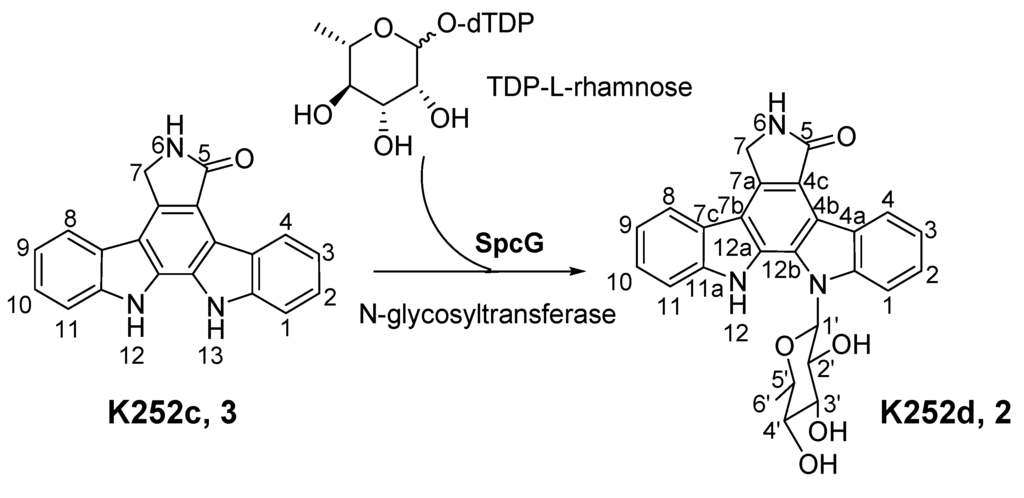
Scheme 2.
Proposed formation of K252d in the ΔspcI mutant LIW602.
2.4. Heterologous Expression of the spc Gene Cluster in Streptomyces coelicolor M1152
Heterologous expression was performed in S. coelicolor M1152, which is well characterized and does not harbor ICZs gene clusters in its genome. pWLI615 was equipped with oriT and φC31 attP/int for conjugation and integration at the attB site at the chromosome, resulting in pWLI617. To test production in a heterologous host, pWLI617 was introduced into S. coelicolor M1152 by conjugation. Apramycin-resistant exconjugants were selected to generate S. coelicolor M1152/pWLI617. HPLC analysis of the fermentation cultures showed that S. coelicolor M1152/pWLI617 produced STA in an excellent yield, in contrast, STA was completely absent from S. coelicolor M1152 (Figure 3b). The identity of STA was confirmed by MS analysis, giving the characteristic molecular ions (m/z for [M + H]+ of 467.2), consistent with the theoretical calculated molecular mass for C28H26N4O3 (Figure S4, Supplementary Materials). Thus, the spc gene cluster was successfully expressed in the heterologous host, demonstrating its integrity for STA biosynthesis.
3. Experimental Section
3.1. Bacterial Strains, Plasmids and Reagents
Bacterial strains and plasmids used and constructed during this study are listed in Table S1. Escherichia coli DH5α was used as the host for general subcloning [32]. E. coli Top10 (Invitrogen, Carlsbad, La Jolla, CA, USA) was used as the transduction host for cosmid library construction. E. coli ET12567/pUZ8002 [33] was used as the cosmid donor host for E. coli-Streptomyces intergeneric conjugation. E. coli BW25113/pIJ790 was used for λRED-mediated PCR-targeting [34]. S. sanyensis FMA wild-type strain has been described previously [4,28]. E. coli strains were grown and manipulated following standard protocols [32,34,35]. S. sanyensis FMA strains were grown at 30 °C on ISP-4 medium for sporulation and conjugation and were cultured in TSB medium for genomic DNA preparation. Common biochemicals and chemicals were purchased from standard commercial sources.
3.2. DNA Manipulation, Sequencing and Bioinformatic Analysis
Plasmid extractions and DNA purification were carried out using commercial kits (OMEGA, BIO-TEK). Genomic DNAs were prepared according to the literature protocol [36]. Both primer synthesis and DNA sequencing were performed at Sunny Biotech Co. Ltd. (Shanghai, China). Orf assignments and their proposed function were accomplished by using the FramePlot 4.0beta [37] and Blast programs [38], respectively.
3.3. Genomic Library Construction
S. sanyensis FMA genomic DNA was partially digested with Sau3AI, and fragments of 40–50 kb were recovered and dephosphorylated with CIAP and then ligated into SuperCos1 that was pretreated with XbaI, dephosphorylated and digested with BamHI. The ligation product was packaged into lambda particles with the MaxPlax Lambda Packaging Extract (Epicenter, Madison, WI, USA), as per the manufacture’s instruction and plated on E. coli Top10. The titer of the primary library was about 2 × 105 cfu per μg of DNA.
3.4. Library Screening
According to the alignment result of RebC, StaC, AtmC and InkE (Figure 1), the FAD-dependent monooxygenases involved in ICZ ring biosynthesis, a pair of degenerate primers were designed as follows: spcCDFP, 5′-GABCTSGGCTGGAAGCTCGCCGC-3′/spcCDRP, 5′-GTCBCCGCGCCAGGCSACGTG-3′. The amplified PCR products were subcloned into pUM-T vector (Bioteke, Beijing, China) and then sequenced, revealing a DNA fragment with the size of 596 bp, according to which another pair of specific primers (spcCSFP, 5′-AGTTGCCGCCCGACATCC-3′/spcCSRP, 5′-GCCCGCTCGTACAGCTTGG-3′) was designed for library screening against 2500 colonies by PCR.
3.5. Gene Inactivation
Gene inactivation in S. sanyensis FMA was performed using the REDIRECT Technology, according to the literature protocol [34,35]. The amplified aac(3)IV-oriT resistance cassette from pIJ773 was transformed into E. coli BW25113/pIJ790/pWLI615 to replace an internal region of the target gene. Mutant cosmids pWLI621 (ΔspcC), pWLI622 (ΔspcI) and pWLI623 (ΔspcR) were constructed (Table S2, Supplementary Materials) and introduced into S. sanyensis FMA by conjugation from E. coli ET12567/pUZ8002, according to the reported procedure [36]. The desired mutants were selected by the apramycin-resistant and kanamycin-sensitive phenotype and were further confirmed by PCR (Table S3, Supplementary Materials).
3.6. Heterologous Expression of the spc Gene Cluster in S. coelicolor M1152
S. coelicolor M1152 was used as the surrogate host for heterologous expression. A DNA fragment from pSET152AB was transformed into E. coli BW25113/pIJ790/pWLI615 to insert the aac(3)IV-oriT -φC31-attP/int into the neomycin resistance gene of SuperCos1. The resulting cosmid pWLI617 was passed through E. coli ET12567/pUZ8002 and then introduced into S. coelicolor M1152 via conjugation, according to the established procedure [36]. Apramycin-resistant exconjugants were selected to afford S. coelicolor M1152/pWLI617. Fermentations of S. coelicolor M1152/pWLI617 and S. coelicolor M1152 were performed under identical conditions as the wild-type S. sanyensis FMA and were analyzed for ICZs production by HPLC with the wild-type strain FMA as a positive control.
