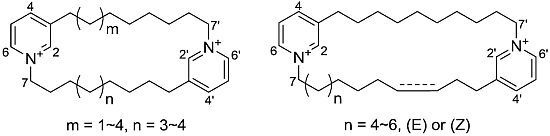
Cyclic Bis-1,3-dialkylpyridiniums from the Sponge Haliclona sp.
Abstract
:1. Introduction
2. Results and Discussion
| Position | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 145.5, | CH | 145.6, | CH | 145.3, | CH | 145.3, | CH | 145.2, | CH | 145.6, | CH | 145.2, | CH | 145.3, | CH |
| 3 | 145.3, | C | 145.3, | C | 145.7, | C | 145.5, | C | 146.7, | C | 145.3, | C | 145.7, | C | 145.5, | C |
| 4 | 147.0, | CH | 146.9, | CH | 146.9, | CH | 147.0, | CH | 146.9, | CH | 146.9, | CH | 146.9, | CH | 147.0, | CH |
| 5 | 129.2, | CH | 129.2, | CH | 129.2, | CH | 128.8, | CH | 129.2, | CH | 129.1, | CH | 129.1, | CH | 129.2, | CH |
| 6 | 143.4, | CH | 143.4, | CH | 143.4, | CH | 143.5, | CH | 143.5, | CH | 143.4, | CH | 143.5, | CH | 143.4, | CH |
| 7 | 62.7, | CH2 | 62.8, | CH2 | 62.8, | CH2 | 62.8, | CH2 | 62.9, | CH2 | 62.9, | CH2 | 62.9, | CH2 | 62.8, | CH2 |
| 8 | 32.2, | CH2 | 32.1, | CH2 | 32.2, | CH2 | 32.3, | CH2 | 24.8, | CH2 | 32.3, | CH2 | 32.3, | CH2 | 32.1, | CH2 |
| 9 | 26.7, | CH2 | 26.5, | CH2 | 26.2, | CH2 | 26.7, | CH2 | 137.6, | CH | 26.9, | CH2 | 26.8, | CH2 | 26.8, | CH2 |
| 10 | 29.5 a, | CH2 | 29.6 b, | CH2 | 29.6, | CH2 | 30.0 d, | CH2 | 124.7, | CH | 29.7 f, | CH2 | 30.5, | CH2 | 30.1 h, | CH2 |
| 11 | 29.8 a, | CH2 | 30.0 b, | CH2 | 32.7, | CH2 | 30.2, | CH2 | 28.1, | CH2 | 30.3 f, | CH2 | 30.7, | CH2 | 30.2 h, | CH2 |
| 12 | 29.9 a, | CH2 | 30.0 b, | CH2 | 132.3, | CH | 28.0, | CH2 | 30.5, | CH2 | 30.1 f, | CH2 | 28.0, | CH2 | 30.2 h, | CH2 |
| 13 | 30.2 a, | CH2 | 30.1 b, | CH2 | 130.7, | CH | 133.3, | CH | 29.2 e, | CH2 | 30.9, | CH2 | 131.1, | CH | 30.7, | CH2 |
| 14 | 29.2, | CH2 | 29.2, | CH2 | 32.4, | CH2 | 128.0, | CH | 29.2 e, | CH2 | 28.3, | CH2 | 130.4, | CH | 28.2, | CH2 |
| 15 | 31.3, | CH2 | 31.2, | CH2 | 31.1, | CH2 | 29.2, | CH2 | 29.6 e, | CH2 | 135.7, | CH | 27.9, | CH2 | 133.5, | CH |
| 16 | 33.2, | CH2 | 33.1, | CH2 | 33.2, | CH2 | 33.2, | CH2 | 31.1, | CH2 | 125.1, | CH | 29.7, | CH2 | 127.8, | CH |
| 17 | - | - | - | - | 33.1, | CH2 | 31.0, | CH2 | 31.3, | CH2 | 29.1, | CH2 | ||||
| 18 | - | - | - | - | - | - | 33.3, | CH | 33.2, | CH2 | ||||||
| 2′ | 145.5, | CH | 145.6, | CH | 145.3, | CH | 145.5, | CH | 145.2, | CH | 144.6, | CH | 145.2, | CH | 145.5, | CH |
