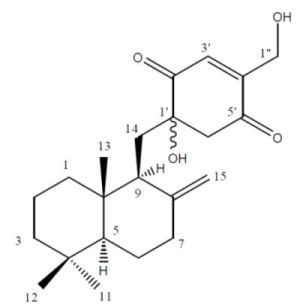
A New Cytotoxic Sesquiterpene Quinone Produced by Penicillium sp. F00120 Isolated from a Deep Sea Sediment Sample
Abstract
:1. Introduction
2. Results and Discussion
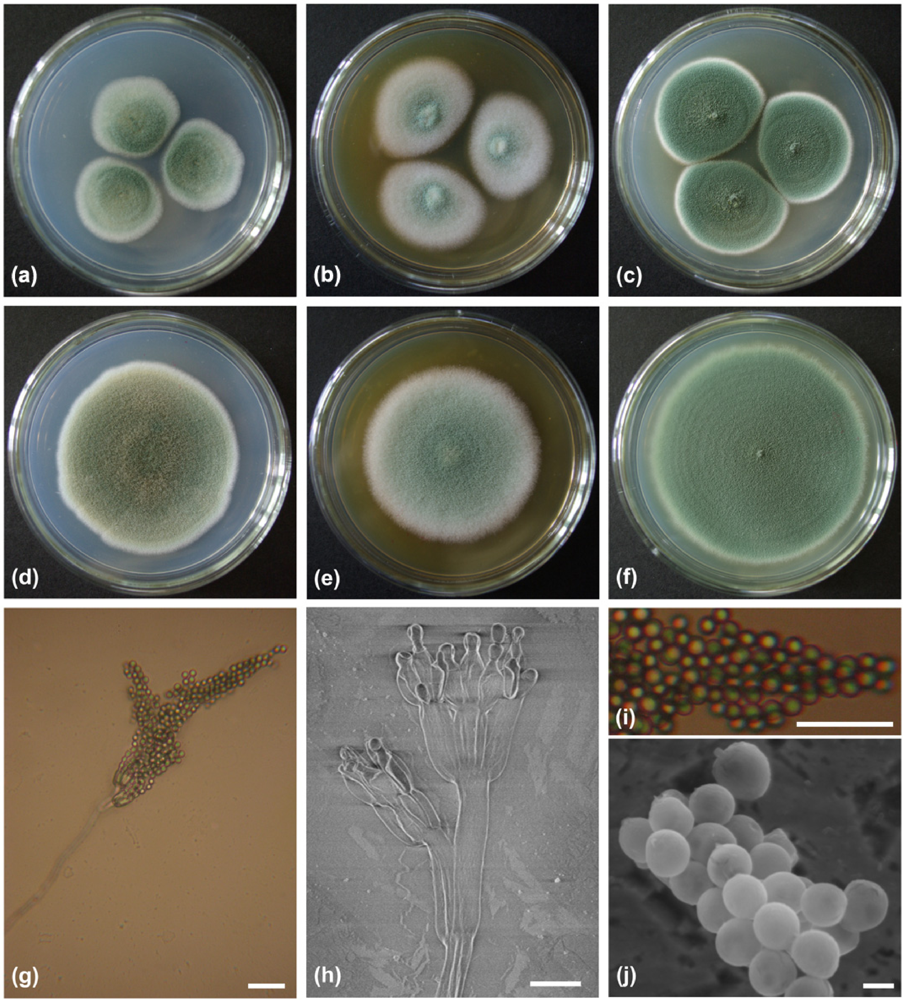
2.1. Characterization and Identification of Isolated Strain F00120

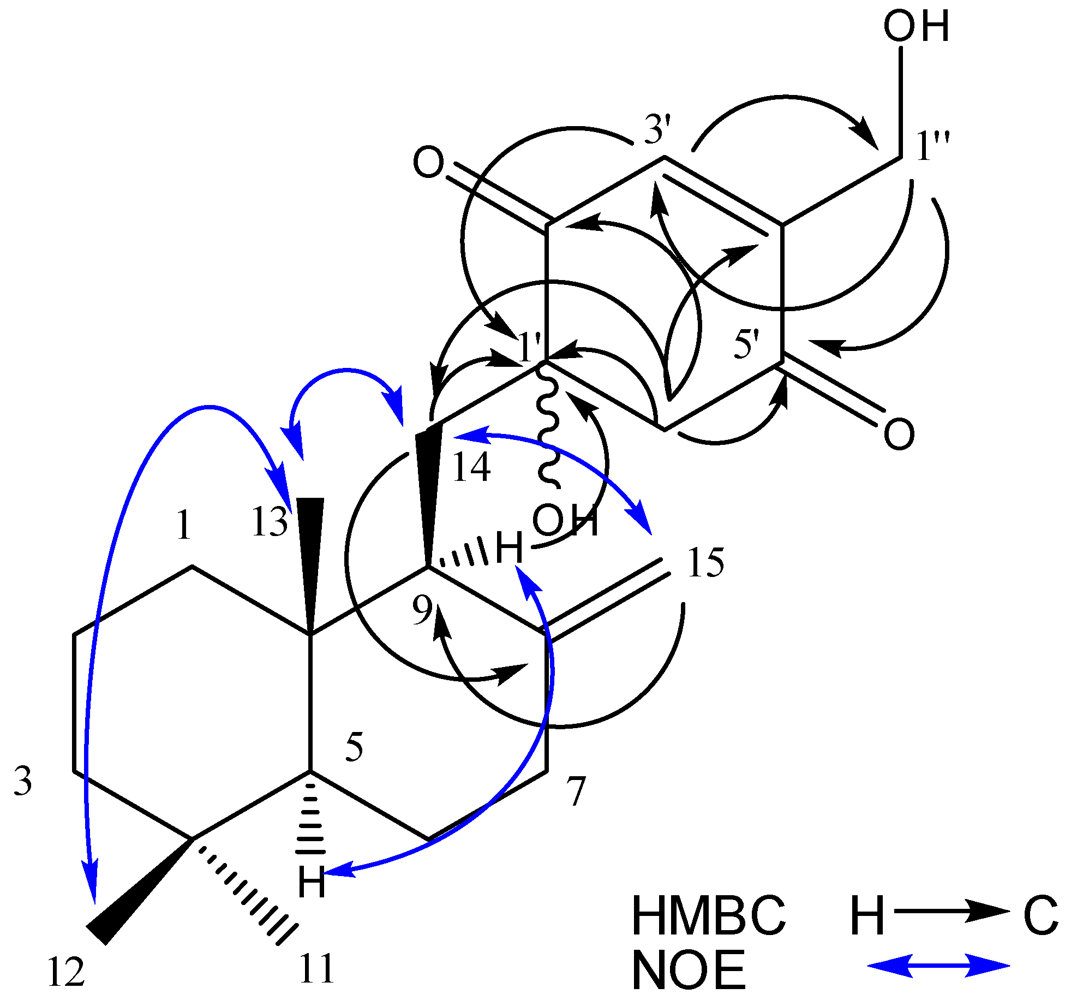
2.2. Structure Elucidation
| Position | δC (m) | δH (m. J Hz) |
|---|---|---|
| 1 | 38.7 CH2 | 1.05 m |
| 1.62 m | ||
| 2 | 19.3 CH2 | 1.49 m |
| 1.52 m | ||
| 3 | 42.0 CH2 | 1.16 m |
| 1.38 m | ||
| 4 | 33.5 C | / |
| 5 | 55.6 CH | 1.12 dd (13, 2.5) |
| 6 | 24.6 CH2 | 1.27 m |
| 1.74 m | ||
| 7 | 38.1 CH2 | 1.89 m a |
| 2.29 ddd (12.5, 6.5, 3.5) | ||
| 8 | 148.9 C | / |
| 9 | 50.5 CH | 1.79 m b |
| 10 | 39.9 C | / |
| 11 | 21.5 CH3 | 0.76 s |
| 12 | 33.6 CH3 | 0.86 s |
| 13 | 15.0 CH3 | 0.58 s |
| 14 | 34.7 CH2 | 1.80 m b |
| 1.89 m a | ||
| 15 | 107.0 CH2 | 4.26 br s |
| 4.75 br s | ||
| 1′ | 77.0 C | / |
| 2′ | 201.1 C | / |
| 3′ | 134.3 CH | 6.82 s |
| 4′ | 150.8 C | / |
| 5′ | 196.5 C | / |
| 6′ | 53.0 CH2 | 2.97 d (1.6) |
| 3.12 d (1.6) | ||
| 1′′ | 59.7 CH2 | 4.44 d (1.7) |
| 4.54 d (1.7) |
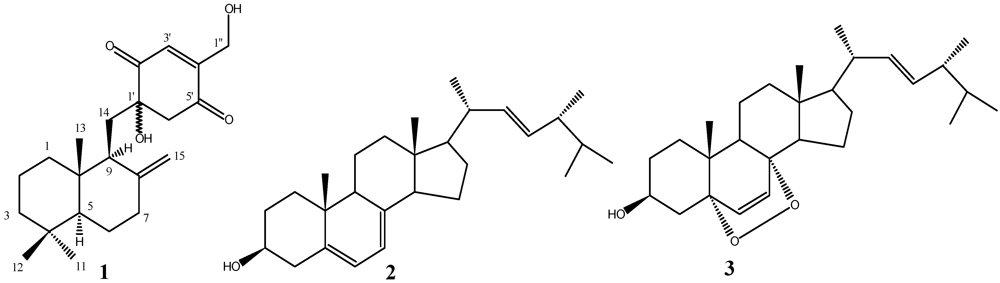
3. Experimental Section
3.1. General Experimental Procedures
3.2. Microbial Material
3.3. Extraction and Isolation
4. Conclusions
Acknowledgments
- Samples Availability: Available from the authors.
References
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E.; Nelson, W.; et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [PubMed]
- Mayer, A.M.; Gustafson, K.R. Marine pharmacology in 2005–2006: Antitumour and cytotoxic compounds. Eur. J. Cancer 2008, 44, 2357–2387. [Google Scholar]