3.7. Production and Analyses of ICZs in S. sanyensis FMA Strains
For the production of ICZs, both seed and production media consisted of 1.5% soybean meal, 0.5% yeast extract, 0.2% soluble starch, 0.2% peptone, 0.4% NaCl, 0.4% CaCO3 and 3.3% sea salt, pH 7.3 [4]. Spores of FMA strains were first inoculated into 50 mL of seed medium in a 250 mL flask and incubated at 28 °C, 220 rpm for 2 days. The resulting seed cultures were used to inoculate the production medium (5 mL into 50 mL of medium in a 250 mL flask for production analysis or 20 mL into 200 mL in a 1 L flask for isolation) and incubated at 28 °C, 220 rpm for another 5 days. The fermentation cultures were harvested by centrifugation, and the supernatant was extracted twice with an equal volume of ethyl acetate. The combined EtOAc extracts were concentrated in vacuo to afford a brown residue. The mycelia were extracted twice with acetone. The combined acetone extracts were concentrated in vacuo to afford the water phase. The resulting water phase was extracted twice with EtOAc. The combined EtOAc extracts were concentrated in vacuo to afford a brown residue. The above residues were dissolved in MeOH, combined, filtered through a 0.2 μm filter and subjected to HPLC. The HPLC system consisted of Agilent 1260 Infinity Quaternary pumps and a 1260 Infinity diode-array detector. Analytical HPLC was performed on an Eclipse C18 column (5 μm, 4.6 × 150 mm) developed with a linear gradient from 30% to 100% MeOH/H2O in 20 min, followed by an additional 10 min at 100% MeOH at flow rate of 1 mL/min and UV detection at 290 nm. Semi-preparative HPLC was conducted using an YMC-Pack ODS-A C18 column (5 μm, 120 nm, 250 × 10 mm). Samples were eluted with a linear gradient from 70% to 95% MeOH/H2O in 25 min, followed by 100% MeOH for 5 min at a flow rate of 2.0 mL/min and UV detection at 290 nm. The identities of STA, K252c and K252d produced by FMA strains were confirmed by MS and NMR analysis. LC-MS was carried out on Agilent 6430 Triple Quadrupole LC mass spectrometer. NMR data was recorded with a Bruker Avance 600 spectrometer.
3.8. Nucleotide Sequence Accession Number
The nucleotide sequence reported in this paper has been deposited in the GenBank database under accession number KC182794.
4. Conclusions
In conclusion, we described the cloning, characterization and heterologous expression of the ICZ gene cluster from the marine-derived S. sanyensis FMA. Inactivation of 3 spc genes confirmed its identity. Although this cluster is highly homologous to the STA gene cluster from Streptomyces sp. TP-A0274, a different phenotype of the aminotransferase gene mutant was observed. The accumulation of both K252c and K252d in spcI mutant revealed the relaxed substrate specificity of the N-glycosyltransferase SpcG. In addition, this cluster was expressed in S. coelicolor M1152 with a comparable yield. The work reported here represents the first cloning and characterization of an ICZ gene cluster from a marine-derived actinomycete strain and would be useful for comprehensive elucidation of the biosynthetic mechanism of ICZ glycosides.
Supplementary Material

Table S1.
Bacteria and plasmids used in this study.
| Strains or plasmids | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli Top10 | Host strain of cosmid vector SuperCos1 | Invitrogen |
| E. coli DH5a | Host strain for general cloning | Stratagene |
| E. coli ET12567/pUZ8002 | Host strain for conjugation | [39] |
| E. coli BW25113/pIJ790 | Host strain for PCR-targeting | [40] |
| Strptomyces sanyensis FMA | Wild type, ICZs producer | [4] |
| LIW601 | Δ spcC inactivation mutant of S. sanyensis FMA | This study |
| LIW602 | Δ spcI inactivation mutant of S. sanyensis FMA | This study |
| LIW603 | Δ spcR inactivation mutant of S. sanyensis FMA | This study |
| Strptomyces coelicolor M1152/pWLI617 | Strptomyces coelicolor M1152 carrying pWLI617 | This study |
| Plasmids | ||
| SuperCosI | Apr, Kmr, cosmid vector | Stratagene |
| pUM-T | Apr, general cloning vector | Bioteke |
| pIJ773 | Aprr, source of acc(3)IV-oriT cassette | [34] |
| pIJ790 | Cmr, λ RED recombination plasmid | [34] |
| pWLI601 | pUM-T cloned with the amplified FAD-dependent monooxygenase gene fragment | This study |
| pWLI611-615 | Genomic library cosmids harboring spc biosynthetic genes from S. sanyensis FMA | This study |
| pWLI621 | pWLI615 derivative where spcC was replaced with acc(3)IV-oriT cassette | This study |
| pWLI622 | pWLI615 derivative where spcI was replaced with acc(3)IV-oriT cassette | This study |
| pWLI623 | pWLI615 derivative where spcR was replaced with acc(3)IV-oriT cassette | This study |
| pWLI617 | pWLI615 derivative which was equipped with oriT and φC31 attP/int | This study |
| pSET152AB | pSET152 derivative | [41] |

Table S2.
The primer pairs used for PCR-targeted mutagenesisa.
| Gene | Primer pairs used for inactivation (5'→3') |
|---|---|
| spcC | spcCMF: GCCGCCCGACATCCTGGTGGACGGGCCCGAGGGCGACCGGattccggggatccgtcgacc |
| spcCMR: CGCTCGTACAGCTTGGCGACCTCCTCGCCGCCCCCGCCGCGtgtaggctggagctgcttc | |
| spcI | spcIMF: CATGACCACGCGAGTATGGGACTACCTGGCGGAGTACGAGattccggggatccgtcgacc |
| spcIMR: CCTCACAGCGTCTCCAGCACCTCGCGCAGCGCGTGGACGACtgtaggctggagctgcttc | |
| spcR | spcRMF: CATGGGTCCTCAGGTACGAGCGTTAGCACCGCTGCGCGGCattccggggatccgtcgacc |
| spcRMR: CCTCAGGTCCGGAAGCGCACCAGAGCGGTGCGGGAGCGTATtgtaggctggagctgcttc |
a Underlined letters represent nucleotides homologous to the DNA regions internal to target genes.

Table S3.
The primer pairs used for PCR confirmation of the mutants.
| Gene | Primer pairs designed to verify the mutant strains (5'→3') | Length of fragment replaced | Length of desired PCR fragments | |
|---|---|---|---|---|
| Wild type | Mutant | |||
| spcC | spcCCF: AGTTGCCGCCCGACATCC | 321 bp | 409 bp | 1470 bp |
| spcCCR: GCCCGCTCGTACAGCTTGG | ||||
| spcI | spcICF: GACCACGCGAGTATGGGACTAC | 1032 bp | 1111 bp | 1467 bp |
| spcICR: GCCTCACAGCGTCTCCAGCA | ||||
| spcR | spcRCF: GGTCCCGTCCCCTTCGACA | 2883 bp | 3001 bp | 1540 bp |
| spcRCR: GGCCTCAGGTCCGGAAGC | ||||

Table S4.
Assignments from 600MHz 1H NMR spectroscopy of STA in CDCl3a.
| Position | δH (numbers of H, multiplicity b, J, Hz) |
|---|---|
| 1 | 7.30 (1H, d, 7.9) |
| 2 | 7.48 (1H, dt, 7.7, 1.1) |
| 3 | 7.36 (1H, dt, 7.5, 0.5) |
| 4 | 9.41 (1H, d, 7.9) |
| 6 | 6.21 (1H, brs) |
| 7 | 5.03 (2H, AB, 16.1) |
| 8 | 7.90 (1H, d, 7.7) |
| 9 | 7.32 (1H, t, 7.5) |
| 10 | 7.42 (1H, dt, 7.7,1.1) |
| 11 | 7.93 (1H, d, 8.4) |
| 3′ | 3.88 (1H, d,3.6) |
| 4′ | 3.35 (1H, m) |
| 5′ | 2.41 (1H, ddd, 14.9,5.7,3.7), 2.75 (1H, ddd, 14.9,4.0,1.2) |
| 6′ | 6.57(1H, dd, 5.7,1.1) |
| 2′-CH3 | 2.36 (1H, s) |
| 3′-OCH3 | 3.41 (1H, s) |
| 4′-NCH3 | 1.54 (1H, s) |
a δTMS = 0. b Multiplicity: s, singlet; d, doublet; dd, doublet of doublets; dt, doublet of triplets; ddd, doublet of doublet of doublets; m, multiplets; AB, AB quartet; brs, broad singlet.