| 3′ | 145.4, | C | 145.3, | C | 145.5, | C | 144.9, | C | 145.7, | C | 144.7, | C | 145.7, | C | 145.0, | C |
| 4′ | 146.9, | CH | 146.9, | CH | 146.9, | CH | 147.2, | CH | 147.0, | CH | 146.7, | CH | 146.9, | CH | 147.3, | CH |
| 5′ | 129.2, | CH | 129.2, | CH | 129.1, | CH | 129.2, | CH | 129.2, | CH | 129.2, | CH | 129.1, | CH | 128.9, | CH |
| 6′ | 143.5, | CH | 143.4, | CH | 143.5, | CH | 143.5, | CH | 143.5, | CH | 143.7, | CH | 143.5, | CH | 143.4, | CH |
| 7′ | 62.7, | CH2 | 62.8, | CH2 | 62.8, | CH2 | 62.9, | CH2 | 62.8, | CH2 | 62.8, | CH2 | 62.8, | CH2 | 62.9, | CH2 |
| 8′ | 31.9, | CH2 | 32.1, | CH2 | 32.2, | CH2 | 32.4, | CH2 | 32.0, | CH2 | 32.2, | CH2 | 32.3, | CH2 | 32.6, | CH2 |
| 9′ | 26.4, | CH2 | 26.5, | CH2 | 26.6, | CH2 | 26.8, | CH2 | 26.5, | CH2 | 26.6, | CH2 | 26.7, | CH2 | 27.0, | CH2 |
| 10′ | 30.3 a, | CH2 | 29.6 b, | CH2 | 29.9 c, | CH2 | 29.7 d, | CH2 | 29.7 e, | CH2 | 29.1 f, | CH2 | 29.6 g, | CH2 | 30.3 h, | CH2 |
| 11′ | 30.4 a, | CH2 | 30.0 b, | CH2 | 29.9 c, | CH2 | 29.9 d, | CH2 | 30.1 e, | CH2 | 29.9 f, | CH2 | 29.9 g, | CH2 | 30.4 h, | CH2 |
| 12′ | 29.0, | CH2 | 30.0 b, | CH2 | 30.0 c, | CH2 | 30.0 d, | CH2 | 30.1 e, | CH2 | 29.9 f, | CH2 | 30.0 g, | CH2 | 30.5 h, | CH2 |
| 13′ | 30.9, | CH2 | 30.1 b, | CH2 | 30.0 c, | CH2 | 30.4 d, | CH2 | 30.2 e, | CH2 | 30.0 f, | CH2 | 30.1 g, | CH2 | 30.7 h, | CH2 |
| 14′ | 33.0, | CH2 | 29.2, | CH2 | 29.2, | CH2 | 28.7, | CH2 | 29.1, | CH2 | 29.1, | CH2 | 29.4, | CH2 | 29.1, | CH2 |
| 15′ | - | 31.2, | CH2 | 31.2, | CH2 | 30.7, | CH2 | 30.9, | CH2 | 30.4, | CH2 | 31.3, | CH2 | 31.1, | CH2 | |
| 16′ | - | 33.1, | CH2 | 33.0, | CH2 | 32.9, | CH2 | 33.2, | CH2 | 33.1, | CH2 | 33.3, | CH2 | 33.1, | CH2 | |
| Position | 3 | 4 | 5 | 6 |
|---|---|---|---|---|
| 2 | 8.96 (1H, s) | 8.98 (1H, s) | 8.95 (1H, s) | 8.99 (1H, s) |
| 3 | - | - | - | - |
| 4 | 8.45 (1H, d, 7.6) | 8.46 (1H, d, 8.0) | 8.46 (1H, d, 8.2) | 8.45 (1H, d, 8.2) |
| 5 | 8.02 (1H, dd, 7.7, 6.2) | 8.02 (1H, dd, 7.7, 6.4) | 8.02 (1H, dd, 7.4, 6.1) | 8.01 (1H, dd, 7.4, 6.6) |
| 6 | 8.83 (1H, dd, 6.0) | 8.85 (1H, dd, 6.0) | 8.83 (1H, br s) | 8.84 (1H, dd, 6.2) |
| 7 | 4.64 (1H, t, 6.8) | 4.65 (2H, t, 6.8) | 4.65 (2H, t, 7.2) | 4.64 (2H, t, 6.9) |
| 8 | 2.00 (2H, quint, 7.3) | 2.01 (2H, tt, 7.4, 6.8) | 2.03 (2H, m) | 2.00 (2H, m) |
| 9 | 1.26 (2H, m) | 1.25 (2H, m) | 1.29 (2H, m) | 1.23 (2H, m) |
