Core Oligosaccharide of Plesiomonas shigelloides PCM 2231 (Serotype O17) Lipopolysaccharide — Structural and Serological Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of Lipopolysaccharide and Core Oligosaccharide
2.2. Chemical Analysis of the Oligosaccharides PSV and OSI
2.3. MALDI-TOF MS Analyses of the PSV, OSI and OSII

2.4. NMR Analysis of the PSV
| Residue | Chemical shifts (ppm) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| H-1 | H-2 | H-3a,b | H-4 | H-5 | H-6a,b | H-7a,b | H-8a,b | ||
| C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | C-7 | C-8, CH3CO | ||
| A | →4)-α- D -Gal p A-(1→ | 5.33 | 3.83 | 4.25 | 4.36 | 4.53 | |||
| 102.1 | 69.2 | 69.0 | 80.1 | 72.3 | 175.6 | ||||
| A′ | →4)-α- D -Gal p A-(1→ | 5.40 | 3.84 | 4.22 | 4.41 | 4.60 | |||
| 102.4 | 69.3 | 69.0 | 80.6 | 72.5 | 175.6 | ||||
| B | α- D -Glc p N-(1→ | 5.24 | 3.22 | 3.91 | 3.50 | 4.11 | 3.81 a | ||
| 95.1 | 55.0 | 70.3 | 70.0 | 73.0 | 60.7 | ||||
| C | →6)-α-D-GlcpN-(1→ | 5.12 | 3.26 | 3.86 | 3.47 | 4.33 | 3.79, 4.08 | ||
| 97.0 | 54.9 | 70.3 | 70.0 | 71.6 | 68.6 | ||||
| D | →3,4)-L-α-D-Hepp-(1→ | 5.11 | 4.04 | 4.13 | 4.23 | 4.17 | 4.17 | 3.72 b | |
| 101.3 | 71.1 | 75.3 | 75.1 | 72.0 | 69.2 | 63.8 | |||
| E | →4)-α-D-GalpA-(1→ | 5.02 | 3.95 | 4.07 | 4.52 | 4.31 | |||
| 99.7 | 68.8 | 69.7 | 77.3 | 70.6 | 176.5 | ||||
| F | →7)-L-α-D-Hepp-(1→ | 4.88 | 4.00 | 3.84 | 3.89 | 3.60 | 4.21 | 3.58, 3.82 | |
| 103.2 | 70.9 | 71.2 | 66.7 | 73.2 | 68.4 | 72.0 | |||
| G | β-D-Glcp-(1→ | 4.59 | 3.22 | 3.51 | 3.45 | 3.59 | 3.78, 3.90 | ||
| 103.5 | 74.0 | 75.4 | 69.9 | 76.2 | 61.3 | ||||
| H | β-d-Galp-(1→ | 4.51 | 3.50 | 3.62 | 3.95 | 3.66 | 3.69-3.74 b | ||
| 104.2 | 72.2 | 73.1 | 71.0 | 75.8 | 62.6 | ||||
| H′ | β-d-Galp-(1→ | 4.45 | 3.55 | 3.64 | 3.91 | 3.64 | 3.69-3.74 b | ||
| 104.1 | 71.7 | 73.2 | 69.6 | 76.0 | 62.6 | ||||
| K | →5)-Kdo | 1.86, 2.22 | 4.12 | 4.13 | 3.70 | 3.86 | 3.60, 3.84 | ||
| nd | nd | 34.4 | 66.4 | 75.3 | 70.0 | 72.1 | 64.1 | ||
| L | →3,7)-L-α-D-Hepp-(1→ | 5.28 | 4.18 | 3.99 | 4.01 | 3.60 | 4.14 | 3.55, 3.95 | |
| 101.8 | 70.2 | 81.9 | 65.6 | 73.2 | 69.1 | 73.5 | |||
| L′ | →2,3,7)-L-α-D-Hepp-(1→ | 5.38 | 4.22 | 4.10 | 4.04 | 3.60 | 4.14 | 3.56, 3.94 | |