- Mayer, A.M.; Rodriguez, A.D.; Berlinck, R.G.; Fusetani, N. Marine pharmacology in 2007–8: Marine compounds with antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous system, and other miscellaneous mechanisms of action. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 191–222. [Google Scholar] [PubMed]
- Mayer, A.M.; Rodriguez, A.D.; Berlinck, R.G.; Hamann, M.T. Marine pharmacology in 2003–4: Marine compounds with anthelmintic antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiplatelet, antiprotozoal, antituberculosis, and antiviral activities; affecting the cardiovascular, immune and nervous systems, and other miscellaneous mechanisms of action. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007, 145, 553–581. [Google Scholar] [PubMed]
- Mayer, A.M.; Rodriguez, A.D.; Berlinck, R.G.; Hamann, M.T. Marine pharmacology in 2005–6: Marine compounds with anthelmintic, antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; affecting the cardiovascular, immune and nervous systems, and other miscellaneous mechanisms of action. Biochim. Biophys. Acta 2009, 1790, 283–308. [Google Scholar]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar]
- Proksch, P.; Putz, A.; Ortlepp, S.; Kjer, J.; Bayer, M. Bioactive natural products from marine sponges and fungal endophytes. Phytochem. Rev. 2010, 9, 475–489. [Google Scholar]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar]
- Saleem, M.; Ali, M.S.; Hussain, S.; Jabbar, A.; Ashraf, M.; Lee, Y.S. Marine natural products of fungal origin. Nat. Prod. Rep. 2007, 24, 1142–1152. [Google Scholar]
- Wijeratne, E.M.; Paranagama, P.A.; Marron, M.T.; Gunatilaka, M.K.; Arnold, A.E.; Gunatilaka, A.A. Sesquiterpene quinones and related metabolites from Phyllosticta spinarum, a fungal strain endophytic in Platycladus orientalis of the Sonoran Desert. J. Nat. Prod. 2008, 71, 218–222. [Google Scholar]