| 10 | 1.25 ~ 1.40 (2H, m) | 1.20 ~ 1.36 (2H, m) | 1.42 (2H, m) | 1.26 (2H, m) |
| 11 | 1.25 ~ 1.40 (2H, m) | 1.20 ~ 1.36 (2H, m) | 2.01 (2H, m) | 1.24 (2H, m) |
| 12 | 1.25 ~ 1.40 (2H, m) | 1.20 ~ 1.36 (2H, m) | 5.41 (1H, br t, 3.6) | 1.74 (2H, m) |
| 13 | 1.25 ~ 1.40 (2H, m) | 1.20 ~ 1.36 (2H, m) | 5.41 (1H, br t, 3.6) | 5.38 (2H, m) |
| 14 | 1.30 (2H, m) | 1.28 (2H, m) | 2.04 (2H, m) | 5.38 (2H, m) |
| 15 | 1.72 (2H, quint, 7.4) | 1.74 (2H, tt, 7.3, 7.3) | 1.78 (2H, m) | 2.46 (2H, dd, 12.9, 6.5) |
| 16 | 2.89 (2H, 7.3) | 2.90 (2H, t, 7.3) | 2.88 (2H, t, 7.6) | 2.95 (2H, t, 6.9) |
| 17 | - | - | - | - |
| 18 | - | - | - | - |
| 2′ | 8.96 (1H, s) | 8.98 (1H, s) | 8.98 (1H, s) | 8.95 (1H, s) |
| 3′ | - | - | - | - |
| 4′ | 8.44 (1H, d, 7.7) | 8.46 (1H, d, 8.0) | 8.45 (1H, d, 6.8) | 8.45 (1H, d, 8.2) |
| 5′ | 8.02 (1H, dd, 7.7, 6.2) | 8.02 (1H, dd, 7.7, 6.4) | 8.01 (1H, dd, 7.4, 6.1) | 8.02 (1H, dd, 7.4, 6.6) |
| 6′ | 8.82 (1H, dd, 6.0) | 8.85 (1H, dd, 6.0) | 8.84 (1H, br s) | 8.85 (1H, dd, 6.2) |
| 7′ | 4.64 (1H, t, 6.5) | 4.65 (2H, t, 6.8) | 4.63 (2H, t, 6.7) | 4.63 (2H, t, 7.5) |
| 8′ | 2.02 (2H, tt, 7.8, 6.9) | 2.01 (2H, tt, 7.4, 6.8) | 2.00 (2H, m) | 1.98 (2H, m) |
| 9′ | 1.20 (2H, m) | 1.25 (2H, m) | 1.28 (2H, m) | 1.24 (2H, m) |
| 10′ | 1.25 ~ 1.40 (2H, m) | 1.20 ~ 1.36 (2H, m) | 1.27 ~ 1.40 (2H, m) | 1.23 ~ 1.35 (2H, m) |
| 11′ | 1.25 ~ 1.40 (2H, m) | 1.20 ~ 1.36 (2H, m) | 1.27 ~ 1.40 (2H, m) | 1.23 ~ 1.35 (2H, m) |
| 12′ | 1.20 (2H, m) | 1.20 ~ 1.36 (2H, m) | 1.27 ~ 1.40 (2H, m) | 1.23 ~ 1.35 (2H, m) |
| 13′ | 1.76 (2H, tt, 7.4, 7.2) | 1.20 ~ 1.36 (2H, m) | 1.27 ~ 1.40 (2H, m) | 1.23 ~ 1.35 (2H, m) |
| 14′ | 2.91 (2H, t, 7.0) | 1.28 (2H, m) | 1.30 (2H, m) | 1.25 (2H, m) |
| 15′ | - | 1.74 (2H, tt, 7.3, 7.3) | 1.73 (2H, m) | 1.77 (2H, m) |
| 16′ | - | 2.90 (2H, t, 7.3) | 2.89 (2H, t, 7.0) | 2.92 (2H, t, 6.9) |
| Position | 7 | 8 | 9 | 10 |
|---|---|---|---|---|
| 2 | 8.99 (1H, s) | 8.87 (1H, s) | 8.99 (1H, s) | 8.93 (1H, s) |
| 3 | - | - | - | - |
| 4 | 8.43 (1H, d, 8.0) | 8.46 (1H, d, 7.6) | 8.45 (1H, d, 8.0) | 8.45 (1H, d, 8.1) |
| 5 | 8.00 (1H, dd, 7.8, 6.3) | 8.03 (1H, dd, 7.8, 6.1) | 8.02 (1H, dd, 7.7, 6.2) | 8.02 (1H, dd, 7.9, 6.3) |
| 6 | 8.80 (1H, dd, 6.2) | 8.85 (1H, dd, 6.0) | 8.82 (1H, dd, 6.1) | 8.82 (1H, dd, 6.5) |
| 7 | 4.62 (2H, t, 7.1) | 4.65 (2H, t, 7.0) | 4.61 (2H, t, 7.1) | 4.64 (2H, t, 6.9) |
