Smenamides A and B, Chlorinated Peptide/Polyketide Hybrids Containing a Dolapyrrolidinone Unit from the Caribbean Sponge Smenospongia aurea. Evaluation of Their Role as Leads in Antitumor Drug Research
Abstract
:1. Introduction
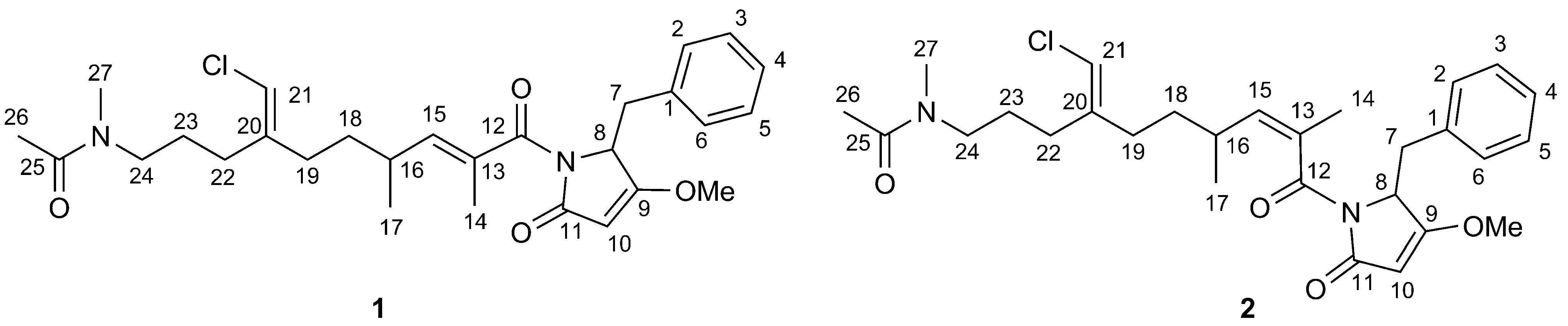
2. Results and Discussion
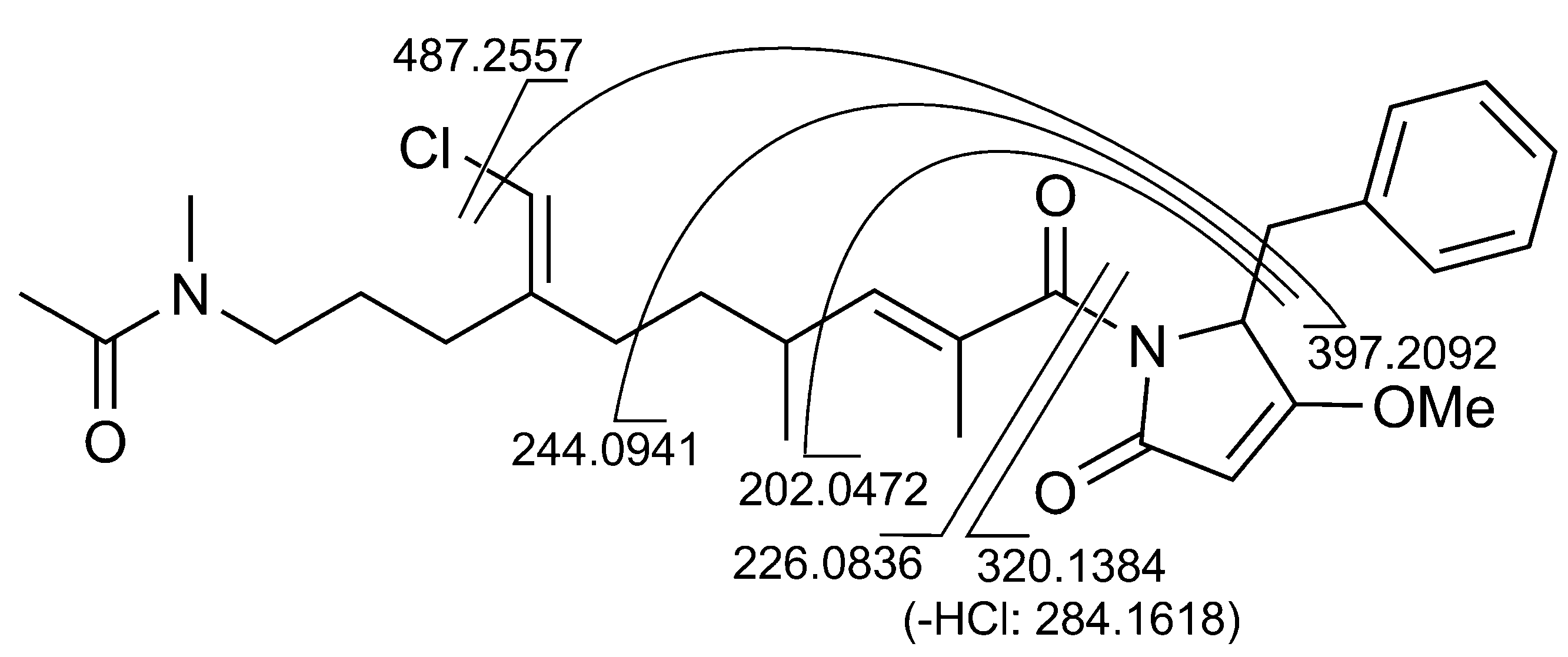
2.1. Structural Elucidation
| Z-Conformer | E-Conformer | ||||||
|---|---|---|---|---|---|---|---|
| Position | δH [Mult., J (Hz)] | δC [Mult.] | δH [Mult., J (Hz)] | δC [Mult.] | COSY | HMBC | |
| 1 | − | 135.6 (C) | − | 135.6 (C) | |||
| 2/6 | 6.99 (m) | 130.8 (CH) | 6.99 (m) | 130.8 (CH) | 3/5 | 4 | |
| 3/5 | 7.23 (ovl) | 129.4 (CH) | 7.23 (ovl) | 129.4 (CH) | 2/6 | 1 | |
| 4 | 7.23 (ovl) | 128.3 (CH) | 7.23 (ovl) | 128.3 (CH) | 2/6 | ||
| 7 | a | 3.37 (ovl) | 34.8 (CH2) | 3.37 (ovl) | 34.8 (CH2) | 7b, 8 | 1, 2/6, 8, 9 |
| b | 3.19 (m) | 3.19 (m) | 7a, 8 | 2/6 | |||
| 8 | 5.02 (ovl) | 60.5 (CH) | 5.02 (ovl) | 60.5 (CH) | 7a, 7b | ||
| 9 | − | 179.5 (C) | − | 179.5 (C) | |||
| 10 | 5.04 (br. s) | 95.5 (CH) | 5.02 (br. s) | 95.5 (CH) | 8, 11 | ||
| 11 | − | 170.7 (C) | − | 170.7 (C) | |||
| 12 | − | 172.3 (C) | − | 172.2 (C) | |||
| 13 | − | 132.1 (C) | − | 132.1 (C) | |||
| 14 | 1.77 (d, 1.5) | 13.7 (CH3) | 1.78 (d, 1.5) | 13.7 (CH3) | 15 | 12, 13, 15 | |
| 15 | 5.36 (br. d, 10.2) | 144.1 (CH) | 5.36 (br. d, 10.2) | 144.1 (CH) | 14, 16 | ||
| 16 | 2.45 (m) | 33.4 (CH) | 2.48 (m) | 33.4 (CH) | 15, 17, 18a | ||
| 17 | 0.98 (d, 6.5) | 20.4 (CH3) | 1.00 (d, 6.5) | 20.6 (CH3) | 16 | 15, 18, 19 | |
| 18 | a | 1.51 (ovl) | 36.1 (CH2) | 1.52 (ovl) | 35.9 (CH2) | 16, 19a, 19b | 19 |
| b | 1.28 (ovl) | 1.30 (ovl) | 19a, 19b | ||||
| 19 | a | 2.19 (ovl) | 33.2 (CH2) | 2.23 (ovl) | 33.2 (CH2) | 18a, 18b, 19b, 21 | 20, 21 |
| b | 2.06 (ovl) | 2.05 (ovl) | 18a, 18b, 19a, 21 | 20 | |||