- Shirane, N.; Takenaka, H.; Ueda, K.; Hashimoto, Y.; Katoh, K.; Ishii, H. Sterol analysis of DMI-resistant and -sensitive strains of Venturia inaequalis. Phytochemistry 1996, 41, 1301–1308. [Google Scholar]
- Krzyczkowski, W.; Malinowska, E.; Suchocki, P.; Kleps, J.; Olejnik, M.; Herold, E. Isolation and quantitative determination of ergosterol peroxide in various edible mushroom species. Food Chemistry 2009, 113, 351–355. [Google Scholar]
- Kawashima, K.; Nakanishi, K.; Tada, M. Structure of tauranin. Tetrahedron Lett. 1964, 5, 1227–1231. [Google Scholar]
- Torigoe, K.; Wakasugi, N.; Sakaizumi, N.; Suzuki, H.; Kojiri, K.; Suda, H. Antitumor substance BE-40644S. Jpn. Patent JPO 07-304762 A, 21 11 1995. [Google Scholar]
- Torigoe, K.; Wakasugi, N.; Sakaizumi, N.; Ikejima, T.; Suzuki, H.; Kojiri, K.; Suda, H. BE-40644, a new human thioredoxin system inhibitor isolated from Actinoplanes sp. A40644. J. Antibiot. 1996, 49, 314–317. [Google Scholar] [PubMed]
- Kawato, M.; Shinobu, R. On Streptomyces herbaricolor sp. nov., supplement: A single technique for microscopical observation. Mem. Osaka Univ. Lib. Arts Educ. B Nat. Sci. 1959, 8, 114–119. [Google Scholar]
- Zhou, Z.H.; Liu, Z.H.; Qian, Y.D.; Kim, S.B.; Goodfellow, M. Saccharopolyspora spinosporotrichia sp. nov., a novel actinomycete from soil. Int. J. Syst. Bacteriol. 1998, 48, 53–58. [Google Scholar] [PubMed]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479–490. [Google Scholar]
- Lin, X.; Wen, Y.; Li, M.; Chen, Z.; Guo, J.; Song, Y.; Li, J. A new strain of Streptomyces avermitilis produces high yield of oligomycin A with potent anti-tumor activity on human cancer cell lines in vitro. Appl. Microbiol. Biotechnol. 2009, 81, 839–845. [Google Scholar]