| 99.7 | 78.9 | 79.3 | 66.4 | 73.2 | 69.1 | 73.6 | |||
| M | →3)-β-d-FucpNAc4N-(1→ | 4.57 | 3.83 | 4.16 | 3.98 | 4.09 | 1.32 | 2.07 | |
| 101.9 | 51.6 | 76.5 | 55.4 | 68.1 | 16.3 | 23.0, 174.7 | |||
| N | →4)-β-D-GlcpNAc-(1→ | 4.48 | 3.75 | 3.68 | 3.65 | 3.49 | 3.63, 3.82 | 2.06 | |
| 102.2 | 55.9 | 73.0 | 79.1 | 75.3 | 60.8 | 23.0, 175.4 | |||
| O | α-l-AltpNAcA | 4.88 | 3.97 | 3.66 | 4.38 | 4.41 | 2.00 | ||
| 101.7 | 52.2 | 68.8 | 69.9 | 78.7 | 175.4 | 23.0, 175.3 | |||
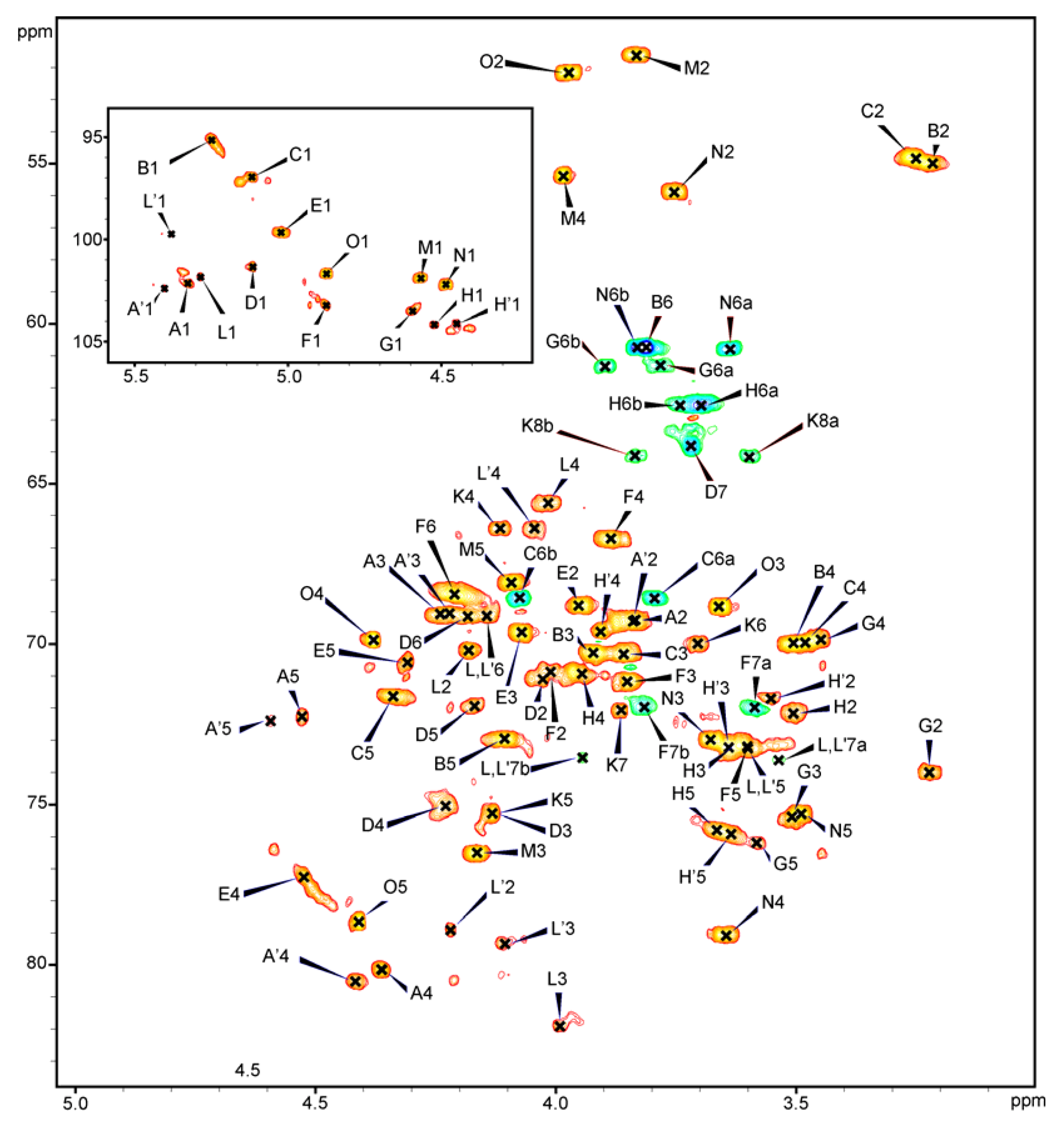
| Residue | Atom H-1/C-1 (ppm) | Connectivities to | Inter-Residue atom/residue | ||
|---|---|---|---|---|---|
| δH | δC | ||||
| A | →4)-α-D-GalpA-(1→ | 5.33/102.1 | 3.99 | nd | H-3 of L |
| B | α-D-GlcpN-(1→ | 5.24/95.1 | 4.52 | nd | H-4 of E |
| C | →6)-α-D-GlcpN-(1→ | 5.12/97.0 | 4.36 | 80.1 | H-4, C-4 of A |
| D | →3,4)-L-α-D-Hepp-(1→ | 5.11/101.3 | 4.13 | 75.3 | H-5, C-5 of K |
| E | →4)-α-D-GalpA-(1→ | 5.02/99.7 | 3.58 | 71.9 | H-7a, C-7 of F |
| F | →7)-L-α-D-Hepp-(1→ | 4.88/103.2 | 3.95 | 73.5 | H-7b, C-7 of L,L′ |
| G | β-D-Glcp-(1→ | 4.59/103.5 | nd | 78.9 | C-2 of L′ |
| H | β-D-Galp-(1→ | 4.51/104.2 | 4.23 | 75.1 | H-4, C-4 of D |
| L | →3,7)-L-α-D-Hepp-(1→ | 5.28/101.2 | 4.13 | 75.3 | H-3, C-3 of D |
| M | →3)-β-D-FucpNAc4N-(1→ | 4.57/101.9 | 3.65 | 79.1 | H-4, C-4 of N |
| N | →4)-β-D-GlcpNAc-(1→ | 4.48/102.2 | 3.79, 4.08 | 68.6 | H-6a, H-6b, C-6 of C |
| O | α-l-AltpNAcA | 4.88/101.7 | 4.16 | 76.5 | H-3, C-3 of M |
2.5. Serological Studies
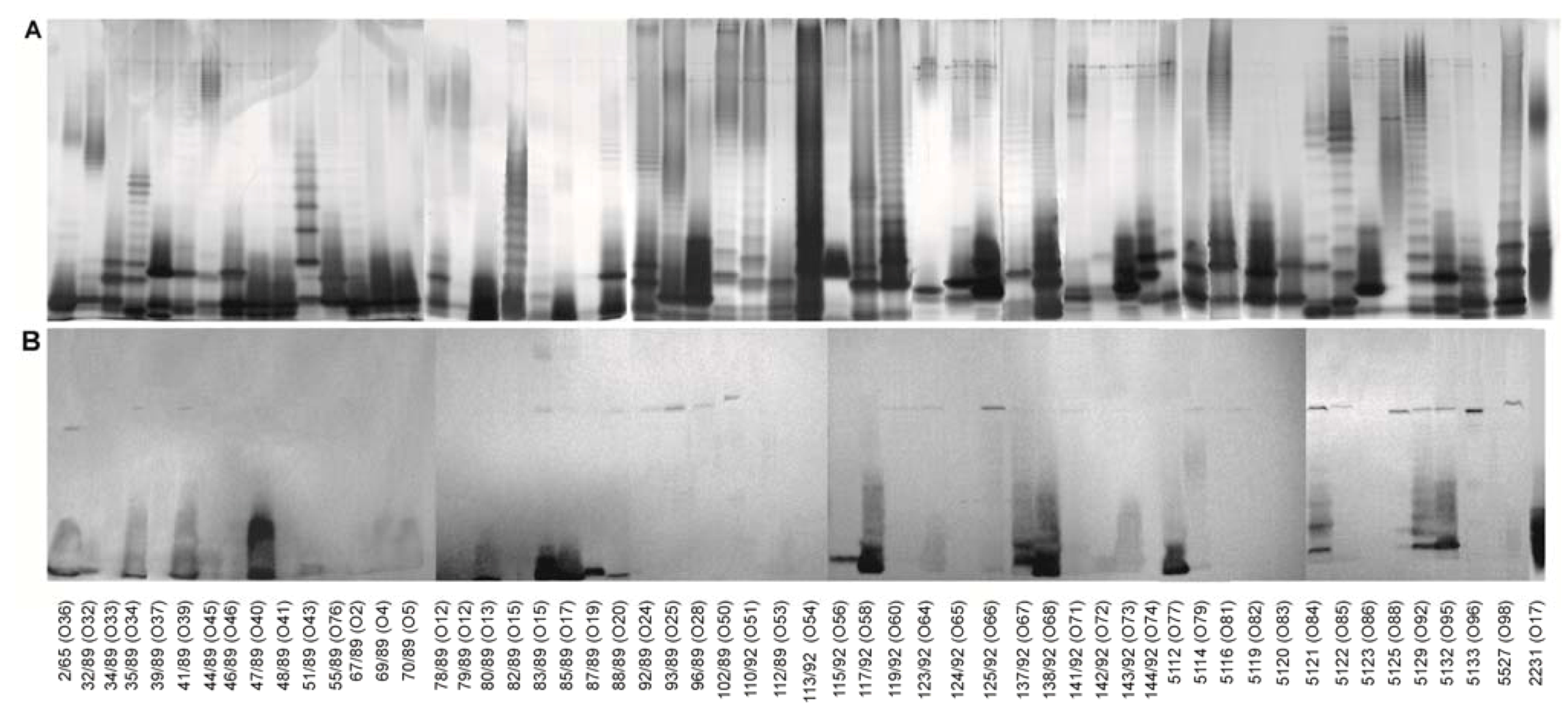
3. Experimental Section
3.1. Bacteria
3.2. Lipopolysaccharide and Core Oligosaccharide
3.3. Analytical Methods
3.4. Mass Spectrometry
3.5. NMR Spectroscopy
3.6. Preparation of Oligosaccharide Conjugate with BSA
3.7. Immunization Procedures
3.8. ELISA
3.9. SDS-PAGE
3.10. Immunoblotting
4. Conclusions
Acknowledgments
Conflict of Interests
References
- Garrity, G.M.; Bell, J.A.; Lilburn, T.G. Bergey’s Taxonomic Outline. Available online: http://dx.doi.org/10.1007/bergeysoutline200310 (accessed on 31 January 2013).
- Stock, I. Plesiomonas shigelloides: An emerging pathogen with unusual properties. Rev. Med. Microbiol. 2004, 15, 129–139. [Google Scholar] [CrossRef]
