Pardaxin, a Fish Antimicrobial Peptide, Exhibits Antitumor Activity toward Murine Fibrosarcoma in Vitro and in Vivo
Abstract
:1. Introduction
2. Results
2.1. Effects of Pardaxin on Cytotoxicity, Clonal Growth, Membrane Structure, and Cell Motility of MN-11 Cells
2.2. Pardaxin Induced Apoptosis of MN-11 Cells
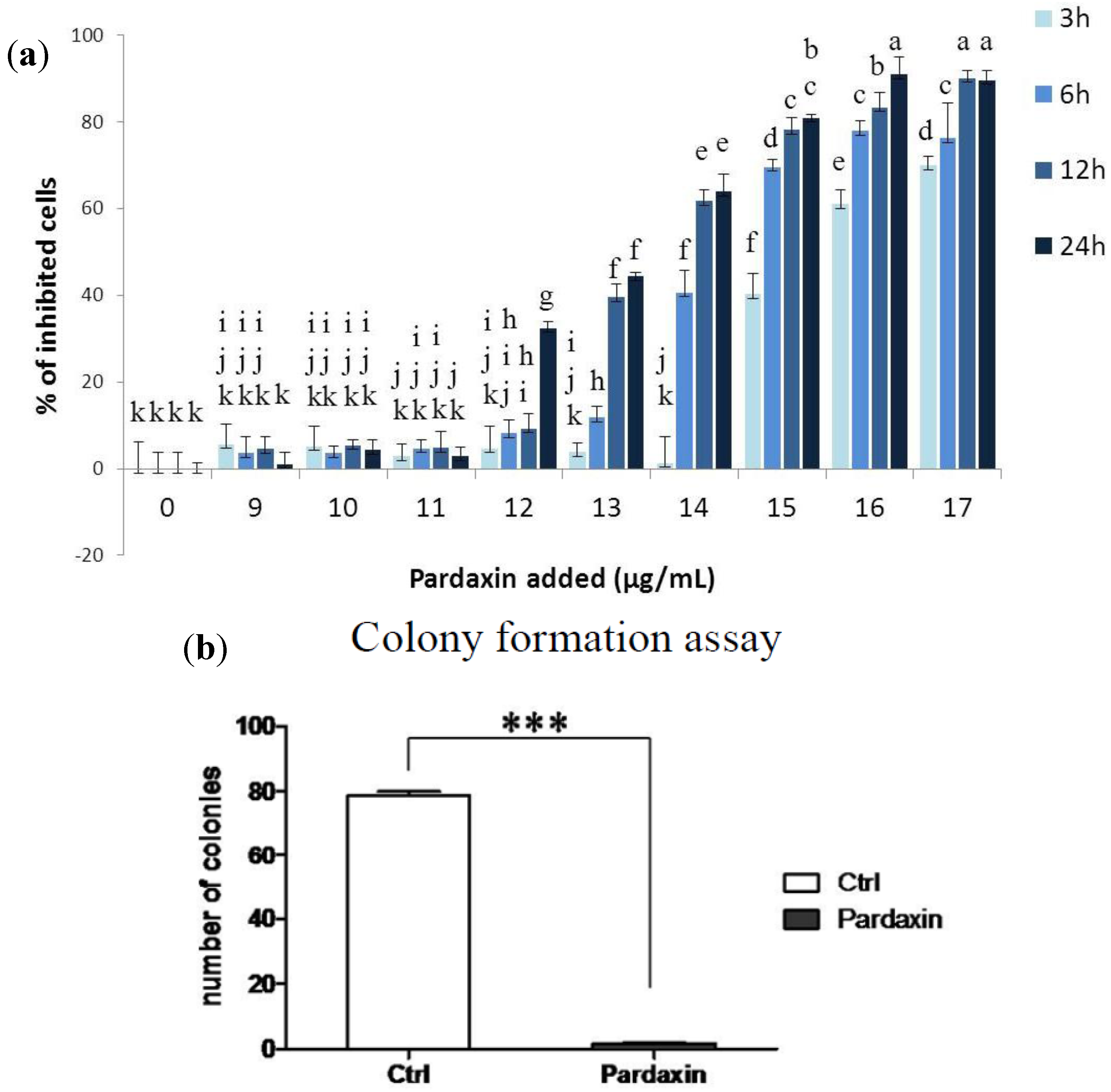
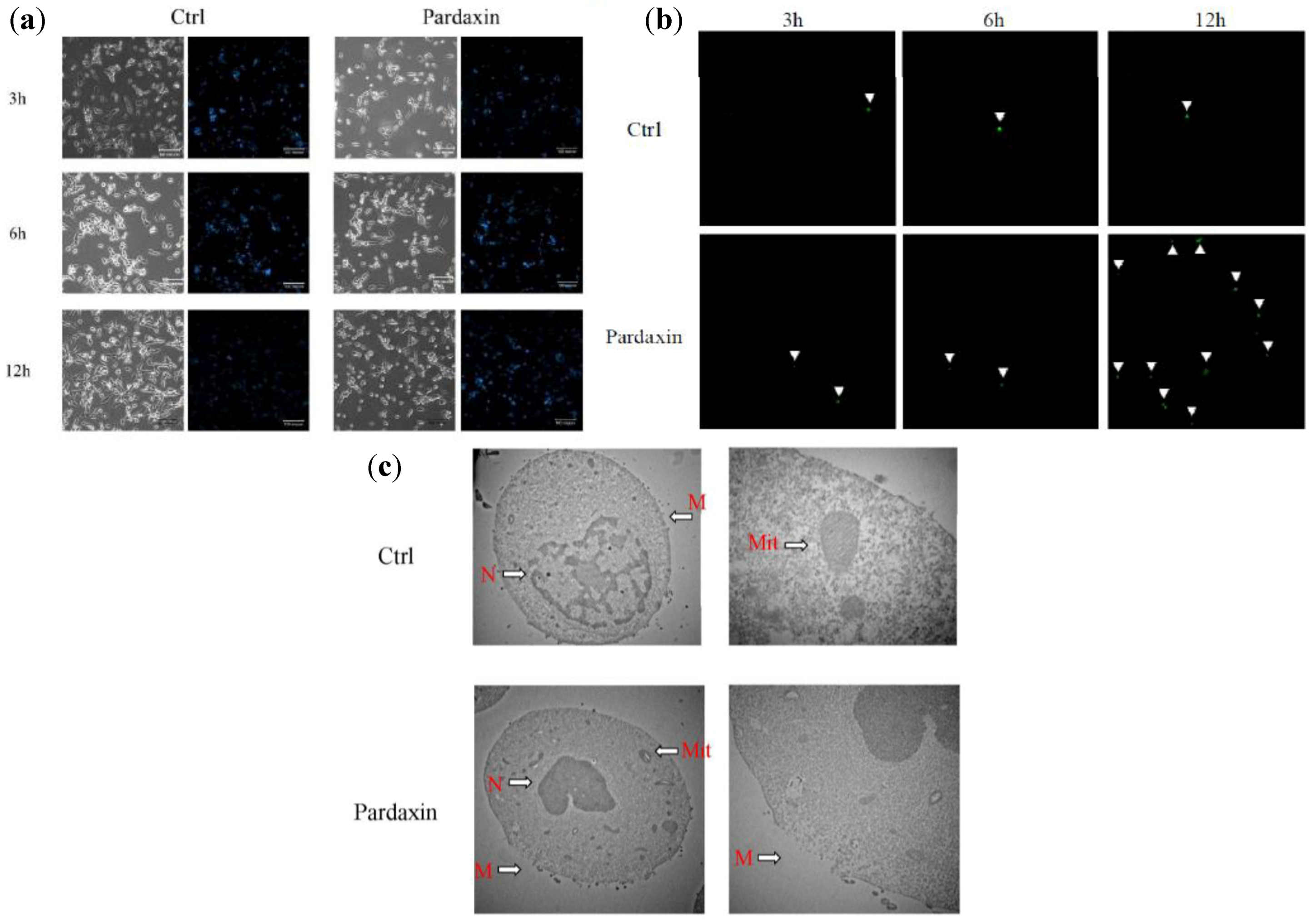
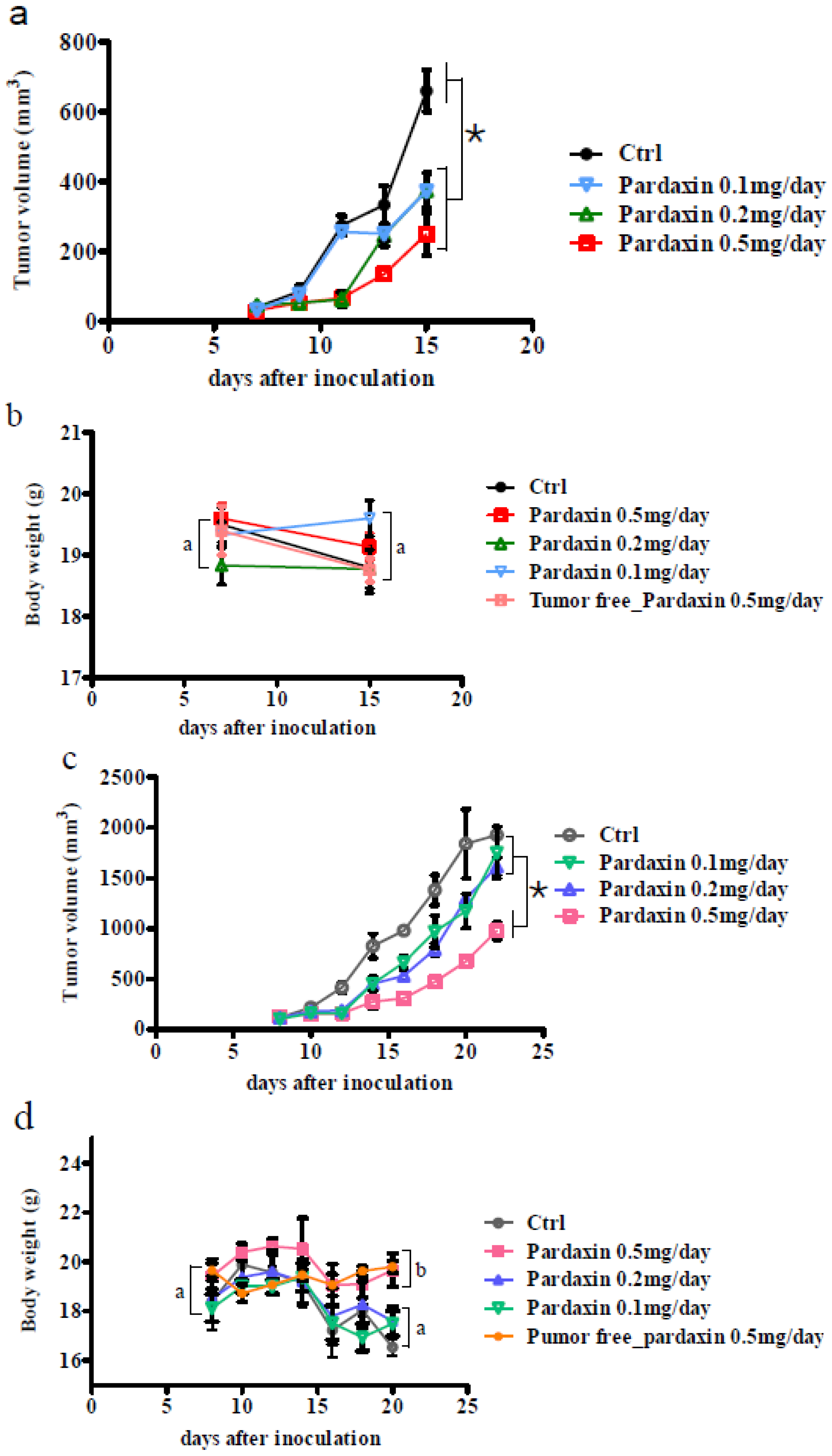
2.3. Pardaxin Inhibits Tumor Growth and Vascularization in Vivo

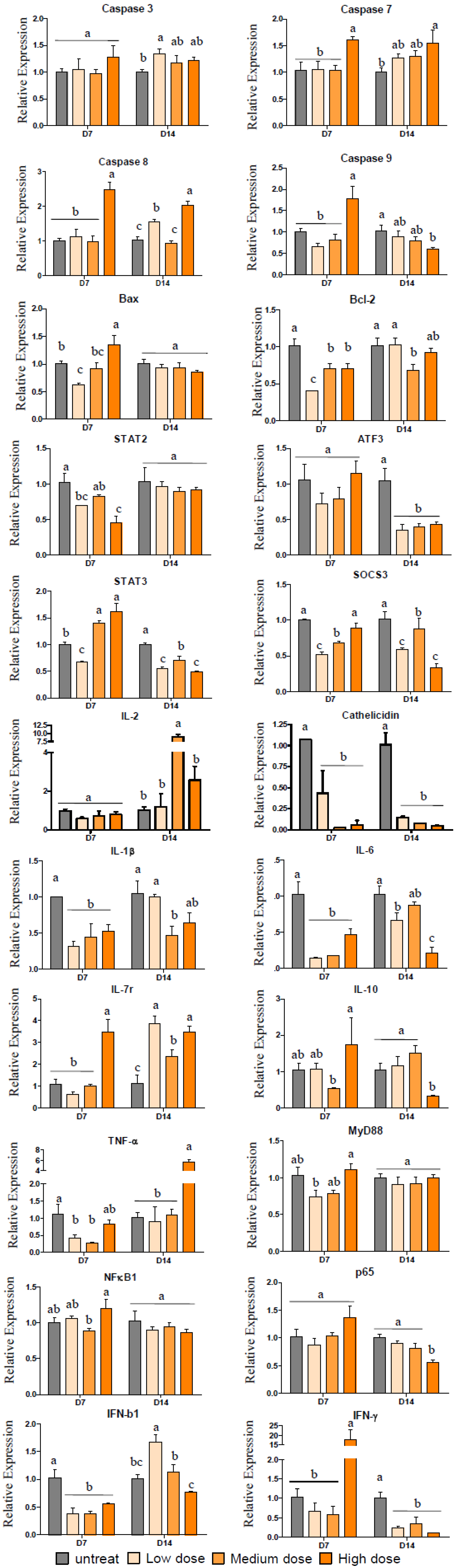
2.4. Gene Expression after Treatment with Pardaxin in Tumor-Bearing Mice
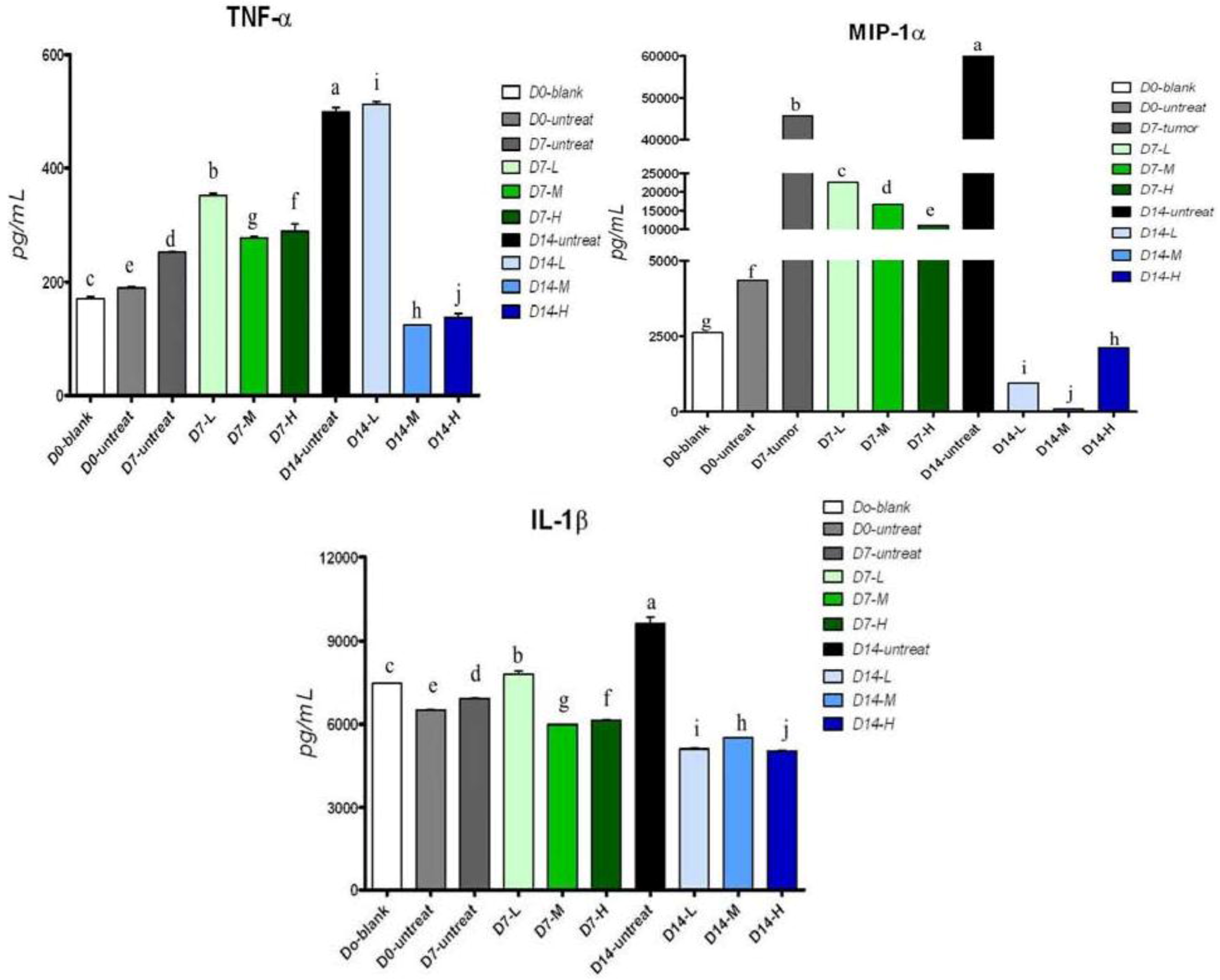
2.5. Cytokine Secretion in Serum of Tumor-Bearing Mice after Treatment with Pardaxin
3. Discussion

4. Experimental Section
4.1. Cells and Cell Culture
4.2. In Vitro Cytotoxicity Assay and Soft-Agar Colony Formation Assay
4.3. Detection of Cell Viability by Acridine Orange (AO)/Ethidium Bromide (EtBr) Staining and Transmission Electron Microscopy (TEM)
4.4. Hoechst 33258 Assay and Caspase-3/7 Activity for Apoptosis
4.5. In Vivo Antitumor Efficacy of Pardaxin in Tumor Xenografts
4.6. Detection of Cytokine Expression Levels and Immunohistochemistry
4.7. RNA Isolation and Reverse-Transcription Qualitative Polymerase Chain Reaction (RT-qPCR)
| gene | primer (5′→3′) | product (bp) | |
|---|---|---|---|
| 1 | mActin F′ | 5′-TTCGTTGCCGGTCCACACCC-3′ | 90 |
| mActin R′ | 5′-GCTTTGCACATGCCGGAGCC-3′ | ||
| 2 | mCamp F′ | 5′-GCCGCTGATTCTTTTGACAT-3′ | 108 |
| mCamp R′ | 5′-AATCTTCTCCCCACCTTTGC-3′ | ||
| 3 | mIL-1β F′ | 5′-TGTAATGAAAGACGGCACACC-3′ | 68 |
| mIL-1β R′ | 5′-TCTTCTTTGGGTATTGCTTGG-3′ | ||
| 4 | mIL-2 F′ | 5′-TCTGAGGAGATGGATAGC-3′ | 78 |
| mIL-2 R′ | 5′-TGTTGTAAGCAGGAGGTA-3′ | ||
| 5 | mIL-4 F′ | 5′-CATCGGCATTTTGAACGAG-3′ | 104 |
| mIL-4 R′ | 5′-CGAGCTCACTCTCTGTGGTG-3′ | ||
| 6 | mIL-6 F′ | 5′-GCTACCAAACTGGATATAATCAGGA-3′ | 85 |
| mIL-6 R′ | 5′-CCAGGTAGCTATGGTACTCCAGAA-3′ | ||
| 7 | mIL-7r F′ | 5′-CGAAACTCCAGAACCCAAGA-3′ | 61 |
| mIL-7r R′ | 5′-AATGGTGACACTTGGCAAGAC-3′ | ||
| 8 | mIL-10 F′ | 5′-CAGAGCCACATGCTCCTAGA-3′ | 78 |
| mIL-10 R′ | 5′-GTCCAGCTGGTCCTTTGTTT-3′ | ||
