Reassembled Biosynthetic Pathway for a Large-scale Synthesis of CMP-Neu5Ac
Abstract
:Introduction
Results and Discussion
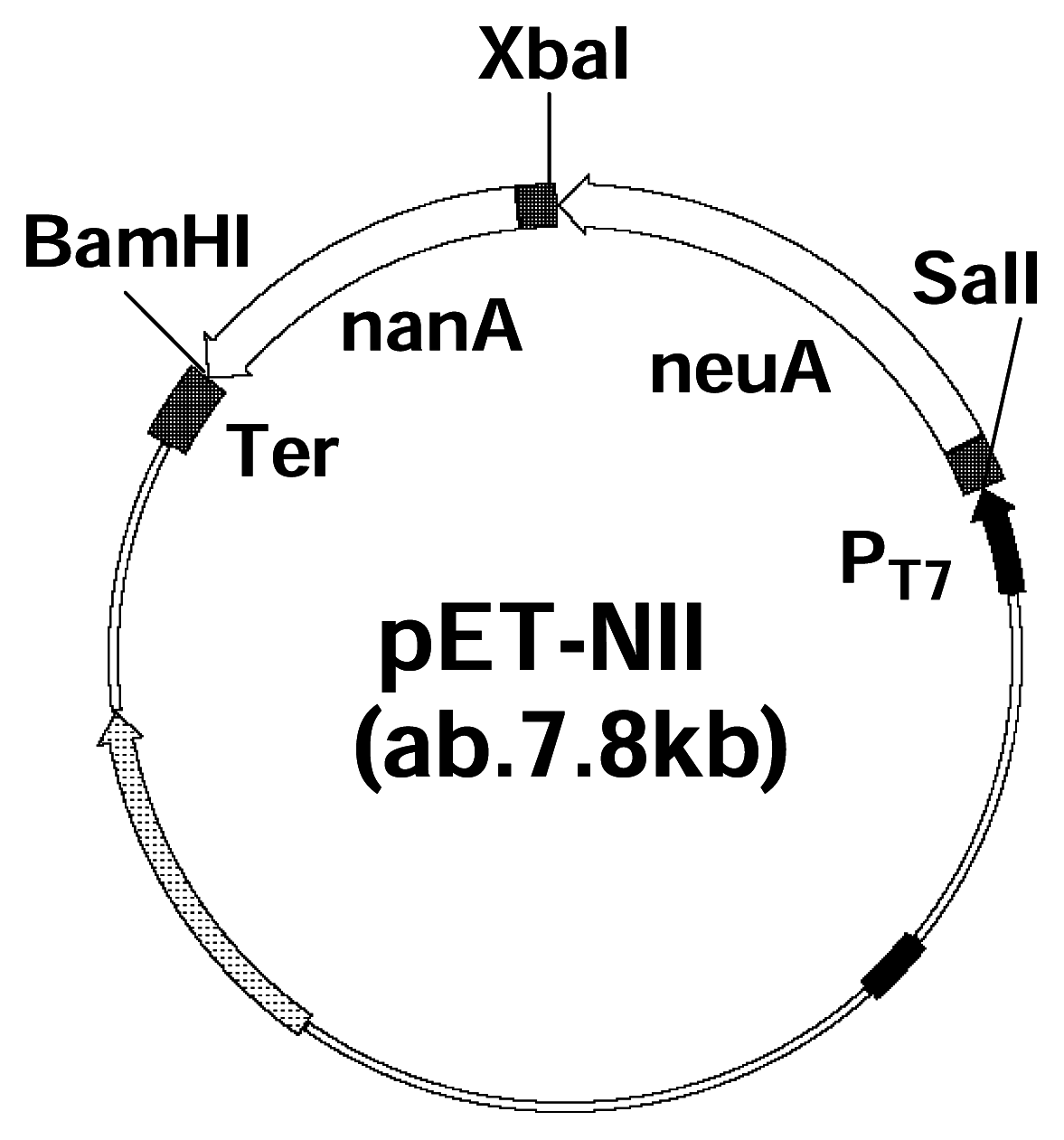
Co-expression and characterization of recombinant enzymes
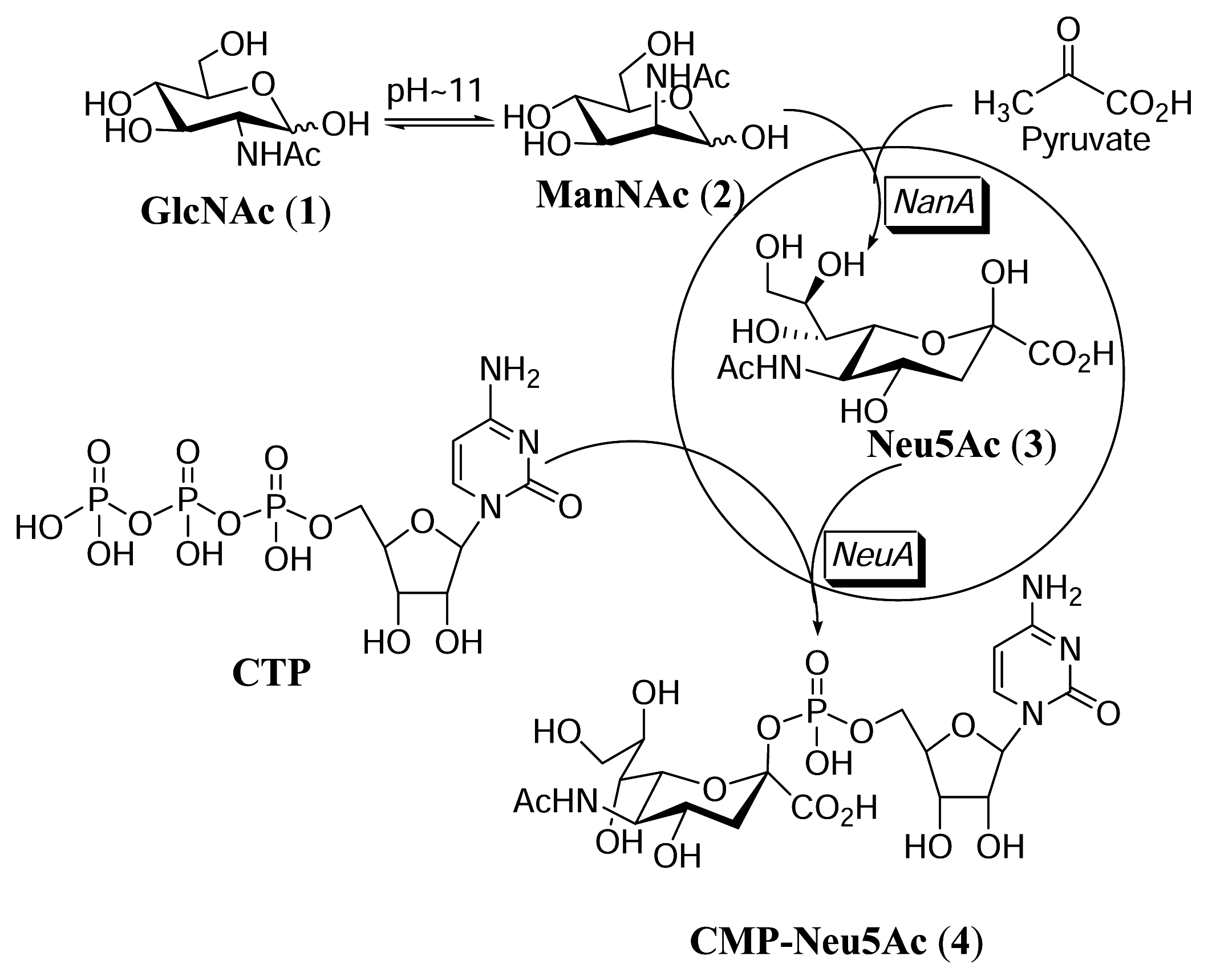