Table S5.
Assignments from 600MHz NMR spectroscopies of K-252c in DMSO a.
| Position | δH(multiplicity b, J, Hz) | δC |
|---|---|---|
| 1 | 7.70 (d, 8.1) | 111.9 |
| 2 | 7.43 (dt, 7.7, 1.1) | 125.5 |
| 3 | 7.22 (dt, 7.5, 0.8) | 119.4 |
| 4 | 9.20(d, 7.9) | 125.3 |
| 6 | 8.44 (brs) | / |
| 7 | 4.96 (s) | 45.7 |
| 8 | 8.04 (d, 7.7) | 121.6 |
| 9 | 7.31 (dt, 7.5, 0.8) | 120.2 |
| 10 | 7.48 (dt, 7.7,1.1) | 125.3 |
| 11 | 7.77 (d, 8.1) | 112.2 |
| 12 | 11.70(brs) | / |
| 13 | 11.52(brs) | / |
a DMSO is used as internal reference. δH = 2.50, δC = 39.9. b Multiplicity: s, singlet; d, doublet; dt, doublet of triplets; brs, broad singlet.

Table S6.
Assignments from 600MHz NMR spectroscopies of K-252d in DMSO a.
| Position | δH(multiplicity b, J, Hz) | δC |
|---|---|---|
| 1 | 7.69 (d, 8.5) | 110.3 |
| 2 | 7.48 (m) | 125.4 |
| 3 | 7.27 (dt, 7.5, 0.8) | 119.7 |
| 4 | 9.45(d, 7.8) | 125.8 |
| 4a | / | 122.9 |
| 4c | / | 118.8 |
| 5 | / | 172.4 |
| 6 | 8.53 (brs) | / |
| 7 | 5.00 (AB,17.5) | 45.6 |
| 7a | / | 134.4 |
| 7b | / | 115.4 |
| 7c | / | 122.5 |
| 8 | 8.06 (d, 7.8) | 121.5 |
| 9 | 7.31 (dt, 7.4, 0.9) | 120.2 |
| 10 | 7.50 (m) | 125.4 |
| 11 | 7.60 (d, 8.1) | 111.8 |
| 11a | / | 139.4 |
| 12 | 11.68 (brs) | / |
| 12b | / | 124.9 |
| 13a | / | 140.5 |
| 1′ | 6.39 (d, 9.6) | 77.3 |
| 2′ | 4.48 (m) | 67.2 |
| 3′ | 4.17 (dd, 3.6, 5.9) | 72.1 |
| 4′ | 4.04 (dd, 1.0, 3.6) | 71.9 |
| 5′ | 4.47 (m) | 76.7 |
| 6′ | 1.69 (d, 7.3) | 15.8 |
a DMSO is used as internal reference. δH = 2.50, δC = 39.9. b Multiplicity: s, singlet; d, doublet; dd, doublet of doublets; dt, doublet of triplets; m, multiplets; brs, broad singlet. /: there is no H or C at this position.
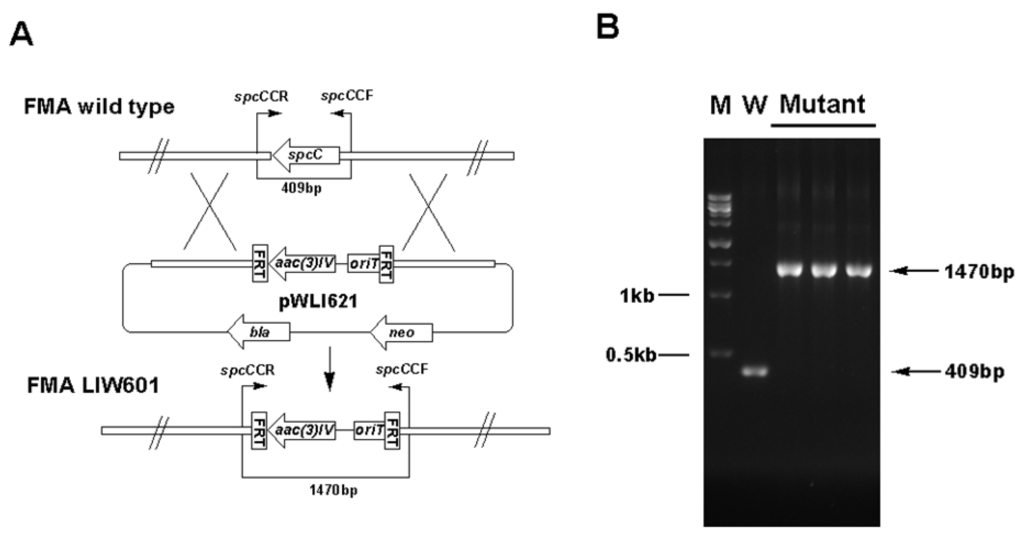
Figure S1.
Inactivation of spcC. (A) Construction of spcC gene inactivation mutant. (B) PCR confirmation of the double-crossover mutant. M: DNA marker; W: S. sanyensis FMA wild type strain; Mutant: spcC gene inactivation mutant.
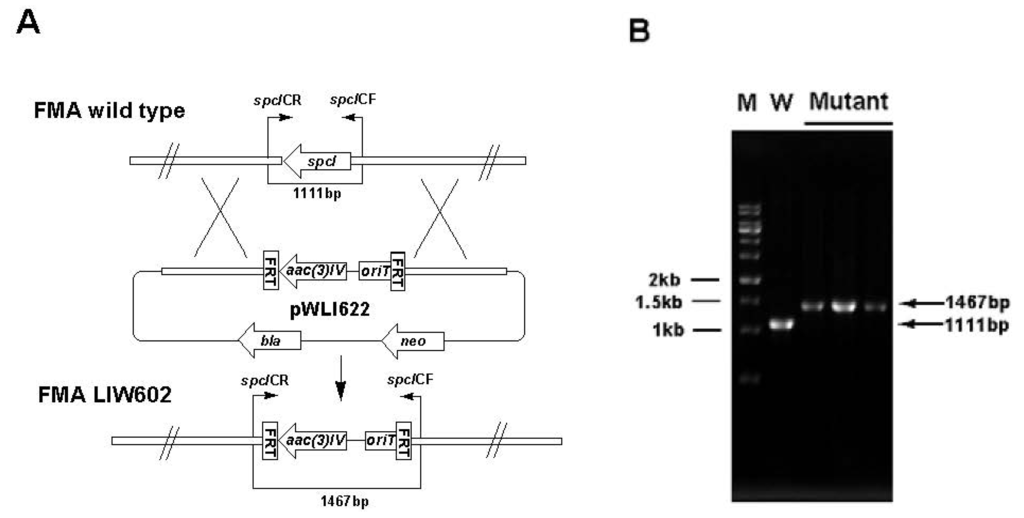
Figure S2.
Inactivation of spcI. (A) Construction of spcI gene inactivation mutant. (B) PCR confirmation of the double-crossover mutant. M: DNA marker; W: S. sanyensis FMA wild type strain; Mutant: spcI gene inactivation mutant.