- Linnerborg, M.; Widmalm, G.; Weintraub, A.; Albert, M.J. Structural elucidation of the O-antigen lipopolysaccharide from two strains of Plesiomonas shigelloides that share a type-specific antigen with Shigella flexneri 6, and the common group 1 antigen with Shigella flexneri spp. and Shigella dysenteriae 1. Eur. J. Biochem. 1995, 231, 839–844. [Google Scholar] [CrossRef]
- Czaja, J.; Jachymek, W.; Niedziela, T.; Lugowski, C.; Aldova, E.; Kenne, L. Structural studies of the O-specific polysaccharide from Plesiomonas shigelloides strain CNCTC 113/92. Eur. J. Biochem. 2000, 267, 1672–1679. [Google Scholar] [CrossRef]
- Lukasiewicz, J.; Dzieciatkowska, M.; Niedziela, T.; Jachymek, W.; Augustyniuk, A.; Lugowski, C.; Kenne, L. Complete lipopolysaccharide of Plesiomonas shigelloides O74:H5 (strain CNCTC 144/92) 2. Lipid A, its structural variability, the linkage to the core oligosaccharide, and the biological activity of lipopolysaccharide. Biochemistry 2006, 45, 10434–10447. [Google Scholar] [CrossRef]
- Lukasiewicz, J.; Niedziela, T.; Jachymek, W.; Kenne, L.; Lugowski, C. Structure of the lipid A-inner core region and biological activity of Plesiomonas shigelloides O54 (strain CNCTC 113/92) lipopolysaccharide. Glycobiology 2006, 16, 538–550. [Google Scholar] [CrossRef]
- Niedziela, T.; Dag, S.; Lukasiewicz, J.; Dzieciatkowska, M.; Jachymek, W.; Lugowski, C.; Kenne, L. Complete lipopolysaccharide of Plesiomonas shigelloides O74:H5 (strain CNCTC 144/92). 1. Structural analysis of the highly hydrophobic lipopolysaccharide, including the O-antigen, its biological repeating unit, the core oligosaccharide, and the linkage between them. Biochemistry 2006, 45, 10422–10433. [Google Scholar]
- Niedziela, T.; Lukasiewicz, J.; Jachymek, W.; Dzieciatkowska, M.; Lugowski, C.; Kenne, L. Core oligosaccharides of Plesiomonas shigelloides O54:H2 (strain CNCTC 113/92)—Structural and serological analysis of the lipopolysaccharide core region, the O-antigen biological repeating unit, and the linkage between them. J. Biol. Chem. 2002, 277, 11653–11663. [Google Scholar]