| 20 | − | 143.1 (C) | − | 142.8 (C) | |||
| 21 | 5.93 (br. s) | 113.9 (CH) | 5.97 (br. s) | 114.1 (CH) | 19a, 19b | 20, 22 | |
| 22 | a | 2.22 (m) | 28.1 (CH2) | 2.26 (m) | 28.0 (CH2) | 22b, 23 | 20, 21 |
| b | 2.15 (m) | 2.18 (m) | 22a, 23 | 20 | |||
| 23 | 1.64 (m) | 25.9 (CH2) | 1.70 (m) | 26.6 (CH2) | 22a, 22b, 24 | ||
| 24 | 3.36 (ovl) | 48.6 (CH2) | 3.33 (ovl) | 51.5 (CH2) | 23 | 22, 23, 25, 27 | |
| 25 | − | 172.9 (C) | − | 172.7 (C) | |||
| 26 | 2.08 (s) | 21.7 (CH3) | 2.07 (s) | 21.1 (CH3) | 27 | 25 | |
| 27 | 3.03 (s) | 36.6 (CH3) | 2.88 (s) | 33.7 (CH3) | 26 | 24, 25 | |
| OMe | 3.97 (s) | 59.7 (CH3) | 3.97 (s) | 59.7 (CH3) | 9 | ||

2.2. 16S rRNA Metagenomic Analysis
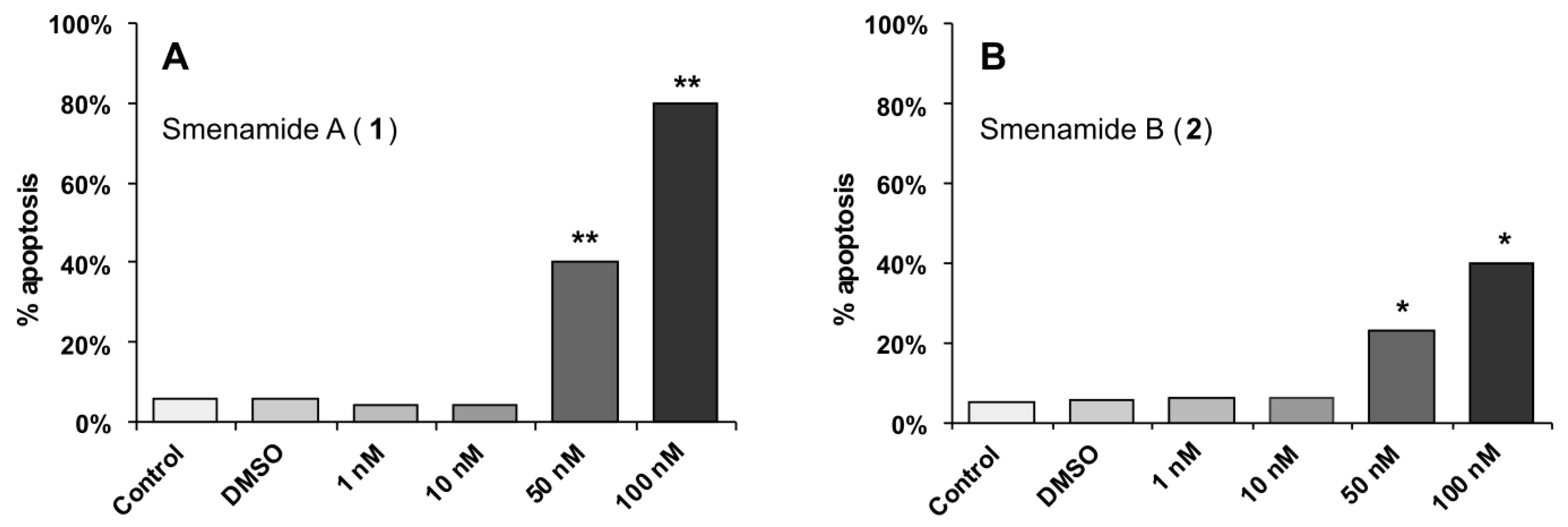
2.3. In Vitro Evaluation of Cytotoxic Activity of Smenamides

3. Experimental Section
3.1. General Experimental Procedures
3.2. Collection, Extraction, and Isolation
3.3. Smenamide A (1)
3.4. Smenamide B (2)
3.5. DNA Isolation
3.6. PCR Amplification and Clone Library Construction
3.7. Cell Culture
3.8. MTT Assay
3.9. Apoptosis assay
3.10. Accession Codes
4. Conclusion
Acknowledgments
Conflicts of Interest
References
- Lamoral-Theys, D.; Fattorusso, E.; Mangoni, A.; Perinu, C.; Kiss, R.; Costantino, V. Evaluation of the Antiproliferative Activity of Diterpene Isonitriles from the Sponge Pseudoaxinella flava in Apoptosis-sensitive and Apoptosis-resistant Cancer Cell Lines. J. Nat. Prod. 2011, 74, 2299–2303. [Google Scholar] [CrossRef]
- Costantino, V.; Fattorusso, E.; Mangoni, A.; Perinu, C.; Cirino, G.; De Gruttola, L.; Roviezzo, F. Tedanol: A potent anti-inflammatory ent-pimarane diterpene from the Caribbean Sponge Tedania ignis. Bioorg. Med. Chem. 2009, 17, 7542–7547. [Google Scholar] [CrossRef]
- Bialonska, D.; Zjawiony, J.K. Aplysinopsins—Marine Indole Alkaloids: Chemistry, Bioactivity and Ecological Significance. Mar. Drugs 2009, 7, 166–183. [Google Scholar] [CrossRef]
