Assessing the Effectiveness of Functional Genetic Screens for the Identification of Bioactive Metabolites
Abstract
:1. Introduction
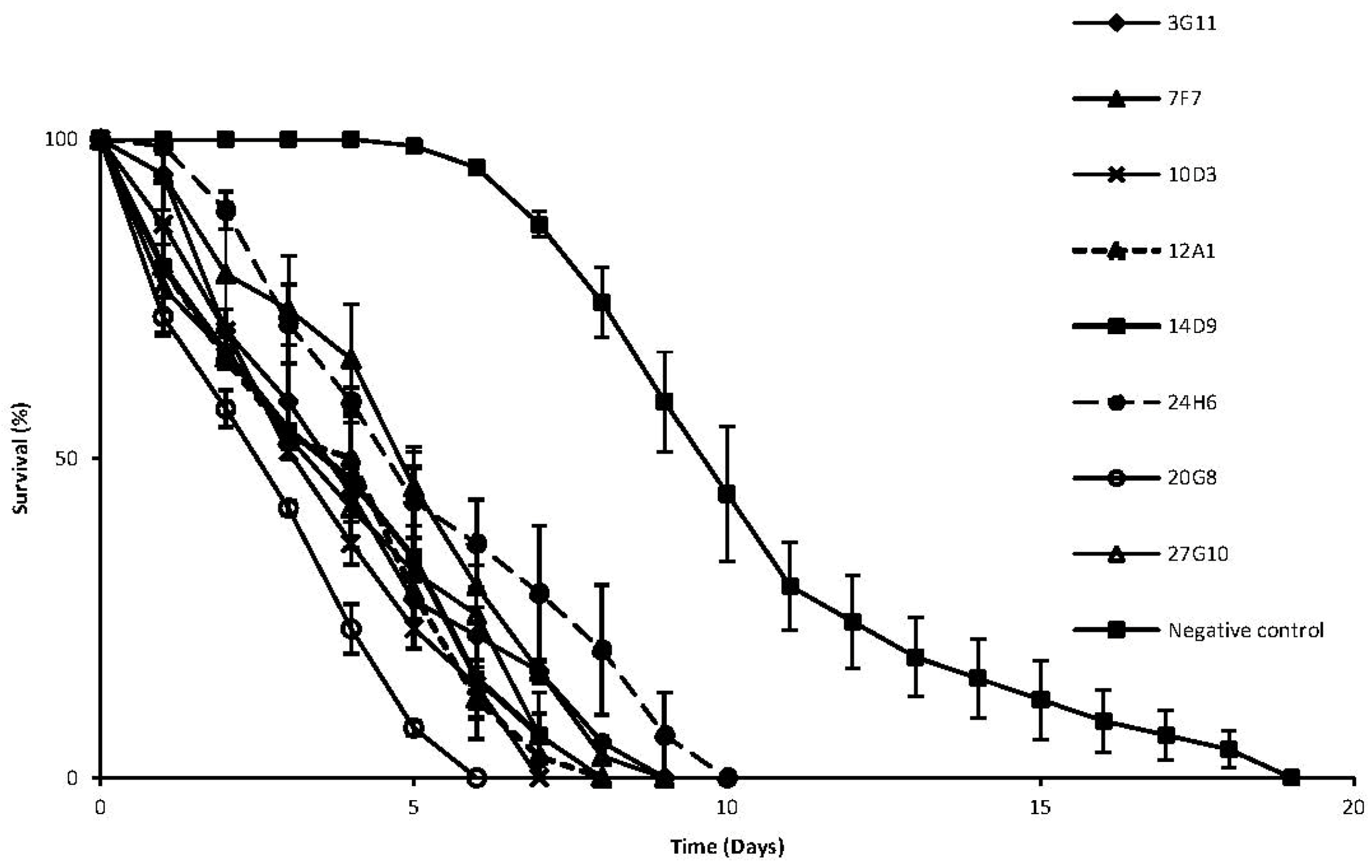
2. Results and Discussion
| Library clone number | GenBank no. | Fosmid size (bp) | Antibacterial activity | Antinematode activity | Parental strain * |
|---|---|---|---|---|---|
| 3G11 | JX523949 | 24 296 | + | + | D250 |
| 7F7 | KC211770 | 34 039 | − | + | U95 |
| 10D3 | JX523951 | 36 314 | + | + | D323 |
| 12A1 | JX523952 | 32 547 | + | + | D250 |
| 14D9 | JX523953 | 19 858 | + | + | D323 |
| 24H6 | KC211769 | 15 952 | − | + | D323 |
| 27G10 | KC211768 | 33 970 | − | + | U95 |
| 20G8 | JX523957 | 25 000 | + | + | D250 |
| 15E10 | JX523954 | 30 000 | + | + | D250 |
| 23H6 | JX523958 | 23 000 | + | + | D250 |
| 19F10 | JX523956 | 29 696 | + | − | U95 |
| 16B12 | JX523955 | 39 325 | + | − | D323 |
| 9E12 | JX523950 | 37 944 | + | − | D323 |
| Strain ID | GenBank no. | Isolation source | Closest relative | Phylum | % Identity |
|---|---|---|---|---|---|
| U95 | FJ440958 | Ulva australis | Uncultured alpha-proteobacterium, JN874385 | Proteobacteria | 98 |
| U140 | FJ440963 | Ulva australis | Micrococcus luteus, JQ795852 | Actinobacteria | 99 |
| U156 | FJ440965 | Ulva australis | Gamma-proteobacterium D261, FJ440978 | Proteobacteria | 99 |
| D250 | FJ440973 | Delisea pulchra | Gamma-proteobacterium D259, FJ440977 | Proteobacteria | 99 |
| D295 | FJ440982 | Delisea pulchra | Flavobacteriaceae bacterium SW058, AF493683 | Bacteroidetes | 98 |
| D323 | FJ440988 | Delisea pulchra | Pseudovibrio sp. Pv348, 1413, HE818384 | Proteobacteria | 100 |
3. Experimental Section
4. Conclusions
Acknowledgments
Supplementary Material
Analysis of Fosmids
Identification of Fosmid Parental Strains
| Fosmid | Primer pairs | Sequence (5′ to 3′) | Product length |
|---|---|---|---|
| 3G11 | 3G11 forward | GGC TAG AGG CGT TGC GTA TTG TGC | 679 bp |
| 3G11 reverse | CTT TAA AGG CGC CGG GCT CCA TCT | ||
| 7F7 | 7F7 forward | AAC CTG CCA GAT ACC AAA CG | 1728 bp |
| 7F7 reverse | GGT CAA CCG GAA CAC AGA GT | ||
| 9E12 | 9E12 forward | TGC TGA AGC GGA AGT GGA GTA TGA | 388 bp |
| 9E12 reverse | CGG CAC GTT GAA GTC GAA GTA GTC | ||
| 10D3 | 10D3 forwards | CTA TGA TCA CGA CCA GCA CAC GAG | 571 bp |
| 10D3 reverse | ACC AGG TCC GAG CCA TCT ACA CAA | ||
| 12A1 | 12A1 forward | ACA GCG GTG GTC ATT ATT GGA ACG | 432 bp |
| 12A1 reverse | GGC GGT GTG AAA GCG GTG ATA GTC | ||
| 14D9 | 14D9 forward | GGC ACA CGG CTC TTC ATC TTC ACA | 532 bp |
| 14D9 reverse | GCC GCG TTC GTT CCC GTC AC | ||
| 24H6 | 24H6 forward | CGT GAA TGT GGA AGG TGT TG | 2228 bp |
| 24H6 reverse | AAA GAA AGC TTG GCG TTG AA | ||
| 15E10 | 15E10 forward | GCT AAA CTG CCT GAC TTC TAC ACG | 509 bp |
| 20G8 | 15E10 reverse | CTG GAT ACT GCT GGT TTG ACT ACG | |
| 23H6 | |||