| 9 | mSTAT2 F′ | 5′-CCTGGTAAGATCCCTTTCTGG-3′ | 70 |
| mSTAT2 R′ | 5′-GATCCTTCAGGTGGTCGTGT-3′ | ||
| 10 | mSTAT3 F′ | 5′-GGAAATAACGGTGAAGGTGCT-3′ | 66 |
| mSTAT3 R′ | 5′-GGAAATAACGGTGAAGGTGCT-3′ | ||
| 11 | mSOCS3 F′ | 5′-ATTTCGCTTCGGGACTAGC-3′ | 126 |
| mSOCS3 R′ | 5′-AACTTGCTGTGGGTGACCAT-3′ | ||
| 12 | mTnfα F′ | 5′-TCTTCTCATTCCTGCTTGTGG-3′ | 128 |
| mTnfα R′ | 5′-GGTCTGGGCCATAGAACTGA-3′ | ||
| 13 | mMyD88 F′ | 5′-TGGCCTTGTTAGACCGTGA-3′ | 73 |
| mMyD88 R′ | 5′-AAGTATTTCTGGCAGTCCTCCTC-3′ | ||
| 14 | mNfkb1 F′ | 5′-CACTGCTCAGGTCCACTGTC-3′ | 78 |
| mNfkb1 R′ | 5′-CTGTCACTATCCCGGAGTTCA-3′ | ||
| 15 | mp65 F′ | 5′-CCCAGACCGCAGTATCCAT-3′ | 68 |
| mp65 R′ | 5′-GCTCCAGGTCTCGCTTCTT-3′ | ||
| 16 | mBcl-2 F′ | 5′-GTACCTGAACCGGCATCTG-3′ | 130 |
| mBcl-2 R′ | 5′-GCTGAGCAGGGTCTTCAGAG-3′ | ||
| 17 | mBax F′ | 5′-CTCCGTGAGCGGCTGCTTGTC-3′ | 82 |
| mBax R′ | 5′-GCCATGTGGGGGTCCCGAAG-3′ | ||
| 18 | mAtf3 F′ | 5′-GCTGGAGTCAGTTACCGTCAA-3′ | 93 |
| mAtf3 R′ | 5′-CGCCTCCTTTTCCTCTCAT-3′ | ||
| 19 | mCaspase 3 F′ | 5′-GAGGCTGACTTCCTGTATGCTT-3′ | 77 |
| mCaspase 3 R′ | 5′-AACCACGACCCGTCCTTT-3′ | ||
| 20 | mCaspase 7 F′ | 5′-CCGTCCACAATGACTGCTC-3′ | 78 |
| mCaspase 7 R′ | 5′-CCGAGTTGCTGTGGTCCT-3′ | ||
| 21 | mCaspase 8 F′ | 5′-TTGAACAATGAGATCCCCAAA-3′ | 70 |
| mCaspase 8 R′ | 5′-CCATTTCTACAAAAATTTCAAGCAG-3′ | ||
| 22 | mCaspase 9 F′ | 5′-TGCAGTCCCTCCTTCTCAG-3′ | 77 |
| mCaspase 9 R′ | 5′-GCTTTTTCCGGAGGAAGTTAAA-3′ | ||
| 23 | mIFNb1 F′ | 5′-CTGGCTTCCATCATGAACAA-3′ | 73 |
| mIFNb1 R′ | 5′-AGAGGGCTGTGGTGGAGAA-3′ | ||
| 24 | mIFNγ F′ | 5′-CACACCTGATTACTACCTTCT-3′ | 75 |
| mIFNγ R′ | 5′-CCTCAAACTTGGCAATACTC-3′ |
5. Conclusions
Acknowledgements
References
- Singer, S.; Corson, J.M.; Demetri, G.D.; Healey, E.A.; Marcus, K.; Eberlein, T.J. Prognostic factors predictive of survival for truncal and retroperitoneal soft-tissue sarcoma. Ann. Surg. 1995, 221, 185–195. [Google Scholar] [CrossRef]
- Daigeler, A.; Chromik, A.M.; Haendschke, K.; Emmelmann, S.; Siepmann, M.; Hensel, K.; Schmitz, G.; Klein-Hitpass, L.; Steinau, H.U.; Lehnhardt, M.; et al. Synergistic effects of sonoporation and taurolidin/TRAIL on apoptosis in human fibrosarcoma. Ultrasound Med. Biol. 2010, 36, 1893–1906. [Google Scholar] [CrossRef]