Isomerization of GlcNAc to ManNAc
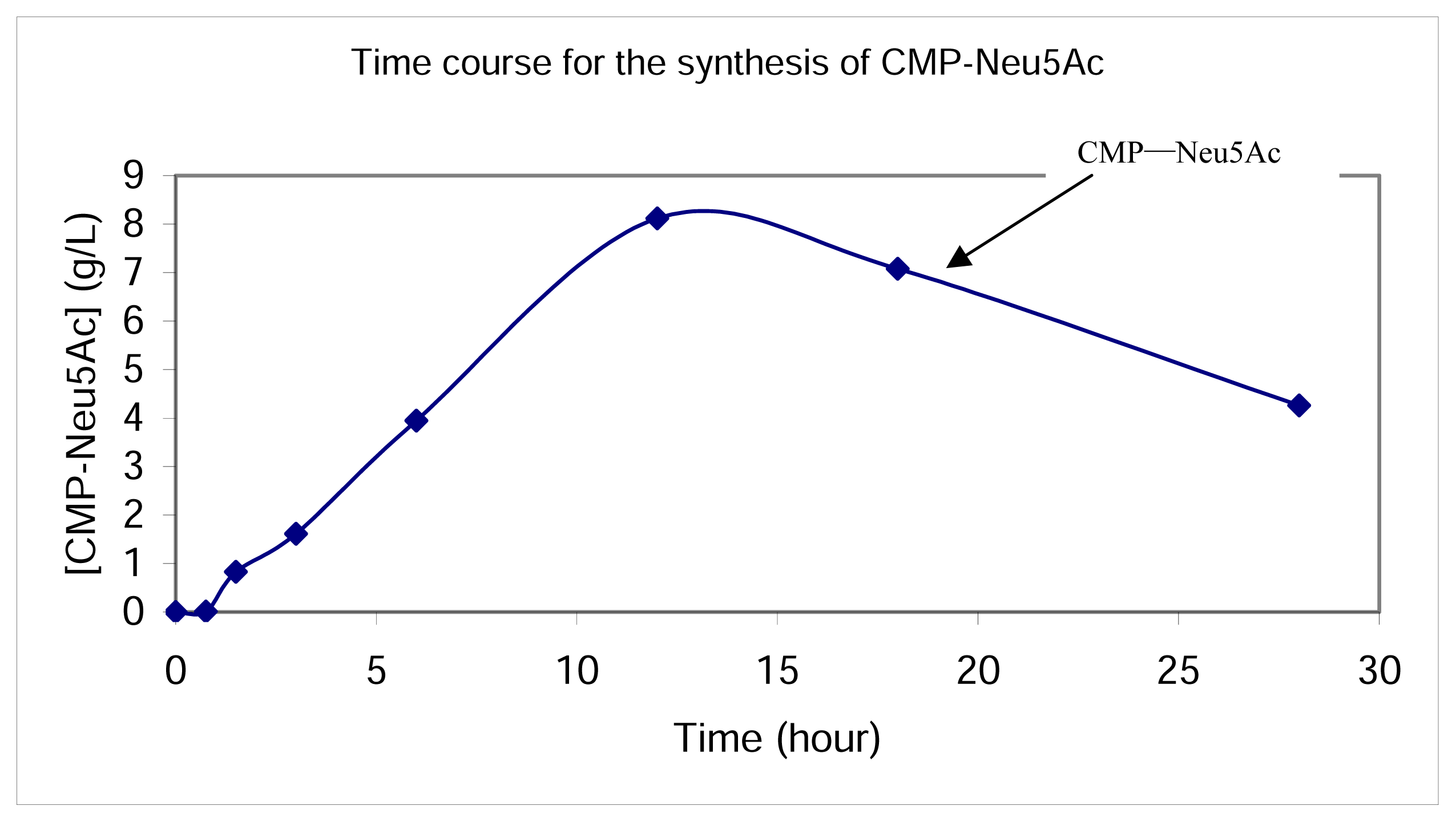
Synthesis of CMP-Neu5Ac in the whole cell reaction
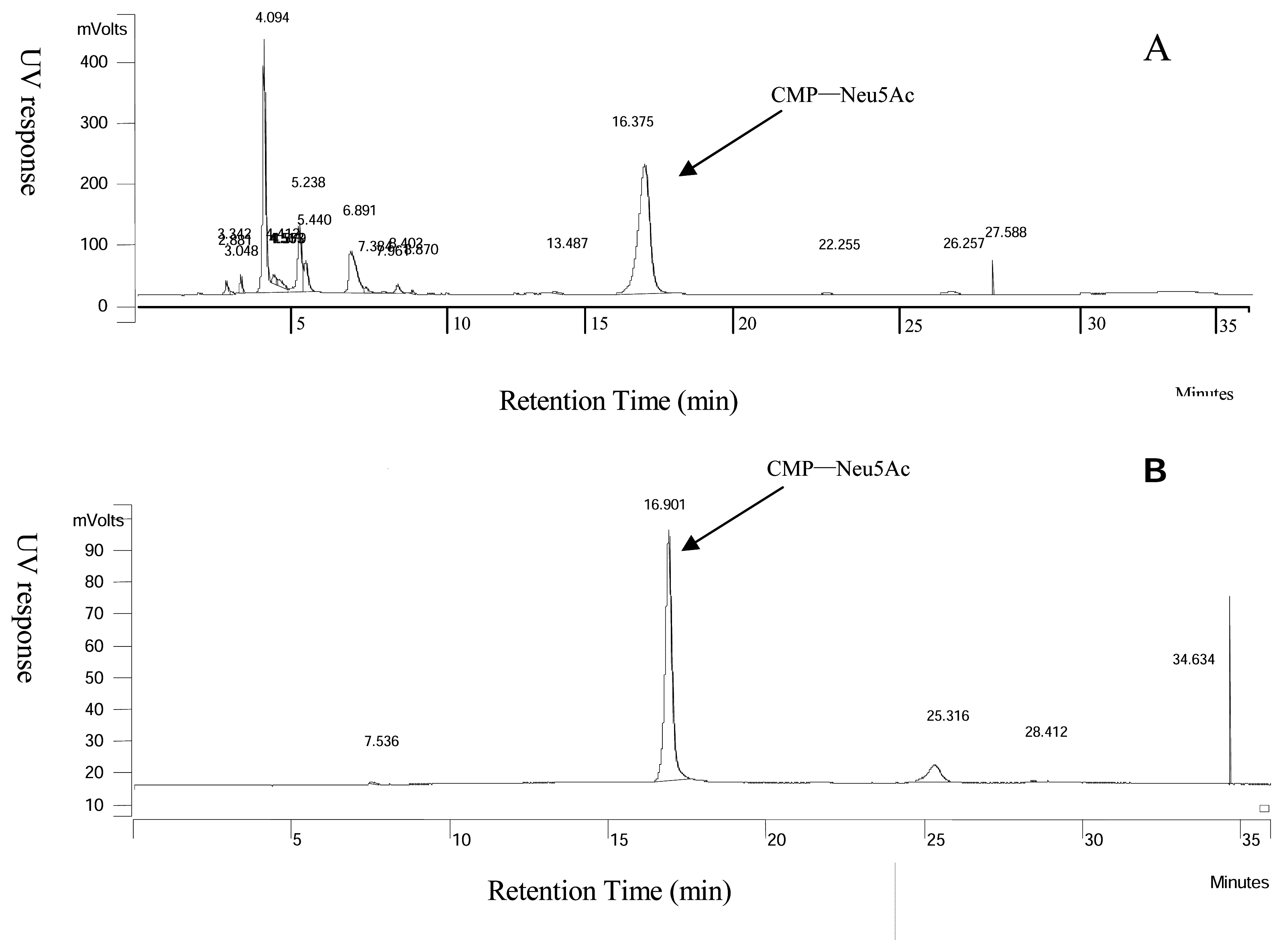
Analysis of CMP-Neu5Ac
Purification of CMP-Neu5Ac
Conclusions
Experimental
General
PCR and Molecular Cloning
Co-expression and purification of Neu5Ac aldolase gene and CMP-Neu5Ac synthetase
Isomerization of N-Acetylglucosamine to N-Acetylmannosamine
Synthesis of CMP-NeuAc in whole cell reaction
Purification of CMP-Neu5Ac
| Alcohols | Solubility (g/L) | ManNAc/GlcNAc | |
|---|---|---|---|
| ManNAc | GlcNAc | ||
| Methanol | 90.6 | 4.2 | 21.6 |
| Ethanol | 8.0 | 0.7 | 11.4 |
| n-Propanol | 1.1 | 0.2 | 5.5 |
| i-Propanol | 1.0 | 0.2 | 5.0 |
- Sample Availability: Available from the authors.