- Song, J.; Jeong, W.; Wang, N.; Lee, H.-S.; Sim, C. J.; Oh, K.-B.; Shin, J. Scalarane Sesterterpenes from the Sponge Smenospongia sp. J. Nat. Prod. 2008, 71, 1866–1871. [Google Scholar] [CrossRef]
- Djura, P.; Stierle, D.B.; Sullivan, B.; Faulkner, D.J.; Arnold, E.V.; Clardy, J. Some metabolites of the marine sponges Smenospongia aurea and Smenospongia (=Polyfibrospongia) echina. J. Org. Chem. 1980, 45, 1435–1441. [Google Scholar] [CrossRef]
- Aiello, A.; Fattorusso, E.; Menna, M. A new antibiotic chloro-sesquiterpene from the Caribbean sponge Smenospongia aurea. Z. Naturforsch. B 1993, 48, 209–212. [Google Scholar]
- Bourguet-Kondracki, M.L.; Martin, M.T.; Guyot, M. Smenoqualone, a novel sesquiterpenoid from the marine sponge Smenospongia sp. Tetrahedron Lett. 1992, 33, 8079–8080. [Google Scholar] [CrossRef]
- Kazlauskas, R.; Murphy, P.T.; Quinn, R.J.; Wells, R.J. Aplysinopsin, a new tryptophan derivative from a sponge. Tetrahedron Lett. 1977, 18, 61–64. [Google Scholar] [CrossRef]
- Hollenbeak, K.H.; Schmitz, F.J. Aplysinopsin: Antineoplastic tryptophan derivative from the marine sponge Verongia spengelii. Lloydia 1977, 10, 479–481. [Google Scholar]
- Pretsch, E.; Büelhmann, P.; Affolter, M. Structure Determination of Organic Compounds. In Tables of Spectral Data, 3rd ed.; Springer-Verlag Berlin: Heidelberg, Germany, 2000; pp. 83 and 225. [Google Scholar]
- Pettit, G.R.; Kamano, Y.; Dufresne, C.; Cerny, R.L.; Herald, C.L.; Schmidt, J.M. Isolation and Structure of the Cytostatic Linear Depsipeptide Dolastatin 15. J. Org. Chem. 1989, 54, 6005–6006. [Google Scholar] [CrossRef]
- Ghisalberti, E.L. Bioactive tetramic acid metabolites. Stud. Nat. Prod. Chem. 2003, 28, 109–163. [Google Scholar] [CrossRef]