| 8 | 2.10 (2H, m) | 1.98 (2H, m) | 2.00 (2H, m) | 2.00 (2H, m) |
| 9 | 5.37 (1H, ddd, 11.4, 6.7, 2.0) | 1.26 (2H, m) | 1.29 (2H, m) | 1.24 (2H, m) |
| 10 | 5.36 (1H, ddd, 11.4, 6.7, 2.0) | 1.20 ~ 1.35 (2H, m) | 1.32 (2H, m) | 1.20 ~ 1.32 (2H, m) |
| 11 | 1.94 (2H, m) | 1.20 ~ 1.35 (2H, m) | 1.28 (2H, m) | 1.20 ~ 1.32 (2H, m) |
| 12 | 1.36 (2H, m) | 1.20 ~ 1.35 (2H, m) | 1.99 (2H, m) | 1.20 ~ 1.32 (2H, m) |
| 13 | 1.23 ~ 1.35 (2H, m) | 1.36 (2H, m) | 5.34 (1H, m) | 1.20 ~ 1.32 (2H, m) |
| 14 | 1.23 ~ 1.35 (2H, m) | 2.15 (2H, m) | 5.32 (1H, m) | 1.70 (2H, m) |
| 15 | 1.23 ~ 1.35 (2H, m) | 5 71 (1H, dt, 10.8, 7.3) | 2.06 (2H, m) | 5.39 (1H, dt, 10.6, 6.7) |
| 16 | 1.78 (2H, m) | 5.66 (1H, dt, 10.8, 7.3) | 1.38 (2H, m) | 5.38 (1H, dt, 10.6, 6.7) |
| 17 | 2.88 (2H, t, 7.2) | 3.68 (2H, t, 7.3) | 1.72 (2H, m) | 2.46 (2H, dt, 13.5, 6.6) |
| 18 | - | - | 2.90 (2H, t, 7.5) | 2.95 (2H, t, 6.8) |
| 2′ | 8.90 (1H, s) | 8.79 (1H, s) | 8.98 (1H, s) | 8.89 (1H, s) |
| 3′ | - | - | - | - |
| 4′ | 8.45 (1H, d, 8.0) | 8.47 (1H, d, 7.6) | 8.45 (1H, d, 8.0) | 8.43 (1H, d, 8.1) |
| 5′ | 8.02 (1H, dd, 7.8, 6.3) | 8.03 (1H, dd, 7.8, 6.1) | 8.01 (1H, dd, 7.7, 6.2) | 8.01 (1H, dd, 7.9, 6.3) |
| 6′ | 8.82 (1H, dd, 6.2) | 8.83 (1H, dd, 6.0) | 8.83 (1H, dd, 5.8) | 8.83 (1H, dd, 6.5) |
| 7′ | 4.60 (2H, t, 6.7) | 4.63 (2H, t, 7.5) | 4.63 (2H, t, 6.8) | 4.61 (2H, t, 6.9) |
| 8′ | 1.99 (2H, m) | 2.02 (2H, m) | 2.01 (2H, m) | 1.98 (2H, m) |
| 9′ | 1.24 (2H, m) | 1.28 (2H, m) | 1.27 (2H, m) | 1.25 (2H, m) |
| 10′ | 1.23 ~ 1.38 (2H, m) | 1.26 ~ 1.35 (2H, m) | 1.26 ~ 1.36 (2H, m) | 1.25 ~ 1.32 (2H, m) |
| 11′ | 1.23 ~ 1.38 (2H, m) | 1.26 ~ 1.35 (2H, m) | 1.26 ~ 1.36 (2H, m) | 1.25 ~ 1.32 (2H, m) |
| 12′ | 1.23 ~ 1.38 (2H, m) | 1.26 ~ 1.35 (2H, m) | 1.26 ~ 1.36 (2H, m) | 1.25 ~ 1.32 (2H, m) |
| 13′ | 1.23 ~ 1.38 (2H, m) | 1.26 ~ 1.35 (2H, m) | 1.26 ~ 1.36 (2H, m) | 1.25 ~ 1.32 (2H, m) |
| 14′ | 1.32 (2H, m) | 1.34 (2H, m) | 1.30 (2H, m) | 1.29 (2H, m) |
| 15′ | 1.73 (2H, m) | 1.74 (2H, m) | 1.73 (2H, m) | 1.75 (2H, m) |
| 16′ | 2.88 (2H, t, 7.0) | 2.90 (2H, t, 7.2) | 2.89 (2H, t, 7.3) | 2.91 (2H, t, 7.0) |
| Compound | MIC (μg/mL) | LC50 (μM) a | |||||
|---|---|---|---|---|---|---|---|
| Gram (+) Bacterium | Gram (−) Bacterium | A549 | |||||
| A | B | C | D | E | F | ||