| 16B12 | 16B12 forward | CTC TTT ACG CCC AGT GAT TCC | 613 bp |
| 16B12 reverse | TTA TTT GCG TGT TCC TCG TCT ATT | ||
| 19F10 | 19F10 forward | ACA TCA TCG CCG CTA AGG TA | 772 bp |
| 19F10 reverse | TAT GGG ATT CTG TTG TTT CGT AA | ||
| 27G10 | 27G10 forward | AGC GGC TTA CCT CAA GAA CA | 1803 bp |
| 27G10 reverse | GCT GAG AAC CCA GAA AGT CG |
References
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef]
- Wang, G.Y.; Graziani, E.; Waters, B.; Pan, W.; Li, X.; McDermott, J.; Meurer, G.; Saxena, G.; Andersen, R.J.; Davies, J. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2000, 2, 2401–2404. [Google Scholar]
- Brady, S.F.; Chao, C.J.; Clardy, J. New natural product families from an environmental DNA (eDNA) gene cluster. J. Am. Chem. Soc. 2002, 124, 9968–9969. [Google Scholar] [CrossRef]
- MacNeil, I.A.; Tiong, C.L.; Minor, C.; August, P.R.; Grossman, T.H.; Loiacono, K.A.; Lynch, B.A.; Phillips, T.; Narula, S.; Sundaramoorthi, R.; et al. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J. Mol. Microbiol. Biotechnol. 2001, 3, 301–308. [Google Scholar]
- Burke, C.; Thomas, T.; Egan, S.; Kjelleberg, S. The use of functional genomics for the identification of a gene cluster encoding for the biosynthesis of an antifungal tambjamine in the marine bacterium Pseudoalteromonas tunicata. Environ. Microbiol. 2007, 9, 814–818. [Google Scholar] [CrossRef]
- Egan, S.; Thomas, T.; Kjelleberg, S. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opin. Microbiol. 2008, 11, 219–225. [Google Scholar]
- Sørensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005, 115, 113–128. [Google Scholar]
- Uchiyama, T.; Miyazaki, K. Functional metagenomics for enzyme discovery: Challenges to efficient screening. Curr. Opin. Biotechnol. 2009, 20, 616–622. [Google Scholar] [CrossRef]
- Ekkers, D.M.; Cretoiu, M.S.; Kielak, A.M.; Elsas, J.D. The great screen anomaly—A new frontier in product discovery through functional metagenomics. Appl. Microbiol. Biotechnol. 2012, 93, 1005–1020. [Google Scholar] [CrossRef]
- Kimelman, A.; Levy, A.; Sberro, H.; Kidron, S.; Leavitt, A.; Amitai, G.; Yoder-Himes, D.R.; Wurtzel, O.; Zhu, Y.; Rubin, E.M.; et al. A vast collection of microbial genes that are toxic to bacteria. Genome Res. 2012, 22, 802–809. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 2003, 14, 303–310. [Google Scholar] [CrossRef]
- Peláez, F. The historical delivery of antibiotics from microbial natural products—Can history repeat? Biochem. Pharmacol. 2006, 71, 981–990. [Google Scholar] [CrossRef]
- Brady, S.F.; Clardy, J. Palmitoylputrescine, an antibiotic isolated from the heterologous expression of DNA extracted from bromeliad tank water. J. Nat. Prod. 2004, 67, 1283–1286. [Google Scholar] [CrossRef]
- Henne, A.; Schmitz, R.A.; Bömeke, M.; Gottschalk, G.; Daniel, R. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 3113–3116. [Google Scholar]