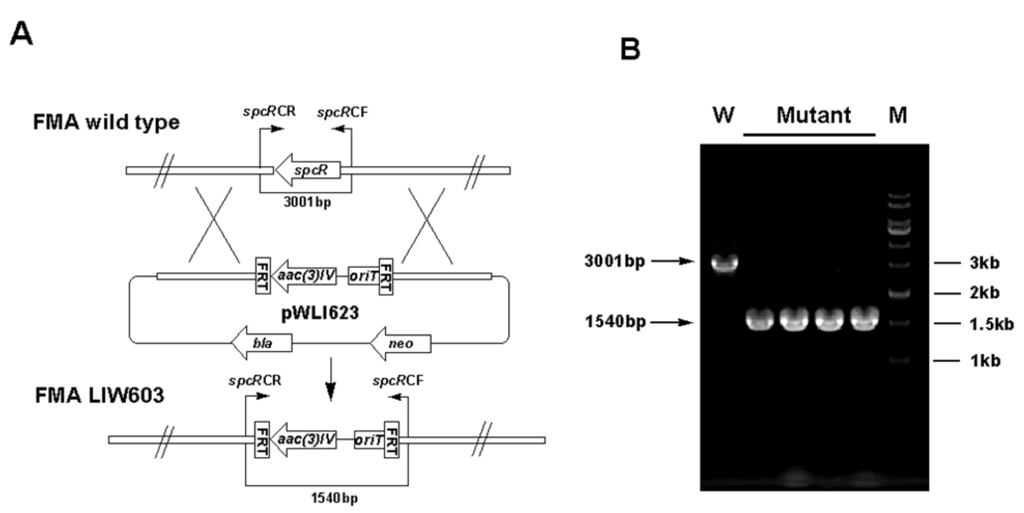
Figure S3.
Inactivation of spcR. (A) Construction of spcR gene inactivation mutant. (B) PCR confirmation of the double-crossover mutant. M: DNA marker; W: S. sanyensis FMA wild type strain; Mutant: spcR gene inactivation mutant.
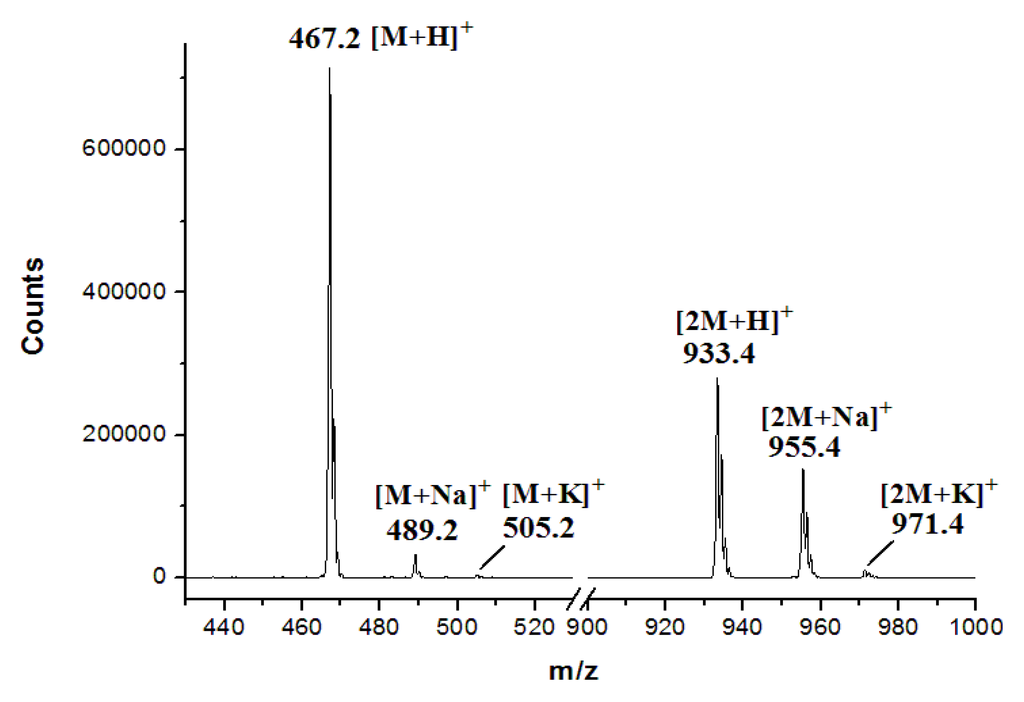
Figure S4.
ESI-MS spectrum of STA.
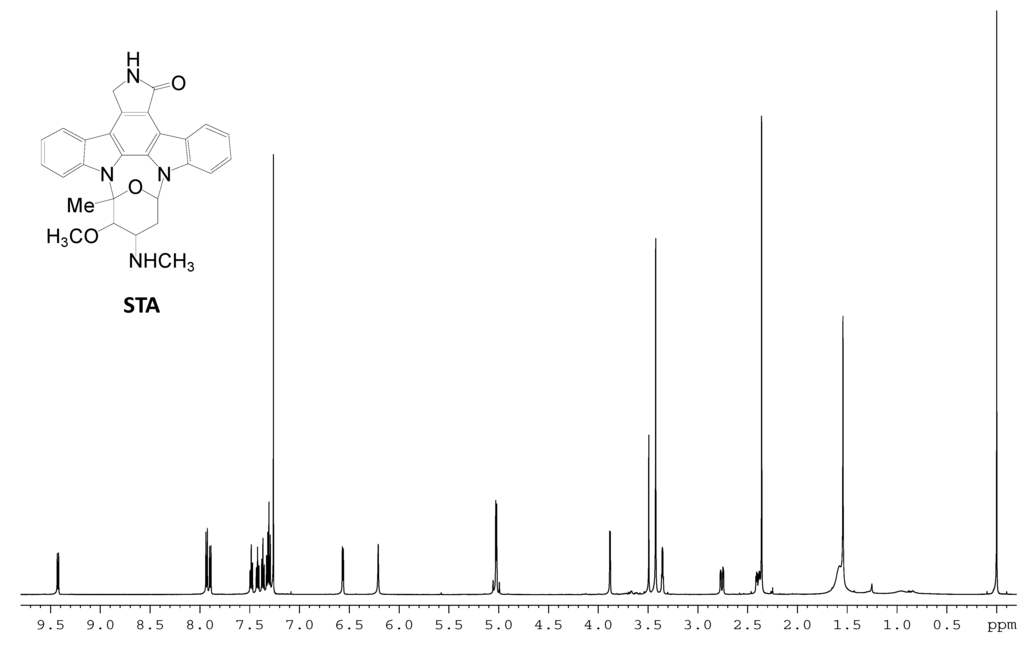
Figure S5.
1H NMR spectrum of STA.
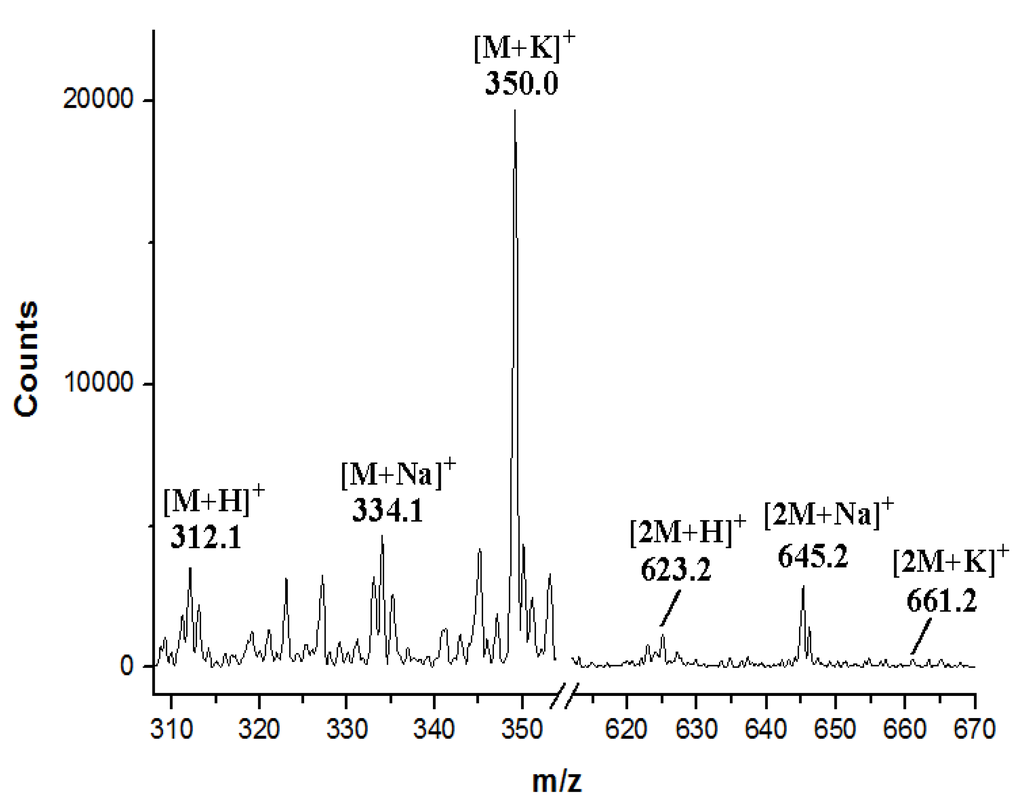
Figure S6.