References and Notes
- Schauer, R. Chemistry, metabolism, and biological functions of sialic acids. Adv. Carbohydr. Chem. Biochem 1982, 40, 131–234. [Google Scholar]
- Angata, T.; Varki, A. Chemical Diversity in the Sialic Acids and Related a-Keto Acids: An Evolutionary Perspective. Chem. Rev. (Washington, D. C.) 2002, 102, 439–469. [Google Scholar]
- Sialic acids. In Essentials of Glycobiology; Varki, A.; Cummings, R.; Esko, J.; Freeze, H.; Harth, G.; Marth, J. (Eds.) Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 1999; pp. 195–209.
- Martin, T. J.; Schmidt, R. R. Convenient chemical synthesis of CMP-N-acetylneuraminate (CMP-Neu-5-Ac). Tetrahedron Lett 1993, 34, 1765–8. [Google Scholar]
- Kittelmann, M.; Klein, T.; Kragl, U.; Wandrey, C.; Ghisalba, O. CMP-N-acetylneuraminic acid synthetase from Escherichia coli. Fermentation production and application for the preparative synthesis of CMP-neuraminic acid. Appl. Microbiol. Biotechnol 1995, 44, 59–67. [Google Scholar]
- Endo, T.; Koizumi, S.; Tabata, K.; Ozaki, A. Large-scale production of CMP-NeuAc and sialylated oligosaccharides through bacterial coupling. Appl. Microbiol. Biotechnol 2000, 53, 257–261. [Google Scholar]
- Vimr, E. R.; Troy, F. A. Regulation of sialic acid metabolism in Escherichia coli: role of Nacylneuraminate pyruvate-lyase. J. Bacteriol 1985, 164, 854–60. [Google Scholar]
- Munster, A. K.; Eckhardt, M.; Potvin, B.; Muhlenhoff, M.; Stanley, P.; Gerardy-Schahn, R. Mammalian cytidine 5′-monophosphate N-acetylneuraminic acid synthetase: a nuclear protein with evolutionarily conserved structural motifs. Proc. Nat. Acad. Sci. U.S.A 1998, 95, 9140–5. [Google Scholar]
- Vann, W. F.; Silver, R. P.; Abeijon, C.; Chang, K.; Aaronson, W.; Sutton, A.; Finn, C. W.; Lindner, W.; Kotsatos, M. Purification, properties, and genetic location of Escherichia coli cytidine 5′-monophosphate N-acetylneuraminic acid synthetase. J. Biol. Chem 1987, 262, 17556–62. [Google Scholar]
- Zapata, G.; Vann, W. F.; Aaronson, W.; Lewis, M. S.; Moos, M. Sequence of the cloned Escherichia coli K1 CMP-N-acetylneuraminic acid synthetase gene. J. Biol. Chem 1989, 264, 14769–74. [Google Scholar]
- Edwards, U.; Frosch, M. Sequence and functional analysis of the cloned Neisseria meningitidis CMP-NeuNAc synthetase. FEMS Microbiol. Lett 1992, 75, 161–6. [Google Scholar]
- Tullius, M. V.; Munson, R. S., Jr; Wang, J.; Gibson, B. W. Purification, cloning, and expression of a cytidine 5′-monophosphate N-acetylneuraminic acid synthetase from Haemophilus ducreyi. J. Biol. Chem 1996, 271, 15373–80. [Google Scholar]
- Stoughton, D. M.; Zapata, G.; Picone, R.; Vann, W. F. Identification of Arg-12 in the active site of Escherichia coli K1 CMP-sialic acid synthetase. Biochem. J 1999, 343 Pt 2, 397–402. [Google Scholar]
- Chen, X.; Fang, J.; Zhang, J.; Liu, Z.; Shao, J.; Kowal, P.; Andreana, P.; Wang, P. G. Sugar nucleotide regeneration beads (superbeads): a versatile tool for the practical synthesis of oligosaccharides. J. Am. Chem. Soc 2001, 123, 2081–2082. [Google Scholar]
- Chen, X.; Liu, Z.; Zhang, J.; Zhang, W.; Kowal, P.; Wang, P. G. Reassembled biosynthetic pathway for large-scale carbohydrate synthesis: a-gal epitope producing “superbug”. Chem BioChem 2002, 3, 47–53. [Google Scholar]