| 1 | 50 | 100 | 50 | 100 | 100 | >100 | 22.1 |
| 2 | 12.5 | 25 | 3.125 | 25 | 25 | >100 | 15.3 |
| 3 | 50 | 50 | 100 | 50 | 25 | >100 | 24.0 |
| 4 | 50 | 100 | 6.25 | 100 | 100 | >100 | 26.3 |
| 5 | 25 | 100 | 25 | 50 | 50 | >100 | 25.9 |
| 6 | 50 | 100 | 50 | 100 | 100 | >100 | 28.9 |
| 7 | 50 | 100 | 100 | 100 | 50 | >100 | 89.4 |
| 8 | 12.5 | 25 | 3.125 | 50 | 50 | >100 | 15.7 |
| 9 | 12.5 | 25 | 3.125 | 25 | 25 | >100 | 14.7 |
| 10 | 12.5 | 25 | 3.125 | 25 | 50 | >100 | 14.8 |
| 4a | 25 | 100 | 50 | 100 | 100 | >100 | 21.9 |
| Ampicillin | 0.39 | 0.78 | 0.39 | 0.78 | 0.39 | 6.25 | |
| Doxorubicin | 3.37 | ||||||
3. Experimental Section
3.1. General Experimental Procedures
3.2. Collection and Taxonomic Identification
3.3. Extraction and Isolation
3.4. Preparation of TFA Salt of Compound 4 (4a)
3.5. Biological Assays
4. Conclusion
Supplementary Files
Acknowledgments
References
- Andersen, R.J.; van Soest, R.W.M.; Kong, F. Alkaloids: Chemical and Biological Perspectives; Pelletier, S.W., Ed.; Pergamon Press/Elsevier Science: New York, NY, USA, 1996; pp. 301–355. [Google Scholar]
- Matzanke, N.; Gregg, R.J.; Weinreb, S.M. Biomimetic and synthetic approaches to marine sponge alkaloids derived from bis-pyridine macrocycles. Org. Prep. Proc. Int. 1998, 30, 3–51. [Google Scholar]
- Turk, T.; Sepčic, K.; Mancini, I.; Guella, G. 3-Akylpyridinium and 3-alkylpyridine compounds from marine sponges, their synthesis, biological activities and potential use. Stud. Nat. Prod. Chem. 2008, 35, 355–397. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2012, 29, 144–222. [Google Scholar] [CrossRef]
- Oku, N.; Nagai, K.; Shindoh, N.; Terada, Y.; van Soest, R.W.M.; Matsunaga, S.; Fusetani, N. Three new cyclostellettamines, which inhibit histonedeacetylase, from a marine sponge of the genus Xestospongia. Bioorg. Med. Chem. Lett. 2004, 14, 2617–2620. [Google Scholar] [CrossRef]
- Pérez-Balado, C.; Nebbioso, A.; Rodríguez-Graña, P.; Minichiello, A.; Miceli, M.; Altucci, L.; de Lera, Á.R. Bispyridinium dienes: Histone deacetylase inhibitors with selective activities. J. Med. Chem. 2007, 50, 2497–2505. [Google Scholar] [CrossRef]
- Davies-Coleman, M.T.; Faulkner, D.J.; Dubowchik, G.M.; Roth, G.P.; Polson, C.; Fairchild, C. A new EGF-active polymeric pyridinium alkaloid from the sponge Callyspongia fibrosa. J. Org. Chem. 1993, 58, 5925–5930. [Google Scholar] [CrossRef]