ESI-MS spectrum of K252c.
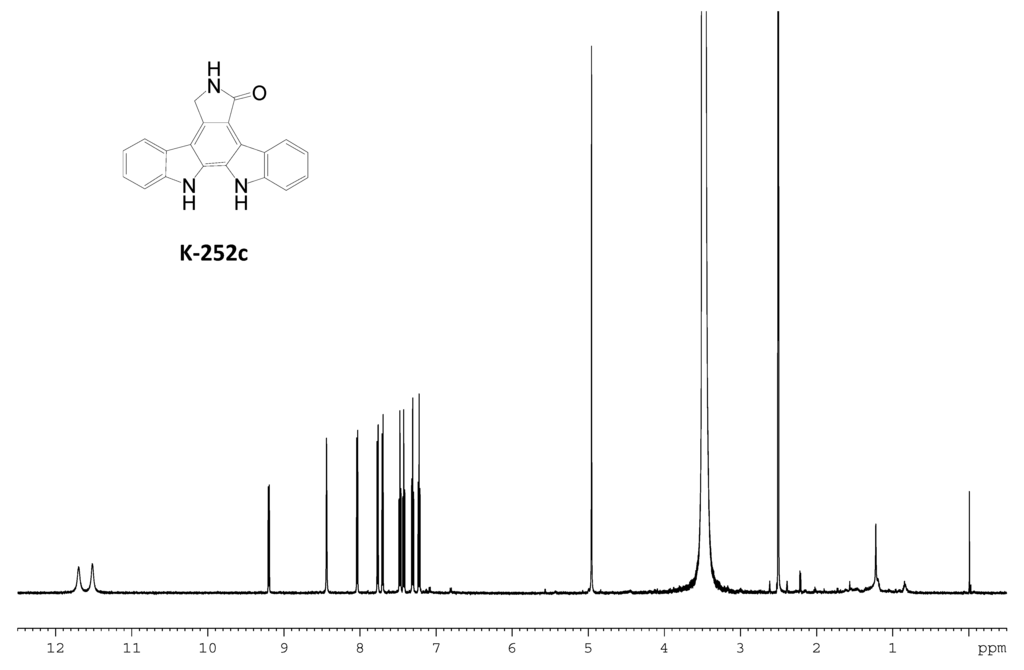
Figure S7.
1H NMR spectrum of K-252c.
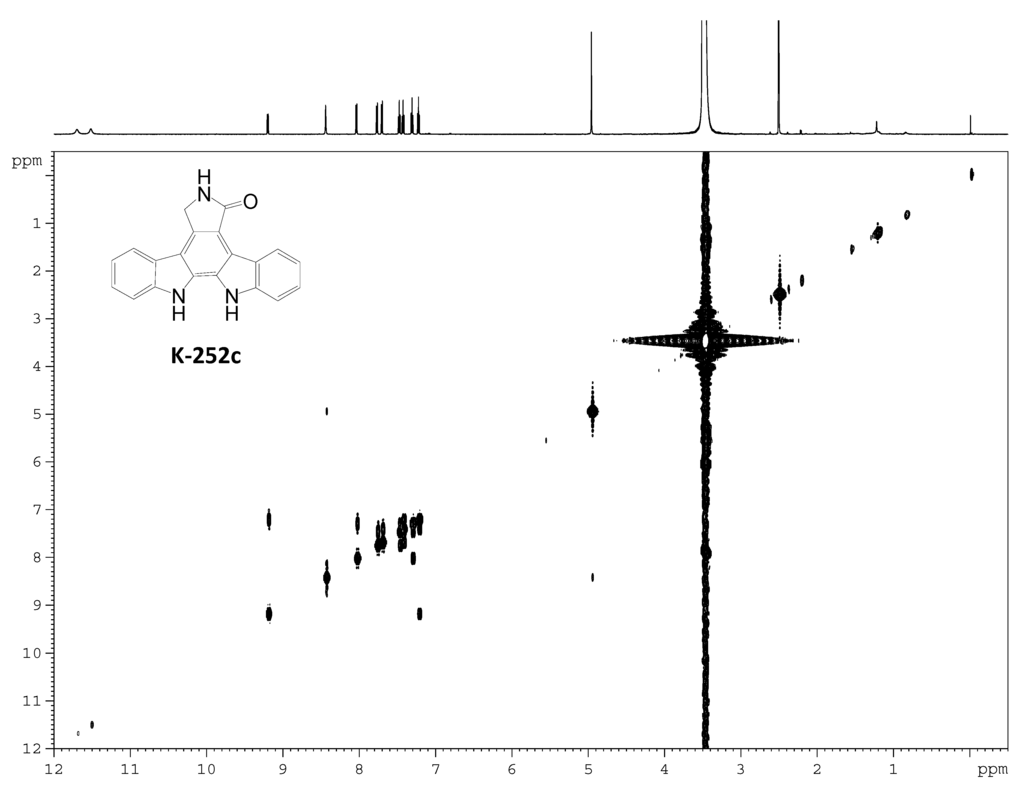
Figure S8.
1H–1H COSY spectrum of K-252c.
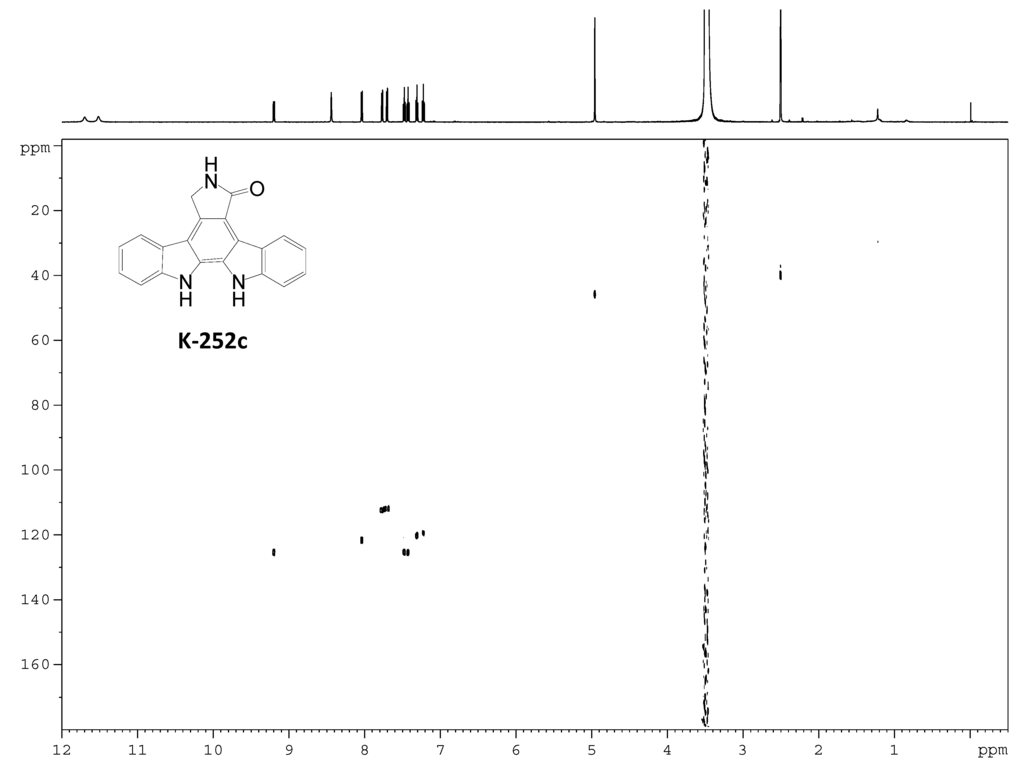
Figure S9.