- Edmonson, J.H.; Petersen, I.A.; Shives, T.C.; Mahoney, M.R.; Rock, M.G.; Haddock, M.G.; Sim, F.H.; Maples, W.J.; O’Connor, M.I.; Gunderson, L.L.; et al. Chemotherapy, irradiation, and surgery for function-preserving therapy of primary extremity soft tissue sarcomas: Initial treatment with ifosfamide, mitomycin, doxorubicin, and cisplatin plus granulocyte macrophage-colony-stimulating factor. Cancer 2002, 94, 786–792. [Google Scholar]
- Daigeler, A.; Brenzel, C.; Bulut, D.; Geisler, A.; Hilgert, C.; Lehnhardt, M.; Steinau, H.U.; Flier, A.; Steinstraesser, L.; Klein-Hitpass, L.; et al. TRAIL and Taurolidine induce apoptosis and decrease proliferation in human fibrosarcoma. J. Exp. Clin. Cancer Res. 2008, 27, 82. [Google Scholar] [CrossRef]
- Steinau, H.U.; Buttemeyer, R.; Vogt, P.; Hussmann, J.; Hebebrand, D. Limb salvage and reconstructive procedures in soft tissue sarcomas of the extremities. Recent Results Cancer Res. 1995, 138, 31–39. [Google Scholar] [CrossRef]
- Daigeler, A.; Lehnhardt, M.; Khadra, A.; Hauser, J.; Steinstraesser, L.; Langer, S.; Goertz, O.; Steinau, H.U. Proximal major limb amputations-a retrospective analysis of 45 oncological cases. World J. Surg. Oncol. 2009, 7, 15. [Google Scholar] [CrossRef]
- Cherix, S.; Speiser, M.; Matter, M.; Raffoul, W.; Lienard, D.; Theumann, N.; Mouhsine, E.; Mirimanoff, R.O.; Leyvraz, S.; Lejeune, F.J.; et al. Isolated limb perfusion with tumor necrosis factor and melphalan for non-resectable soft tissue sarcomas: Long-term results on efficacy and limb salvage in a selected group of patients. J. Surg. Oncol. 2008, 98, 148–155. [Google Scholar] [CrossRef]
- Grunhagen, D.J.; de Wilt, J.H.; Graveland, W.J.; Verhoef, C.; van Geel, A.N.; Eggermont, A.M. Outcome and prognostic factor analysis of 217 consecutive isolated limb perfusions with tumor necrosis factor-alpha and melphalan for limb-threatening soft tissue sarcoma. Cancer 2006, 106, 1776–1784. [Google Scholar] [CrossRef]
- Lazarovici, P.; Primor, N.; Loew, L.M. Purification and pore-forming activity of two hydrophobic polypeptides from the secretion of the Red Sea Moses sole (Pardachirus marmoratus). J. Biol. Chem. 1986, 261, 16704–16713. [Google Scholar]