- Ondeykal, J.G.; Herath, K.B.; Jayasuriya, H.; Polishook, J.D.; Bills, G.F.; Dombrowski, A.W.; Mojena, M.; Koch, G.; DiSalvo, J.; DeMartino, J.; Guan, Z.; Nanakorn, W.; Morenberg, C.M.; Balick, M.J.; Stevenson, D.W.; Slattery, M.; Borris, R.P.; Singh, S.B. Discovery of structurally diverse natural product antagonists of chemokine receptor CXCR3. Mol. Divers. 2005, 9, 123–129. [Google Scholar] [CrossRef]
- Fusetani, N.; Asai, N.; Matsunaga, S.; Honda, K.; Yasumuro, K. Cyclostellettamines A–F, pyridine alkaloids which inhibit binding of methyl quinuclidinylbenzilate (QNB) to muscarinic acetylcholine receptors, from the marine sponge, Stelletta maxia. Tetrahedron Lett. 1994, 35, 3967–3970. [Google Scholar] [CrossRef]
- Anan, H.; Seki, N.; Noshiro, O.; Honda, K.; Yasumuro, K.; Ozasa, T.; Fusetani, N. Total synthesis of cyclostellettamine C, a bispyridiniummacrocyclic alkaloid having muscarinic acetylcholine receptor antagonistic activity. Tetrahedron 1996, 52, 10849–10860. [Google Scholar] [CrossRef]
- Jang, K.H.; Kang, G.W.; Jeon, J.; Lim, C.; Lee, H.-S.; Sim, C.J.; Oh, K.-B.; Shin, J. Haliclonin A, a new macrocyclicdiamide from the sponge Haliclona sp. Org. Lett. 2009, 11, 1713–1716. [Google Scholar]
- Casapullo, A.; Pinto, O.C.; Marzocco, S.; Autore, G.; Riccio, R. 3-Alkylpyridinium alkaloids from the pacific sponge Haliclona sp. J. Nat. Prod. 2009, 72, 301–303. [Google Scholar] [CrossRef]
- Schmidt, G.; Timm, C.; Köck, M. New haliclamines E and F from the arctic sponge Haliclona viscosa. Org. Biomol. Chem. 2009, 7, 3061–3064. [Google Scholar] [CrossRef]
- Grube, A.; Timm, C.; Köck, M. Synthesis and mass spectrometric analysis of cyclostellettamines H, I, K and L. Eur. J. Org. Chem. 2006, 5, 1285–1295. [Google Scholar]
- de Haan, J.W.; van de Ven, L.J.M. Configurations and conformations in acyclic, unsaturated hydrocarbons. A 13C NMR study. A 13C NMR study. Org. Magn. Reson. 1973, 5, 147–153. [Google Scholar]
- de Oliveira, J.H.H.L.; Seleghim, M.H.R.; Timm, C.; Grube, A.; Köck, M.; Nascimento, G.G.F.; Martins, A.C.T.; Silva, E.G.O.; de Souza, A.O.; Minarini, P.R.R.; et al. Antimicrobial and antimycobacterial activity of cyclostellettamine alkaloids from sponge Pachychalina sp. Mar. Drugs 2006, 4, 1–8. [Google Scholar] [CrossRef]