- Phuwapraisirisan, P.; Matsunaga, S.; Fusetani, N. Mycapolyols A–F, New Cytotoxic Metabolites of Mixed Biogenesis from the Marine Sponge Mycale izuensis. Organic Lett. 2005, 7, 2233–2236. [Google Scholar] [CrossRef]
- Simmons, T.L.; McPhail, K.L.; Ortega-Barría, E.; Mooberry, S.L.; Gerwick, W.H. Belamide A, a new antimitotic tetrapeptide from a Panamanian marine cyanobacterium. Tetrahedron Lett. 2006, 47, 3387–3390. [Google Scholar] [CrossRef]
- Edwards, D.J.; Marquez, B.L.; Nogle, L.M.; McPhail, K.; Goeger, D.E.; Roberts, M.A.; Gerwick, W.H. Structure and Biosynthesis of the Jamaicamides, New Mixed Polyketide-Peptide Neurotoxins from the Marine Cyanobacterium Lyngbya majuscula. Chem. Biol. 2004, 11, 817–833. [Google Scholar] [CrossRef]
- Prawat, H.; Mahidol, C.; Kaweetripob, W.; Wittayalai, S.; Ruchirawat, S. Iodo-sesquiterpene hydroquinone and brominated indole alkaloids from the Thai sponge Smenospongia sp. Tetrahedron 2012, 68, 6881–6886. [Google Scholar] [CrossRef]
- Boutte, C.; Grubisic, S.; Balthasart, P.; Wilmotte, A. Testing of primers for the study of cyanobacterial molecular diversity by DGGE. J. Microbiol. Methods 2006, 65, 542–550. [Google Scholar] [CrossRef]
- Erwin, P.M.; Thacker, R.W. Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Mol. Ecol. 2008, 17, 2937–2947. [Google Scholar] [CrossRef]
- Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N.J.; van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; et al. Defensive Bacteriome Symbiont with a Drastically Reduced Genome. Curr. Biol. 2013, 23, 1478–1484. [Google Scholar] [CrossRef]
- Erwin, P.M.; López-Legentil, S.; Turon, X. Ultrastructure, molecular phylogenetics, and chlorophyll a content of novel cyanobacterial symbionts in temperate sponges. Microb. Ecol. 2012, 64, 771–783. [Google Scholar] [CrossRef] [Green Version]