- Bhunia, A.; Domadia, P.N.; Torres, J.; Hallock, K.J.; Ramamoorthy, A.; Bhattacharjya, S. NMR structure of pardaxin, a pore-forming antimicrobial peptide, in lipopolysaccharide micelles: Mechanism of outer membrane permeabilization. J. Biol. Chem. 2010, 285, 3883–3895. [Google Scholar]
- Vad, B.S.; Bertelsen, K.; Johansen, C.H.; Pedersen, J.M.; Skrydstrup, T.; Nielsen, N.C.; Otzen, D.E. Pardaxin permeabilizes vesicles more efficiently by pore formation than by disruption. Biophys. J. 2010, 98, 576–585. [Google Scholar]
- Hsu, J.C.; Lin, L.C.; Tzen, J.T.; Chen, J.Y. Pardaxin-induced apoptosis enhances antitumor activity in HeLa cells. Peptides 2011, 32, 1110–1116. [Google Scholar] [CrossRef]
- Huang, T.C.; Lee, J.F.; Chen, J.Y. Pardaxin, an antimicrobial peptide, triggers caspase-dependent and ROS-mediated apoptosis in HT-1080 cells. Mar. Drugs 2011, 9, 1995–2009. [Google Scholar] [CrossRef]
- Birnboim, H.C.; Wilkinson, D.; Sandhu, J.K.; McLean, J.R.; Ross, W. Mutatect: A mouse tumour model for detecting radiation-induced mutations in vivo. Mutat. Res. 1999, 430, 275–280. [Google Scholar] [CrossRef]
- Sandhu, J.K.; Privora, H.F.; Wenckebach, G.; Birnboim, H.C. Neutrophils, nitric oxide synthase, and mutations in the mutatect murine tumor model. Am. J. Pathol. 2000, 156, 509–518. [Google Scholar] [CrossRef]
- Nakamoto, T.; Yoshimura, H.; Honda, T.; Nakata, K.; Taniguchi, Y.; Yoshida, A.; Uenobe, M.; Yoshioka, N.; Yamaguchi, T.; Inagawa, H.; et al. Treatments for the activating macrophages that reduces surgical stress and postoperative mortalities from bacterial infections and tumor metastases. In Vivo 2007, 21, 357–364. [Google Scholar]
- Pan, C.Y.; Wu, J.L.; Hui, C.F.; Lin, C.H.; Chen, J.Y. Insights into the antibacterial and immunomodulatory functions of the antimicrobial peptide, epinecidin-1, against Vibrio vulnificus infection in zebrafish. Fish Shellfish Immunol. 2011, 31, 1019–1025. [Google Scholar] [CrossRef]
- Pan, C.Y.; Lee, S.C.; Rajanbabu, V.; Lin, C.H.; Chen, J.Y. Insights into the antibacterial and immunomodulatory functions of tilapia hepcidin (TH)2-3 against Vibrio vulnificus infection in mice. Dev. Comp. Immunol. 2012, 36, 166–173. [Google Scholar] [CrossRef]