- Pieretti, G.; Corsaro, M.M.; Lanzetta, R.; Parrilli, M.; Canals, R.; Merino, S.; Tomás, J.M. Structural studies of the O-chain polysaccharide from Plesiomonas shigelloides strain 302–73 (serotype O1). Eur. J. Org. Chem. 2008, 2008, 3149–3155. [Google Scholar]
- Pieretti, G.; Corsaro, M.M.; Lanzetta, R.; Parrilli, M.; Vilches, S.; Merino, S.; Tomás, J.M. Structure of the core region from the lippolysaccharide of Plesiomonas shigelloides strain 302–73 (serotype O1). Eur. J. Org. Chem. 2009, 2009, 1365–1371. [Google Scholar] [CrossRef]
- Pieretti, G.; Carillo, S.; Lindner, B.; Lanzetta, R.; Parrilli, M.; Jimenez, N.; Regue, M.; Tomas, J.M.; Corsaro, M.M. The complete structure of the core of the LPS from Plesiomonas shigelloides 302–73 and the identification of its O-antigen biological repeating unit. Carbohydr. Res. 2010, 345, 2523–2528. [Google Scholar] [CrossRef]
- Maciejewska, A.; Lukasiewicz, J.; Niedziela, T.; Szewczuk, Z.; Lugowski, C. Structural analysis of the O-specific polysaccharide isolated from Plesiomonas shigelloides O51 lipopolysaccharide. Carbohydr. Res. 2009, 344, 894–900. [Google Scholar] [CrossRef]
- Sawen, E.; Ostervall, J.; Landersjo, C.; Edblad, M.; Weintraub, A.; Ansaruzzaman, M.; Widmalm, G. Structural studies of the O-antigenic polysaccharide from Plesiomonas shigelloides strain AM36565. Carbohydr. Res. 2012, 348, 99–103. [Google Scholar] [CrossRef]
- Kubler-Kielb, J.; Mocca, C.; Vinogradov, E. The elucidation of the structure of the core part of the LPS from Plesiomonas shigelloides serotype O17 expressing O-polysaccharide chain identical to the Shigella sonnei O-chain. Carbohydr. Res. 2008, 343, 3123–3127. [Google Scholar] [CrossRef]
- Batta, G.; Liptak, A.; Schneerson, R.; Pozsgay, V. Conformational stabilization of the altruronic acid residue in the O-specific polysaccharide of Shigella sonnei/Plesiomonas shigelloides. Carbohydr. Res. 1997, 305, 93–99. [Google Scholar] [CrossRef]
- Kenne, L.; Lindberg, B.; Petersson, C.; Katzenellenbogen, E.; Romanowska, E. Structural studies of the O-specific side chains of the Shigella sonnei phase I lipopolysaccharide. Carbohydr. Res. 1980, 78, 119–126. [Google Scholar] [CrossRef]