HSQC spectrum of K-252c.
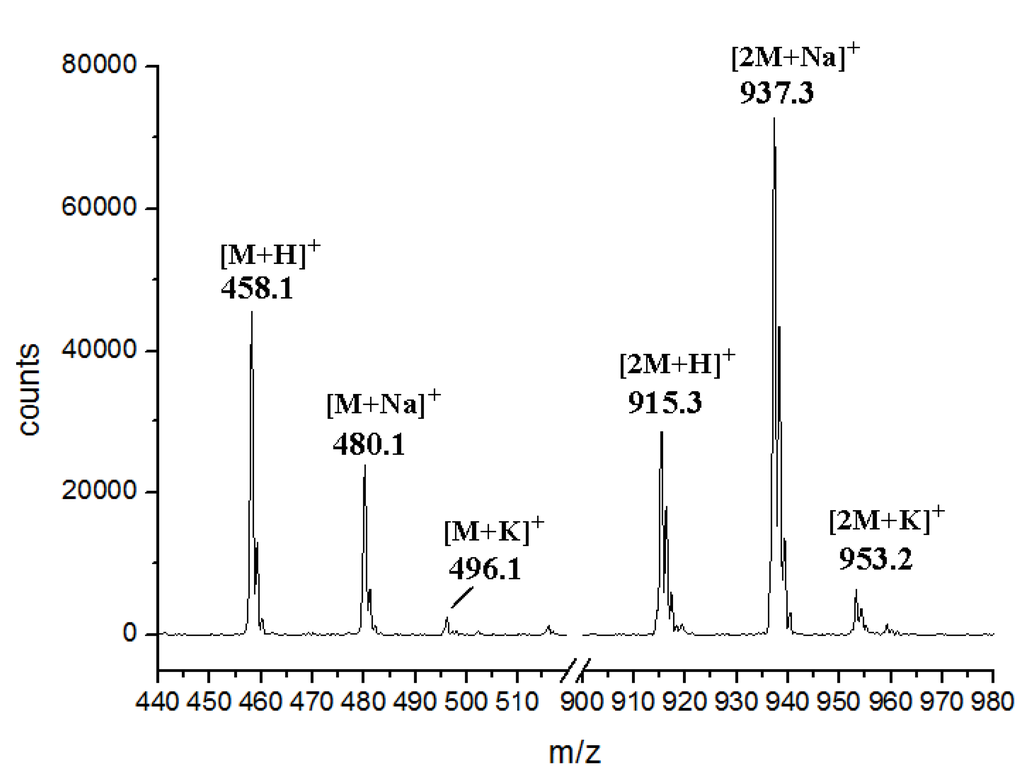
Figure S10.
ESI-MS spectrum of K-252d.
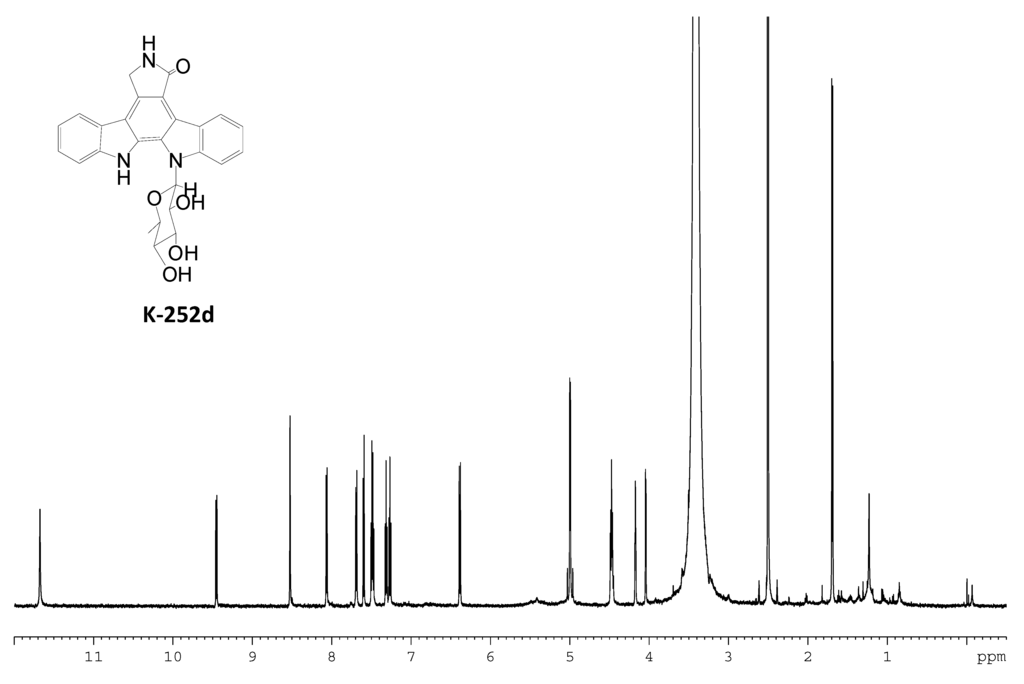
Figure S11.
1H NMR spectrum of K-252d.
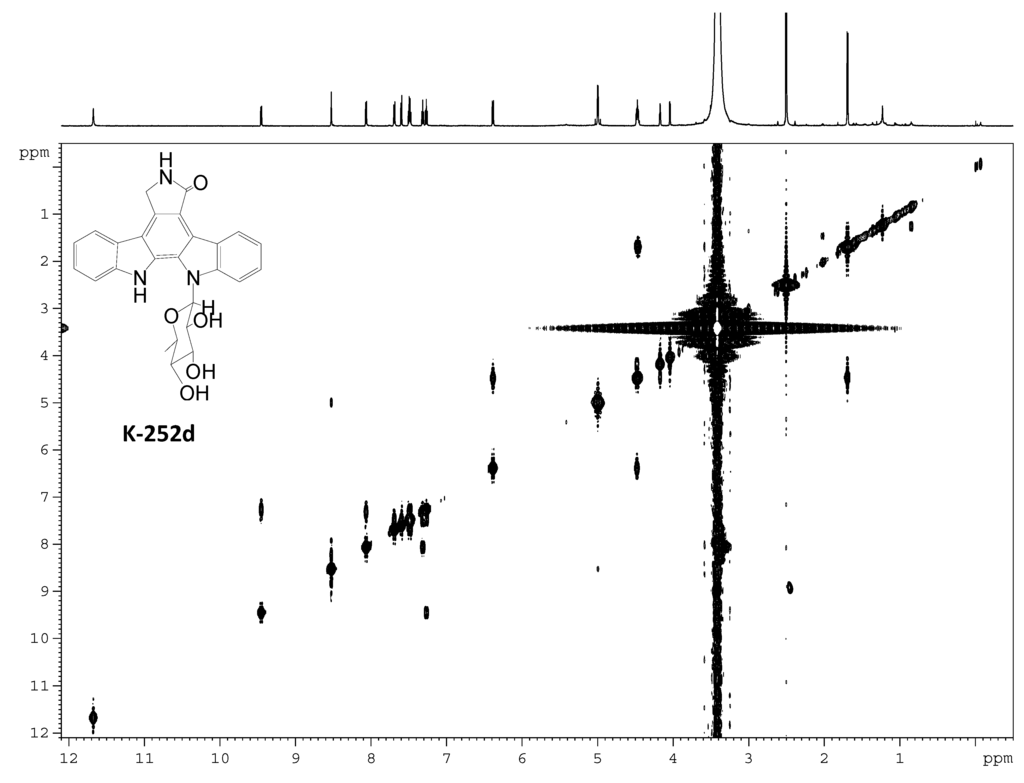
Figure S12.