- Rajanbabu, V.; Pan, C.Y.; Lee, S.C.; Lin, W.J.; Lin, C.C.; Li, C.L.; Chen, J.Y. Tilapia hepcidin 2-3 peptide modulates lipopolysaccharide-induced cytokines and inhibits tumor necrosis factor-alpha through cyclooxygenase-2 and phosphodiesterase 4D. J. Biol. Chem. 2010, 285, 30577–30586. [Google Scholar]
- Pal, R.; Barenholz, Y.; Wagner, R.R. Transcription of vesicular stomatitis virus activated by pardaxin, a fish toxin that permeabilizes the virion membrane. J. Virol. 1981, 39, 641–645. [Google Scholar]
- Primor, N.; Tu, A.T. Conformation of pardaxin, the toxin of the flatfish Pardachirus marmoratus. Biochim. Biophys. Acta 1980, 626, 299–306. [Google Scholar] [CrossRef]
- Shi, Y.L.; Edwards, C.; Lazarovici, P. Ion selectivity of the channels formed by pardaxin, an ionophore, in bilayer membrane. Nat. Toxins 1995, 3, 151–155. [Google Scholar] [CrossRef]
- Oren, Z.; Hong, J.; Shai, Y. A repertoire of novel antibacterial diastereomeric peptides with selective cytolytic activity. J. Biol. Chem. 1997, 272, 14643–14649. [Google Scholar]
- Hallock, K.J.; Lee, D.K.; Omnaas, J.; Mosberg, H.I.; Ramamoorthy, A. Membrane composition determines pardaxin’s mechanism of lipid bilayer disruption. Biophys. J. 2002, 83, 1004–1013. [Google Scholar] [CrossRef]
- Mader, J.S.; Hoskin, D.W. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert Opin. Investig. Drugs 2006, 15, 933–946. [Google Scholar] [CrossRef]
- Kim, Y.J.; Varki, A. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj. J. 1997, 14, 569–576. [Google Scholar] [CrossRef]
- Brugger, B.; Erben, G.; Sandhoff, R.; Wieland, F.T.; Lehmann, W.D. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 1997, 94, 2339–2344. [Google Scholar]
- Porcelli, F.; Buck, B.; Lee, D.K.; Hallock, K.J.; Ramamoorthy, A.; Veglia, G. Structure and orientation of pardaxin determined by NMR experiments in model membranes. J. Biol. Chem. 2004, 279, 45815–45823. [Google Scholar]
- Epand, R.F.; Ramamoorthy, A.; Epand, R.M. Membrane lipid composition and the interaction of pardaxin: The role of cholesterol. Protein Pept. Lett. 2006, 13, 1–5. [Google Scholar]