- Aldova, E. Serovars of Plesiomonas shigelloides. Zentralbl. Bakteriol. 1994, 281, 38–44. [Google Scholar]
- Aldova, E. The importance of serotyping Plesiomonas shigelloides. Epidemiol. Mikrobiol. Immunol. 1995, 44, 147–154. [Google Scholar]
- Gorin, P.A.J.; Mazurek, M. Further studies on the assignment of signals in 13C magnetic resonance spectra of aldoses and derived methyl glycosides. Can. J. Chem. 1975, 53, 1212–1223. [Google Scholar] [CrossRef]
- Boratynski, J.; Roy, R. High temperature conjugation of proteins with carbohydrates. Glycoconj. J. 1998, 15, 131–138. [Google Scholar] [CrossRef]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
- Petersson, C.; Niedziela, T.; Jachymek, W.; Kenne, L.; Zarzecki, P.; Lugowski, C. Structural studies of the O-specific polysaccharide of Hafnia alvei strain PCM 1206 lipopolysaccharide containing D-allothreonine. Eur. J. Biochem. 1997, 244, 580–586. [Google Scholar]
- Westphal, O.; Jann, K. Bacterial lipopolysacharides: Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 1965, 5, 83–89. [Google Scholar]
- Niedziela, T.; Petersson, C.; Helander, A.; Jachymek, W.; Kenne, L.; Lugowski, C. Structural studies of the O-specific polysaccharide of Hafnia alvei strain 1209 lipopolysaccharide. Eur. J. Biochem. 1996, 237, 635–641. [Google Scholar]
- Gerwig, G.J.; Kamerling, J.P.; Vliegenthart, J.F.G. Determination of the D and L configuration of neutral monosaccharides by high-resolution capillary GLC. Carbohydr. Res. 1978, 62, 349–357. [Google Scholar] [CrossRef]
- Gerwig, G.J.; Kamerling, J.P.; Vliegenthart, J.F.G. Determination of the absolute configuration of monosaccharides in complex carbohydrates by capillary GLC. Carbohydr. Res. 1979, 77, 1–7. [Google Scholar] [CrossRef]
- Taylor, R.L.; Shively, J.E.; Conrad, H.E. Stoichiometric reduction of uronic acid carboxyl groups in polysaccharides. Methods Carbohydr. Chem. 1976, 7, 149–151. [Google Scholar]
- Hakomori, S. A rapid permethylation of glycolipid and polysaccharide catalyzed by methylsulphinyl carbanion in dimethyl sulphoxide. J. Biochem. 1964, 55, 205–208. [Google Scholar]
- Goddard, T.D.; Kneller, D.G. Sparky, 3rd ed; University of California: San Francisco, CA, USA, 2001. [Google Scholar]