- Penesyan, A.; Marshall-Jones, Z.; Holmstrom, C.; Kjelleberg, S.; Egan, S. Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol. Ecol. 2009, 69, 113–124. [Google Scholar] [CrossRef]
- Ballestriero, F.; Thomas, T.; Burke, C.; Egan, S.; Kjelleberg, S. Identification of compounds with bioactivity against the nematode Caenorhabditis elegans by a screen based on the functional genomics of the marine bacterium Pseudoalteromonas tunicata D2. Appl. Environ. Microbiol. 2010, 76, 5710–5717. [Google Scholar] [CrossRef]
- Yung, P.Y.; Burke, C.; Lewis, M.; Kjelleberg, S.; Thomas, T. Novel antibacterial proteins from the microbial communities associated with the sponge Cymbastela concentrica and the green alga Ulva australis. Appl. Environ. Microbiol. 2011, 77, 1512–1515. [Google Scholar] [CrossRef]
- August, P.; Grossman, T.; Minor, C.; Draper, M.; MacNeil, I.A.; Pemberton, J.; Call, K.; Holt, D.; Osburne, M. Sequence analysis and functional characterization of the violacein biosynthetic pathway from Chromobacterium violaceum. J. Mol. Microbiol. Biotechnol. 2000, 2, 513–519. [Google Scholar]
- Thomas, T.; Evans, F.F.; Schleheck, D.; Mai-Prochnow, A.; Burke, C.; Penesyan, A.; Dalisay, D.S.; Stelzer-Braid, S.; Saunders, N.; Johnson, J.; et al. Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS One 2008, 3, e3252. [Google Scholar]
- Penesyan, A.; Breider, S.; Schumann, P.; Tindall, B.J.; Egan, S.; Brinkhoff, T. Epibacterium ulvae gen. nov. sp. nov., epibiotic bacteria isolated from the surface of a marine alga. Int. J. Syst. Evol. Microbiol. 2012. [Google Scholar] [CrossRef]
- Takahashi, H.; Kumagai, T.; Kitani, K.; Mori, M.; Matoba, Y.; Sugiyama, M. Cloning and characterization of a Streptomyces single module type non-ribosomal peptide synthetase catalyzing a blue pigment synthesis. J. Biol. Chem. 2007, 12, 9073–9081. [Google Scholar]
- Reverchon, S.; Rouanet, C.; Expert, D.; Nasser, W. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J. Bacteriol. 2002, 184, 654–665. [Google Scholar] [CrossRef]
- Finking, R.; Marahiel, M.A. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 2004, 58, 453–488. [Google Scholar] [CrossRef]
- McMahon, M.D.; Guan, C.; Handelsman, J.; Thomas, M.G. Metagenomic analysis of Streptomyces lividans reveals host-dependent functional expression. Appl. Environ. Microbiol. 2012, 78, 3622–3629. [Google Scholar] [CrossRef]
- Knight, V.; Sanglier, J.J.; DiTullio, D.; Braccili, S.; Bonner, P.; Waters, J.; Hughes, D.; Zhang, L. Diversifying microbial natural products for drug discovery. Appl. Microbiol. Biotechnol. 2003, 62, 446–458. [Google Scholar] [CrossRef]
- Schiewe, H.J.; Zeeck, A. Cineromycins, γ-butyrolactones and ansamycins by analysis of the secondary metabolite pattern created by a single strain of Streptomyces. J. Antibiot. 1999, 52, 635–642. [Google Scholar] [CrossRef]