- Ramamoorthy, A.; Lee, D.K.; Narasimhaswamy, T.; Nanga, R.P. Cholesterol reduces pardaxin’s dynamics-a barrel-stave mechanism of membrane disruption investigated by solid-state NMR. Biochim. Biophys. Acta 2010, 1798, 223–227. [Google Scholar]
- Davis, A.J.; Tannock, J.F. Repopulation of tumour cells between cycles of chemotherapy: A neglected factor. Lancet Oncol. 2000, 1, 86–93. [Google Scholar] [CrossRef]
- Corry, J.; Rischin, D. Strategies to overcome accelerated repopulation and hypoxia-what have we learned from clinical trials? Semin. Oncol. 2004, 31, 802–808. [Google Scholar] [CrossRef]
- Spugnini, E.P.; Renaud, S.M.; Buglioni, S.; Carocci, F.; Dragonetti, E.; Murace, R.; Cardelli, P.; Vincenzi, B.; Baldi, A.; Citro, G. Electrochemotherapy with cisplatin enhances local control after surgical ablation of fibrosarcoma in cats: An approach to improve the therapeutic index of highly toxic chemotherapy drugs. J. Transl. Med. 2011, 9, 152. [Google Scholar]
- Mir, L.M.; Devauchelle, P.; Quintin-Colonna, F.; Delisle, F.; Doliger, S.; Fradelizi, D.; Belehradek, J., Jr.; Orlowski, S. First clinical trial of cat soft-tissue sarcomas treatment by electrochemotherapy. Br. J. Cancer 1997, 76, 1617–1622. [Google Scholar] [CrossRef]
- Spugnini, E.P.; Porrello, A. Potentiation of chemotherapy in companion animals with spontaneous large neoplasms by application of biphasic electric pulses. J. Exp. Clin. Cancer Res. 2003, 22, 571–580. [Google Scholar]
- Spugnini, E.P.; Citro, G.; D’Avino, A.; Baldi, A. Potential role of electrochemotherapy for the treatment of soft tissue sarcoma: First insights from preclinical studies in animals. Int. J. Biochem. Cell Biol. 2008, 40, 159–163. [Google Scholar] [CrossRef]
- Spugnini, E.P.; Baldi, A.; Vincenzi, B.; Bongiorni, F.; Bellelli, C.; Citro, G.; Porrello, A. Intraoperative versus postoperative electrochemotherapy in high grade soft tissue sarcomas: A preliminary study in a spontaneous feline model. Cancer Chemother. Pharmacol. 2007, 59, 375–381. [Google Scholar]