- Solis, P.N.; Wright, C.W.; Anderson, M.M.; Gupta, M.P.; Phillipson, J.D. A microwell cytotoxicity assay using Artemia salina (brine shrimp). Planta. Med. 1993, 59, 250–252. [Google Scholar]
- Schmidtke, M.; Makarov, V.A.; Riabova, O.B.; Granik, V.G.; Wutzler, P. Novel [(biphenyloxy)propyl]isoxazole derivatives for inhibition of human rhinovirus 2 and coxsackievirus B3 replication. J. Antimicrob. Chemother. 2005, 55, 483–488. [Google Scholar]
- Schmidtke, M.; Schnittler, U.; Jahn, B.; Dahse, H.M.; Stelzner, A. A rapid assay for evaluation of antiviral activity against coxsackie virus B3, influenza virus A, and herpes simplex virus type 1. J. Virol. Methods 2001, 95, 133–143. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lin, X.; Zhou, X.; Wang, F.; Liu, K.; Yang, B.; Yang, X.; Peng, Y.; Liu, J.; Ren, Z.; Liu, Y. A New Cytotoxic Sesquiterpene Quinone Produced by Penicillium sp. F00120 Isolated from a Deep Sea Sediment Sample. Mar. Drugs 2012, 10, 106-115. https://doi.org/10.3390/md10010106
Lin X, Zhou X, Wang F, Liu K, Yang B, Yang X, Peng Y, Liu J, Ren Z, Liu Y. A New Cytotoxic Sesquiterpene Quinone Produced by Penicillium sp. F00120 Isolated from a Deep Sea Sediment Sample. Marine Drugs. 2012; 10(1):106-115. https://doi.org/10.3390/md10010106
Chicago/Turabian StyleLin, Xiuping, Xuefeng Zhou, Fazuo Wang, Kaisheng Liu, Bin Yang, Xianwen Yang, Yan Peng, Juan Liu, Zhe Ren, and Yonghong Liu. 2012. "A New Cytotoxic Sesquiterpene Quinone Produced by Penicillium sp. F00120 Isolated from a Deep Sea Sediment Sample" Marine Drugs 10, no. 1: 106-115. https://doi.org/10.3390/md10010106
APA StyleLin, X., Zhou, X., Wang, F., Liu, K., Yang, B., Yang, X., Peng, Y., Liu, J., Ren, Z., & Liu, Y. (2012). A New Cytotoxic Sesquiterpene Quinone Produced by Penicillium sp. F00120 Isolated from a Deep Sea Sediment Sample. Marine Drugs, 10(1), 106-115. https://doi.org/10.3390/md10010106