- Tillett, D.; Neilan, B.A. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 2000, 36, 251–258. [Google Scholar] [CrossRef]
- Whitworth, D.E. Genomes and knowledge—A questionable relationship? Trends Microbiol. 2008, 16, 512–519. [Google Scholar] [CrossRef]
- Whitworth, D.E.; Cock, P.J.A. Evolution of prokaryotic two-component systems: Insights from comparative genomics. Amino Acids 2009, 37, 459–466. [Google Scholar] [CrossRef]
- Allen, H.K.; Cloud-Hansen, K.A.; Wolinski, J.M.; Guan, C.; Greene, S.; Lu, S.; Boeyink, M.; Broderick, N.A.; Raffa, K.F.; Handelsman, J. Resident microbiota of the gypsy moth midgut harbors antibiotic resistance determinants. DNA Cell Biol. 2009, 28, 109–117. [Google Scholar] [CrossRef]
- Gordon, D.; Abajian, C.; Green, P. Consed: A graphical tool for sequence finishing. Genome Res. 1998, 8, 195–202. [Google Scholar]
- Noguchi, H.; Park, J.; Takagi, T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006, 34, 5623–5630. [Google Scholar] [CrossRef]
- MetaGene. Available online: http://metagene.cb.k.u-tokyo.ac.jp/metagene/ (accessed on 10 January 2009).
- Thomas, T.; Rusch, D.; DeMaere, M.Z.; Yung, P.Y.; Lewis, M.; Halpern, A.; Heidelberg, K.B.; Egan, S.; Steinberg, P.D.; Kjelleberg, S. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 2010, 4, 1557–1567. [Google Scholar] [CrossRef]
- ExPASy. Available online: http://expasy.org/sprot/ (accessed on 10 January 2009).
- TIGRRAMS. Available online: http://www.tigr.org/TIGRFAMs/ (accessed on 10 January 2009).
- KEGG. Available online: http://www.genome.jp/kegg/ (accessed on 10 January 2009).
- GOG. Available online: http://www.ncbi.nlm.nih.gov/COG/ (accessed on 10 January 2009).
- Samples Availability: Available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Penesyan, A.; Ballestriero, F.; Daim, M.; Kjelleberg, S.; Thomas, T.; Egan, S. Assessing the Effectiveness of Functional Genetic Screens for the Identification of Bioactive Metabolites. Mar. Drugs 2013, 11, 40-49. https://doi.org/10.3390/md11010040
Penesyan A, Ballestriero F, Daim M, Kjelleberg S, Thomas T, Egan S. Assessing the Effectiveness of Functional Genetic Screens for the Identification of Bioactive Metabolites. Marine Drugs. 2013; 11(1):40-49. https://doi.org/10.3390/md11010040
Chicago/Turabian StylePenesyan, Anahit, Francesco Ballestriero, Malak Daim, Staffan Kjelleberg, Torsten Thomas, and Suhelen Egan. 2013. "Assessing the Effectiveness of Functional Genetic Screens for the Identification of Bioactive Metabolites" Marine Drugs 11, no. 1: 40-49. https://doi.org/10.3390/md11010040
APA StylePenesyan, A., Ballestriero, F., Daim, M., Kjelleberg, S., Thomas, T., & Egan, S. (2013). Assessing the Effectiveness of Functional Genetic Screens for the Identification of Bioactive Metabolites. Marine Drugs, 11(1), 40-49. https://doi.org/10.3390/md11010040