- Sitaram, N.; Nagaraj, R. Interaction of antimicrobial peptides with biological and model membranes: Structural and charge requirements for activity. Biochim. Biophys. Acta 1999, 1462, 29–54. [Google Scholar]
- Thennarasu, S.; Nagaraj, R. Specific antimicrobial and hemolytic activities of 18-residue peptides derived from the amino terminal region of the toxin pardaxin. Protein Eng. 1996, 9, 1219–1224. [Google Scholar] [CrossRef]
- Lin, M.C.; Lin, S.B.; Chen, J.C.; Hui, C.F.; Chen, J.Y. Shrimp anti-lipopolysaccharide factor peptide enhances the antitumor activity of cisplatin in vitro and inhibits HeLa cells growth in nude mice. Peptides 2010, 31, 1019–1025. [Google Scholar] [CrossRef]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta 2008, 1778, 357–375. [Google Scholar]
- Thennarasu, S.; Tan, A.; Penumatchu, R.; Shelburne, C.E.; Heyl, D.L.; Ramamoorthy, A. Antimicrobial and membrane disrupting activities of a peptide derived from the human cathelicidin antimicrobial peptide LL37. Biophys. J. 2010, 98, 248–257. [Google Scholar]
- Ramamoorthy, A.; Thennarasu, S.; Tan, A.; Lee, D.K.; Clayberger, C.; Krensky, A.M. Cell selectivity correlates with membrane-specific interactions: A case study on the antimicrobial peptide G15 derived from granulysin. Biochim. Biophys. Acta 2006, 1758, 154–163. [Google Scholar]
- Chai, W.S.; Zhu, X.M.; Li, S.H.; Fan, J.X.; Chen, B.Y. Role of Bcl-2 family members in caspase-3/9-dependent apoptosis during Pseudomonas aeruginosa infection in U937 cells. Apoptosis 2008, 13, 833–843. [Google Scholar] [CrossRef]
- Brunelle, J.K.; Letai, A. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 2009, 122, 437–441. [Google Scholar] [CrossRef]
- Medan, D.; Luanpitpong, S.; Azad, N.; Wang, L.; Jiang, B.H.; Davis, M.E.; Barnett, J.B.; Guo, L.; Rojanasakul, Y. Multifunctional Role of Bcl-2 in Malignant Transformation and Tumorigenesis of Cr(VI)-Transformed Lung Cells. PLoS One 2012, 7, e37045. [Google Scholar]
- Hsu, J.C.; Lin, L.C.; Tzen, J.T.; Chen, J.Y. Characteristics of the antitumor activities in tumor cells and modulation of the inflammatory response in RAW264.7 cells of a novel antimicrobial peptide, chrysophsin-1, from the red sea bream (Chrysophrys major). Peptides 2011, 32, 900–910. [Google Scholar] [CrossRef]