- Jennings, H.J.; Lugowski, C. Immunochemistry of groups A, B, and C meningococcal polysaccharide-tetanus toxoid conjugates. J. Immunol. 1981, 127, 1011–1018. [Google Scholar]
- Lugowski, C.; Romanowska, E. Enterobacterial common antigen: Isolation from Shigella sonnei, purification and immunochemical characterization. Eur. J. Biochem. 1978, 91, 89–97. [Google Scholar] [CrossRef]
- Voller, A.; Draper, C.; Bidwell, D.E.; Bartlett, A. Microplate enzyme-linked immunosorbent assay for chagas’ disease. Lancet 1975, 1, 426–428. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Lugowski, C.; Jachymek, W.; Niedziela, T.; Rowinski, S. Serological characterisation of anti-endotoxin sera directed against the conjugates of oligosaccharide core of Escherichia coli type R1, R2, R3, J5 and Salmonella Ra with tetanus toxoid. FEMS Immunol. Med. Microbiol. 1996, 16, 21–30. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Maciejewska, A.; Lukasiewicz, J.; Kaszowska, M.; Man-Kupisinska, A.; Jachymek, W.; Lugowski, C. Core Oligosaccharide of Plesiomonas shigelloides PCM 2231 (Serotype O17) Lipopolysaccharide — Structural and Serological Analysis. Mar. Drugs 2013, 11, 440-454. https://doi.org/10.3390/md11020440
Maciejewska A, Lukasiewicz J, Kaszowska M, Man-Kupisinska A, Jachymek W, Lugowski C. Core Oligosaccharide of Plesiomonas shigelloides PCM 2231 (Serotype O17) Lipopolysaccharide — Structural and Serological Analysis. Marine Drugs. 2013; 11(2):440-454. https://doi.org/10.3390/md11020440
Chicago/Turabian StyleMaciejewska, Anna, Jolanta Lukasiewicz, Marta Kaszowska, Aleksandra Man-Kupisinska, Wojciech Jachymek, and Czeslaw Lugowski. 2013. "Core Oligosaccharide of Plesiomonas shigelloides PCM 2231 (Serotype O17) Lipopolysaccharide — Structural and Serological Analysis" Marine Drugs 11, no. 2: 440-454. https://doi.org/10.3390/md11020440
APA StyleMaciejewska, A., Lukasiewicz, J., Kaszowska, M., Man-Kupisinska, A., Jachymek, W., & Lugowski, C. (2013). Core Oligosaccharide of Plesiomonas shigelloides PCM 2231 (Serotype O17) Lipopolysaccharide — Structural and Serological Analysis. Marine Drugs, 11(2), 440-454. https://doi.org/10.3390/md11020440