- Maniatis, T.; Fritsch, E. F.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 1982; pp. 1–35. [Google Scholar]
- Keppler, O. T.; Hinderlich, S.; Langner, J.; Schwartz-Albiez, R.; Reutter, W.; Pawlita, M. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science 1999, 284, 1372–6. [Google Scholar]
- Maru, I.; Ohnishi, J.; Ohta, Y.; Tsukada, Y. Simple and large-scale production of N-acetylneuraminic acid from N-acetyl-D-glucosamine and pyruvate using N-acyl-D-glucosamine 2-epimerase and N-acetylneuraminate lyase. Carbohydr. Res 1998, 306, 575–8. [Google Scholar]
- Muhmoudian, M.; Noble, D.; Drake, C. S.; Middleton, R. F.; Montgomery, D. S.; Piercey, J. E.; Ramlakhan, D.; Todd, M.; Dawson, M. J. An efficient process for production of N-acetylneuraminic acid using N-acetylneuraminic acid aldolase. Enzyme Microb. Technol 1997, 20, 393–400. [Google Scholar]
- Uchida, Y.; Tsukada, Y.; Sugimori, T. Purification and properties of N-acetylneuraminate lyase from Escherichia coli. J. Biochem 1984, 96, 507–22. [Google Scholar]
- Simon, E. S.; Bednarski, M. D.; Whitesides, G. M. Synthesis of CMP-NeuAc from N-acetylglucosamine: generation of CTP from CMP using adenylate kinase. J. Am. Chem. Soc 1988, 110, 7159–63. [Google Scholar]
- Koeller, K. M.; Wong, C.-H. Synthesis of Complex Carbohydrates and Glycoconjugates: Enzyme-Based and Programmable One-Pot Strategies. Chem. Rev. (Washington, D. C.) 2000, 100, 4465–4493. [Google Scholar]
- Palcic, M. M. Biocatalytic synthesis of oligosaccharides. Curr. Opin. Biotechnol 1999, 10, 616–624. [Google Scholar]
- Fang, J.; Li, J.; Chen, X.; Zhang, Y.; Wang, J.; Guo, Z.; Zhang, W.; Yu, L.; Brew, K.; Wang, P. G. Highly Efficient Chemoenzymic Synthesis of a-Galactosyl Epitopes with a Recombinant a(1- >3)-Galactosyltransferase. J. Am. Chem. Soc 1998, 120, 6635–6638. [Google Scholar]
- Seitz, O.; Wong, C.-H. Chemoenzymic solution- and solid-phase synthesis of O-glycopeptides of the Mucin domain of MAdCAM-1. A general route to O-LacNAc, O-Sialyl-LacNAc, and O-Sialyl- Lewis-X peptides. J. Am. Chem. Soc 1997, 119, 8766–8776. [Google Scholar]
- Baisch, G.; Ohrlein, R.; Streiff, M.; Kolbinger, F. Enzymatic synthesis of sialyl-Lewis(a)- libraries with two non-natural monosaccharide units. Bioorg. Med. Chem. Lett 1998, 8, 755–8. [Google Scholar]
© 2003 by MDPI Reproduction is permitted for noncommercial purposes.
Share and Cite
Song, J.; Zhang, H.; Wu, B.; Zhang, Y.; Li, H.; Xiao, M.; Wang, P.G. Reassembled Biosynthetic Pathway for a Large-scale Synthesis of CMP-Neu5Ac. Mar. Drugs 2003, 1, 34-45. https://doi.org/10.3390/md101034
Song J, Zhang H, Wu B, Zhang Y, Li H, Xiao M, Wang PG. Reassembled Biosynthetic Pathway for a Large-scale Synthesis of CMP-Neu5Ac. Marine Drugs. 2003; 1(1):34-45. https://doi.org/10.3390/md101034
Chicago/Turabian StyleSong, Jing, Hesheng Zhang, Bingyuan Wu, Yingxin Zhang, Hanfen Li, Min Xiao, and Peng George Wang. 2003. "Reassembled Biosynthetic Pathway for a Large-scale Synthesis of CMP-Neu5Ac" Marine Drugs 1, no. 1: 34-45. https://doi.org/10.3390/md101034
APA StyleSong, J., Zhang, H., Wu, B., Zhang, Y., Li, H., Xiao, M., & Wang, P. G. (2003). Reassembled Biosynthetic Pathway for a Large-scale Synthesis of CMP-Neu5Ac. Marine Drugs, 1(1), 34-45. https://doi.org/10.3390/md101034