- Pan, C.Y.; Chen, J.Y.; Lin, T.L.; Lin, C.H. In vitro activities of three synthetic peptides derived from epinecidin-1 and an anti-lipopolysaccharide factor against Propionibacterium acnes, Candida albicans, and Trichomonas vaginalis. Peptides 2009, 30, 1058–1068. [Google Scholar] [CrossRef]
- Tomayko, M.M.; Reynolds, C.P. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother. Pharmacol. 1989, 24, 148–154. [Google Scholar]
- Lee, S.C.; Pan, C.Y.; Chen, J.Y. The antimicrobial peptide, epinecidin-1, mediates secretion of cytokines in the immune response to bacterial infection in mice. Peptides 2012, 36, 100–108. [Google Scholar] [CrossRef]
- Peng, K.C.; Pan, C.Y.; Chou, H.N.; Chen, J.Y. Using an improved Tol2 transposon system to produce transgenic zebrafish with epinecidin-1 which enhanced resistance to bacterial infection. Fish Shellfish Immunol. 2010, 28, 905–917. [Google Scholar] [CrossRef]
- Samples Availability: Available from the authors.
Supplementary Files
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wu, S.-P.; Huang, T.-C.; Lin, C.-C.; Hui, C.-F.; Lin, C.-H.; Chen, J.-Y. Pardaxin, a Fish Antimicrobial Peptide, Exhibits Antitumor Activity toward Murine Fibrosarcoma in Vitro and in Vivo. Mar. Drugs 2012, 10, 1852-1872. https://doi.org/10.3390/md10081852
Wu S-P, Huang T-C, Lin C-C, Hui C-F, Lin C-H, Chen J-Y. Pardaxin, a Fish Antimicrobial Peptide, Exhibits Antitumor Activity toward Murine Fibrosarcoma in Vitro and in Vivo. Marine Drugs. 2012; 10(8):1852-1872. https://doi.org/10.3390/md10081852
Chicago/Turabian StyleWu, Shu-Ping, Tsui-Chin Huang, Ching-Chun Lin, Cho-Fat Hui, Cheng-Hui Lin, and Jyh-Yih Chen. 2012. "Pardaxin, a Fish Antimicrobial Peptide, Exhibits Antitumor Activity toward Murine Fibrosarcoma in Vitro and in Vivo" Marine Drugs 10, no. 8: 1852-1872. https://doi.org/10.3390/md10081852
APA StyleWu, S.-P., Huang, T.-C., Lin, C.-C., Hui, C.-F., Lin, C.-H., & Chen, J.-Y. (2012). Pardaxin, a Fish Antimicrobial Peptide, Exhibits Antitumor Activity toward Murine Fibrosarcoma in Vitro and in Vivo. Marine Drugs, 10(8), 1852-1872. https://doi.org/10.3390/md10081852