- Pettit, G.R.; Kamano, Y.; Herald, C.L.; Tuinman, A.A.; Boettner, F.E.; Kizu, H.; Schmidt, J.M.; Baczynskyj, L.; Tomer, K.B.; Bontems, R.J. The isolation and structure of a remarkable marine animal antineoplastic constituent: Dolastatin 10. J. Am. Chem. Soc. 1987, 109, 6883–6885. [Google Scholar] [CrossRef]
- Newman, D.J.; Giddings, L.A. Natural products as leads to antitumor drugs. Phytochem. Rev. 2013, 12, 293–304. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Teta, R.; Irollo, E.; Della Sala, G.; Pirozzi, G.; Mangoni, A.; Costantino, V. Smenamides A and B, Chlorinated Peptide/Polyketide Hybrids Containing a Dolapyrrolidinone Unit from the Caribbean Sponge Smenospongia aurea. Evaluation of Their Role as Leads in Antitumor Drug Research. Mar. Drugs 2013, 11, 4451-4463. https://doi.org/10.3390/md11114451
Teta R, Irollo E, Della Sala G, Pirozzi G, Mangoni A, Costantino V. Smenamides A and B, Chlorinated Peptide/Polyketide Hybrids Containing a Dolapyrrolidinone Unit from the Caribbean Sponge Smenospongia aurea. Evaluation of Their Role as Leads in Antitumor Drug Research. Marine Drugs. 2013; 11(11):4451-4463. https://doi.org/10.3390/md11114451
Chicago/Turabian StyleTeta, Roberta, Elena Irollo, Gerardo Della Sala, Giuseppe Pirozzi, Alfonso Mangoni, and Valeria Costantino. 2013. "Smenamides A and B, Chlorinated Peptide/Polyketide Hybrids Containing a Dolapyrrolidinone Unit from the Caribbean Sponge Smenospongia aurea. Evaluation of Their Role as Leads in Antitumor Drug Research" Marine Drugs 11, no. 11: 4451-4463. https://doi.org/10.3390/md11114451
APA StyleTeta, R., Irollo, E., Della Sala, G., Pirozzi, G., Mangoni, A., & Costantino, V. (2013). Smenamides A and B, Chlorinated Peptide/Polyketide Hybrids Containing a Dolapyrrolidinone Unit from the Caribbean Sponge Smenospongia aurea. Evaluation of Their Role as Leads in Antitumor Drug Research. Marine Drugs, 11(11), 4451-4463. https://doi.org/10.3390/md11114451