1H–1H COSY spectrum of K-252d.
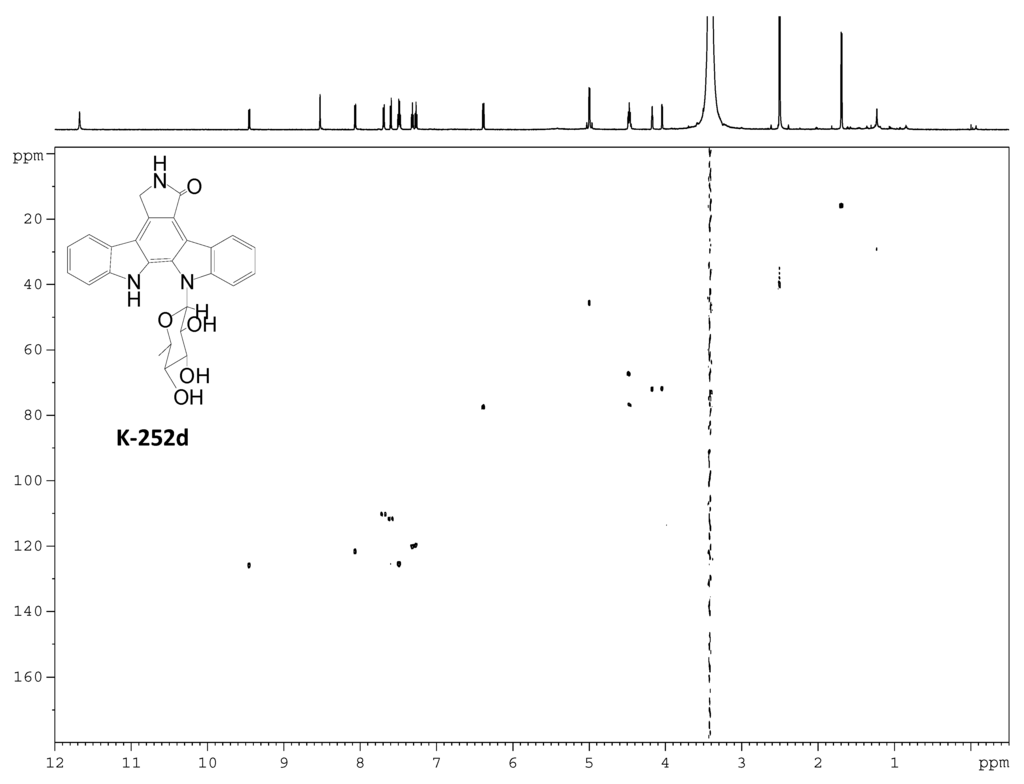
Figure S13.
HSQC spectrum of K-252d.
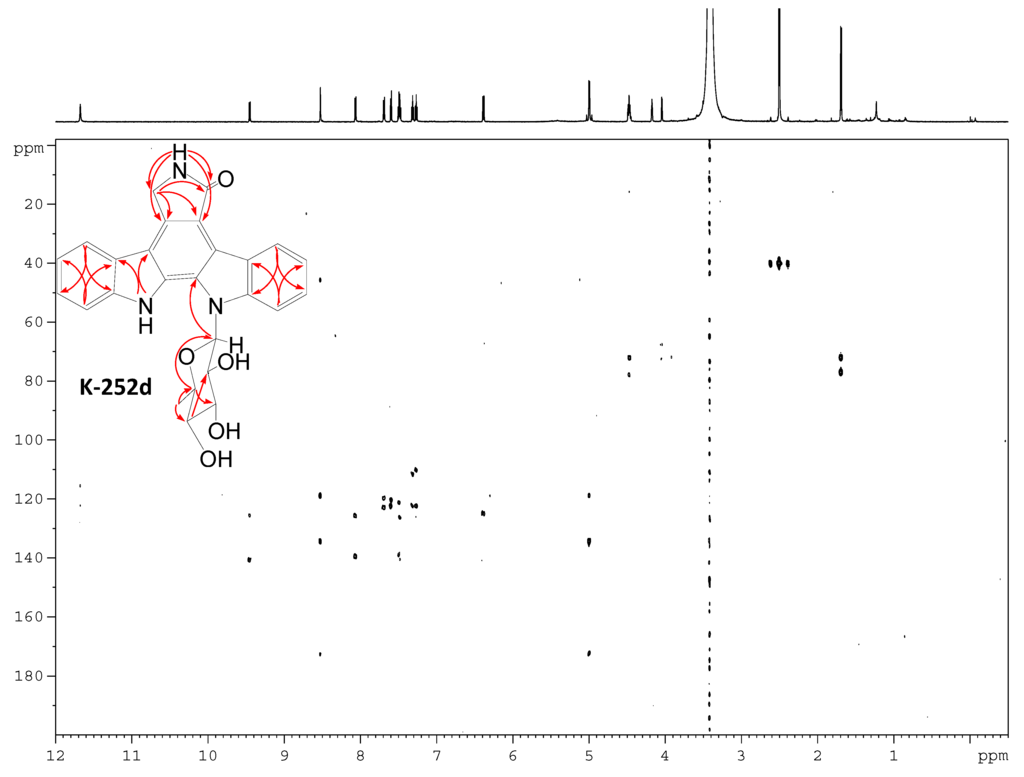
Figure S14.
HMBC spectrum of K-252d.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31070072 and 31171201), the National High Technology Research and Development Program of China (No. 2012AA092201), the Program for New Century Excellent Talents in University (NCET-09-0717) and the State Key Laboratory of Microbial Resources Program, Institute of Microbiology, Chinese Academy of Sciences (No. SKLMR-20110601). We would like to thank Mervyn J. Bibb (John Innes Centre, UK) and Jianhua Ju (South Sea Institute of Oceanology, Chinese Academy of Sciences, China) for kindly providing us S. coelicolor M1152 and pSET152AB, respectively.
References
- Omura, S.; Iwai, Y.; Hirano, A.; Nakagawa, A.; Awaya, J.; Tsuchya, H.; Takahashi, Y.; Masuma, R. A new alkaloid AM-2282 of Streptomyces. origin. Taxonomy, fermentation, isolation and preliminary characterization. J. Antibiot. (Tokyo) 1977, 30, 275–282. [Google Scholar] [CrossRef]
- Sanchez, C.; Mendez, C.; Salas, J.A. Indolocarbazole natural products: Occurrence, biosynthesis, and biological activity. Nat. Prod. Rep. 2006, 23, 1007–1045. [Google Scholar] [CrossRef]
- Reyes, F.; Fernandez, R.; Rodriguez, A.; Bueno, S.; de Eguilior, C.; Francesch, A.; Cuevas, C. Cytotoxic staurosporines from the marine ascidian Cystodytes solitus. J. Nat. Prod. 2008, 71, 1046–1048. [Google Scholar] [CrossRef]
- Fu, P.; Yang, C.; Wang, Y.; Liu, P.; Ma, Y.; Xu, L.; Su, M.; Hong, K.; Zhu, W. Streptocarbazoles A and B, two novel indolocarbazoles from the marine-derived actinomycete strain Streptomyces sp. FMA. Org. Lett. 2012, 14, 2422–2425. [Google Scholar] [CrossRef]
- Acero, N.; Brana, M.F.; Anorbe, L.; Dominguez, G.; Munoz-Mingarro, D.; Mitjans, F.; Piulats, J. Synthesis and biological evaluation of novel indolocarbazoles with anti-angiogenic activity. Eur. J. Med. Chem. 2012, 48, 108–113. [Google Scholar] [CrossRef]
- Liu, J.; Hu, Y.; Waller, D.L.; Wang, J.; Liu, Q. Natural products as kinase inhibitors. Nat. Prod. Rep. 2012, 29, 392–403. [Google Scholar] [CrossRef]
- Prudhomme, M. Biological targets of antitumor indolocarbazoles bearing a sugar moiety. Curr. Med. Chem. Anticancer Agents 2004, 4, 509–521. [Google Scholar] [CrossRef]
- Kurosu, M.; Begari, E. Bacterial protein kinase inhibitors. Drug Dev. Res. 2010, 71, 168–187. [Google Scholar]
- Gani, O.A.; Engh, R.A. Protein kinase inhibition of clinically important staurosporine analogues. Nat. Prod. Rep. 2010, 27, 489–498. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Available online: http://www.clinicaltrials.gov/ (accessed on 21 November 2012).