- de Oliveira, J.H.H.L.; Grube, A.; Kock, M.; Berlinck, R.G.; Macedo, M.L.; Ferreira, A.G.; Hajdu, E. Ingenamine G and cyclostellettamines G–I, K, and L from the new brazilian species of marine sponge Pachychalina sp. J. Nat. Prod. 2004, 67, 1685–1689. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Ulukaya, E.; Ozdikicioglu, F.; Oral, A.Y.; Demirci, M. The MTT assay yields a relatively lower result of growth inhibition than the ATP assay depending on the chemotherapeutic drugs tested. Toxicol. In Vitro 2008, 22, 232–239. [Google Scholar] [CrossRef]
- Oh, K.-B.; Lee, J.H.; Chung, S.-C.; Shin, J.; Shin, H.J.; Kim, H.-K.; Lee, H.-S. Antimicrobial activities of the bromophenols from the red alga Odonthalia corymbifera and some synthetic derivatives. Bioorg. Med. Chem. Lett. 2008, 18, 104–108. [Google Scholar] [CrossRef]
- Lee, H.-S.; Lee, T.-H.; Yang, S.H.; Shin, H.J.; Shin, J.; Oh, K.-B. Sesterterpene sulfates as isocitratelyase inhibitors from tropical sponge Hippospongia sp. Bioorg. Med. Chem. Lett. 2007, 17, 2483–2486. [Google Scholar] [CrossRef]
- Johansson, M.; Karlsson, L.; Wennergren, M.; Jansson, T.; Powell, T.L. Activity and protein expression of Na+/K+ ATPase are reduced in microvilloussyncytiotrophoblast plasma membranes isolated from pregnancies complicated by intrauterine growth restriction. J. Clin. Endocrinol. Metab. 2003, 88, 2831–2837. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lee, Y.; Jang, K.H.; Jeon, J.-e.; Yang, W.-Y.; Sim, C.J.; Oh, K.-B.; Shin, J. Cyclic Bis-1,3-dialkylpyridiniums from the Sponge Haliclona sp. Mar. Drugs 2012, 10, 2126-2137. https://doi.org/10.3390/md10092126
Lee Y, Jang KH, Jeon J-e, Yang W-Y, Sim CJ, Oh K-B, Shin J. Cyclic Bis-1,3-dialkylpyridiniums from the Sponge Haliclona sp. Marine Drugs. 2012; 10(9):2126-2137. https://doi.org/10.3390/md10092126
Chicago/Turabian StyleLee, Yoonyeong, Kyoung Hwa Jang, Ju-eun Jeon, Woo-Young Yang, Chung J. Sim, Ki-Bong Oh, and Jongheon Shin. 2012. "Cyclic Bis-1,3-dialkylpyridiniums from the Sponge Haliclona sp." Marine Drugs 10, no. 9: 2126-2137. https://doi.org/10.3390/md10092126
APA StyleLee, Y., Jang, K. H., Jeon, J.-e., Yang, W.-Y., Sim, C. J., Oh, K.-B., & Shin, J. (2012). Cyclic Bis-1,3-dialkylpyridiniums from the Sponge Haliclona sp. Marine Drugs, 10(9), 2126-2137. https://doi.org/10.3390/md10092126