- Onaka, H.; Taniguchi, S.; Igarashi, Y.; Furumai, T. Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP-A0274 and its heterologous expression in Streptomyces lividans. J. Antibiot. (Tokyo) 2002, 55, 1063–1071. [Google Scholar] [CrossRef]
- Sanchez, C.; Butovich, I.A.; Brana, A.F.; Rohr, J.; Mendez, C.; Salas, J.A. The biosynthetic gene cluster for the antitumor rebeccamycin: Characterization and generation of indolocarbazole derivatives. Chem. Biol. 2002, 9, 519–531. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, J.S.; Chae, C.S.; Hyun, C.G.; Choi, B.W.; Shin, J.; Oh, K.B. Genetic organization of the biosynthetic gene cluster for the indolocarbazole K-252a in Nonomuraea longicatena JCM 11136. Appl. Microbiol. Biotechnol. 2007, 75, 1119–1126. [Google Scholar] [CrossRef]
- Chiu, H.T.; Chen, Y.L.; Chen, C.Y.; Jin, C.; Lee, M.N.; Lin, Y.C. Molecular cloning, sequence analysis and functional characterization of the gene cluster for biosynthesis of K-252a and its analogs. Mol. Biosyst. 2009, 5, 1180–1191. [Google Scholar]
- Gao, Q.; Zhang, C.; Blanchard, S.; Thorson, J.S. Deciphering indolocarbazole and enediyne aminodideoxypentose biosynthesis through comparative genomics: Insights from the AT2433 biosynthetic locus. Chem. Biol. 2006, 13, 733–743. [Google Scholar] [CrossRef]
- Howard-Jones, A.R.; Walsh, C.T. Enzymatic generation of the chromopyrrolic acid scaffold of rebeccamycin by the tandem action of RebO and RebD. Biochemistry 2005, 44, 15652–15663. [Google Scholar] [CrossRef]
- Onaka, H. Biosynthesis of indolocarbazole and goadsporin, two different heterocyclic antibiotics produced by actinomycetes. Biosci. Biotechnol. Biochem. 2009, 73, 2149–2155. [Google Scholar] [CrossRef]
- Asamizu, S.; Shiro, Y.; Igarashi, Y.; Nagano, S.; Onaka, H. Characterization and functional modification of StaC and RebC, which are involved in the pyrrole oxidation of indolocarbazole biosynthesis. Biosci. Biotechnol. Biochem. 2011, 75, 2184–2193. [Google Scholar] [CrossRef]
- Goldman, P.J.; Ryan, K.S.; Hamill, M.J.; Howard-Jones, A.R.; Walsh, C.T.; Elliott, S.J.; Drennan, C.L. An unusual role for a mobile flavin in StaC-like indolocarbazole biosynthetic enzymes. Chem. Biol. 2012, 19, 855–865. [Google Scholar] [CrossRef]
- Groom, K.; Bhattacharya, A.; Zechel, D.L. Rebeccamycin and staurosporine biosynthesis: Insight into the mechanisms of the flavin-dependent monooxygenases RebC and StaC. ChemBioChem 2011, 12, 396–400. [Google Scholar] [CrossRef]
- Ryan, K.S.; Drennan, C.L. Divergent pathways in the biosynthesis of bisindole natural products. Chem. Biol. 2009, 16, 351–364. [Google Scholar] [CrossRef]
- Sanchez, C.; Zhu, L.; Brana, A.F.; Salas, A.P.; Rohr, J.; Mendez, C.; Salas, J.A. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc. Natl. Acad. Sci. USA 2005, 102, 461–466. [Google Scholar]
- Howard-Jones, A.R.; Walsh, C.T. Staurosporine and rebeccamycin aglycones are assembled by the oxidative action of StaP, StaC, and RebC on chromopyrrolic acid. J. Am. Chem. Soc. 2006, 128, 12289–12298. [Google Scholar] [CrossRef]
- Makino, M.; Sugimoto, H.; Shiro, Y.; Asamizu, S.; Onaka, H.; Nagano, S. Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton. Proc. Natl. Acad. Sci. USA 2007, 104, 11591–11596. [Google Scholar] [CrossRef]
- Olano, C.; Mendez, C.; Salas, J.A. Antitumor compounds from marine actinomycetes. Mar. Drugs 2009, 7, 210–248. [Google Scholar]
- Hernandez, L.M.; Blanco, J.A.; Baz, J.P.; Puentes, J.L.; Millan, F.R.; Vazquez, F.E.; Fernandez-Chimeno, R.I.; Gravalos, D.G. 4′-N-methyl-5′-hydroxystaurosporine and 5′-hydroxystaurosporine, new indolocarbazole alkaloids from a marine Micromonospora sp. strain. J. Antibiot. (Tokyo) 2000, 53, 895–902. [Google Scholar] [CrossRef]
- Liu, R.; Zhu, T.; Li, D.; Gu, J.; Xia, W.; Fang, Y.; Liu, H.; Zhu, W.; Gu, Q. Two indolocarbazole alkaloids with apoptosis activity from a marine-derived actinomycete Z(2)039-2. Arch. Pharm. Res. 2007, 30, 270–274. [Google Scholar]
- Sui, J.L.; Xu, X.X.; Qu, Z.; Wang, H.L.; Lin, H.P.; Xie, Q.Y.; Ruan, J.S.; Hong, K. Streptomyces sanyensis sp. nov., isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 2010, 61, 1632–1637. [Google Scholar]
- Kui, H. Wuhan University: Wuhan, China, Unpublished work. 2012.
- Meksuriyen, D.; Cordell, G.A. Biosynthesis of staurosporine, 1. 1H- and 13C-NMR assignments. J. Nat. Prod. 1988, 51, 884–892. [Google Scholar] [CrossRef]
- Onaka, H. Biosynthesis of heterocyclic antibiotics in actinomycetes and an approach to synthesize the natural compounds. Actinomycetologica 2006, 20, 62–71. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Paget, M.S.; Chamberlin, L.; Atrih, A.; Foster, S.J.; Buttner, M.J. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 1999, 181, 204–211. [Google Scholar]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef]
- Gust, B.; Chandra, G.; Jakimowicz, D.; Yuqing, T.; Bruton, C.J.; Chater, K.F. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 2004, 54, 107–128. [Google Scholar] [CrossRef]
- Kieser, T.; Bib, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Ishikawa, J.; Hotta, K. FramePlot: A new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 1999, 174, 251–253. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- MacNeil, D.J.; Gewain, K.M.; Ruby, C.L.; Dezeny, G.; Gibbons, P.H.; MacNeil, T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 1992, 111, 61–68. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, H.; Chen, Q.; Luo, M.; Sun, A.; Song, Y.; Ma, J.; Ju, J. South Sea Institute of Oceanology, Chinese Academy of Sciences: Guangzhou, China, Unpublished work. 2013.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).