Identification of Insulin Resistance Biomarkers in Metabolic Syndrome Detected by UHPLC-ESI-QTOF-MS
Abstract
:1. Introduction
2. Results
2.1. Participant and Blood Sample Characteristics
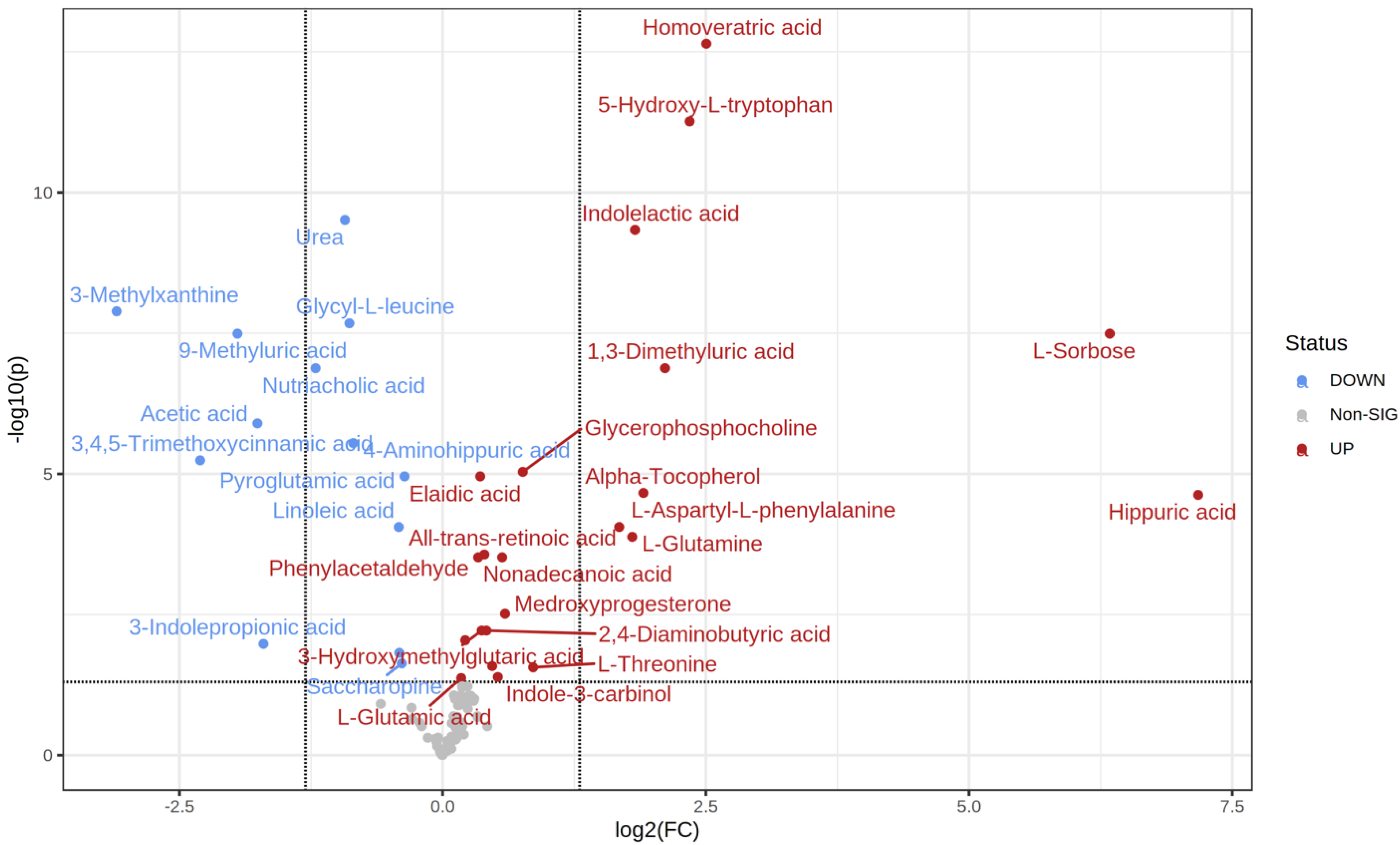
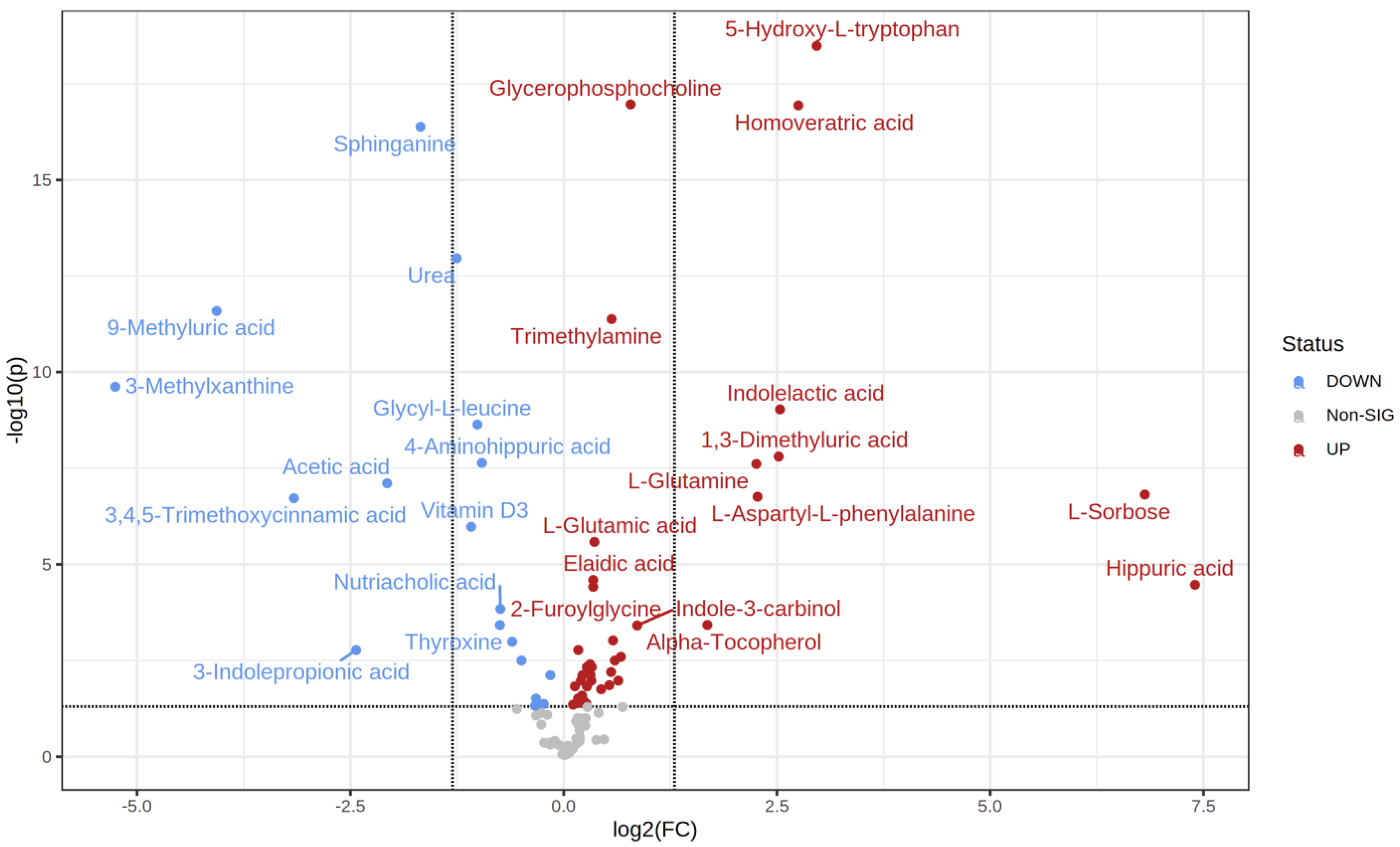
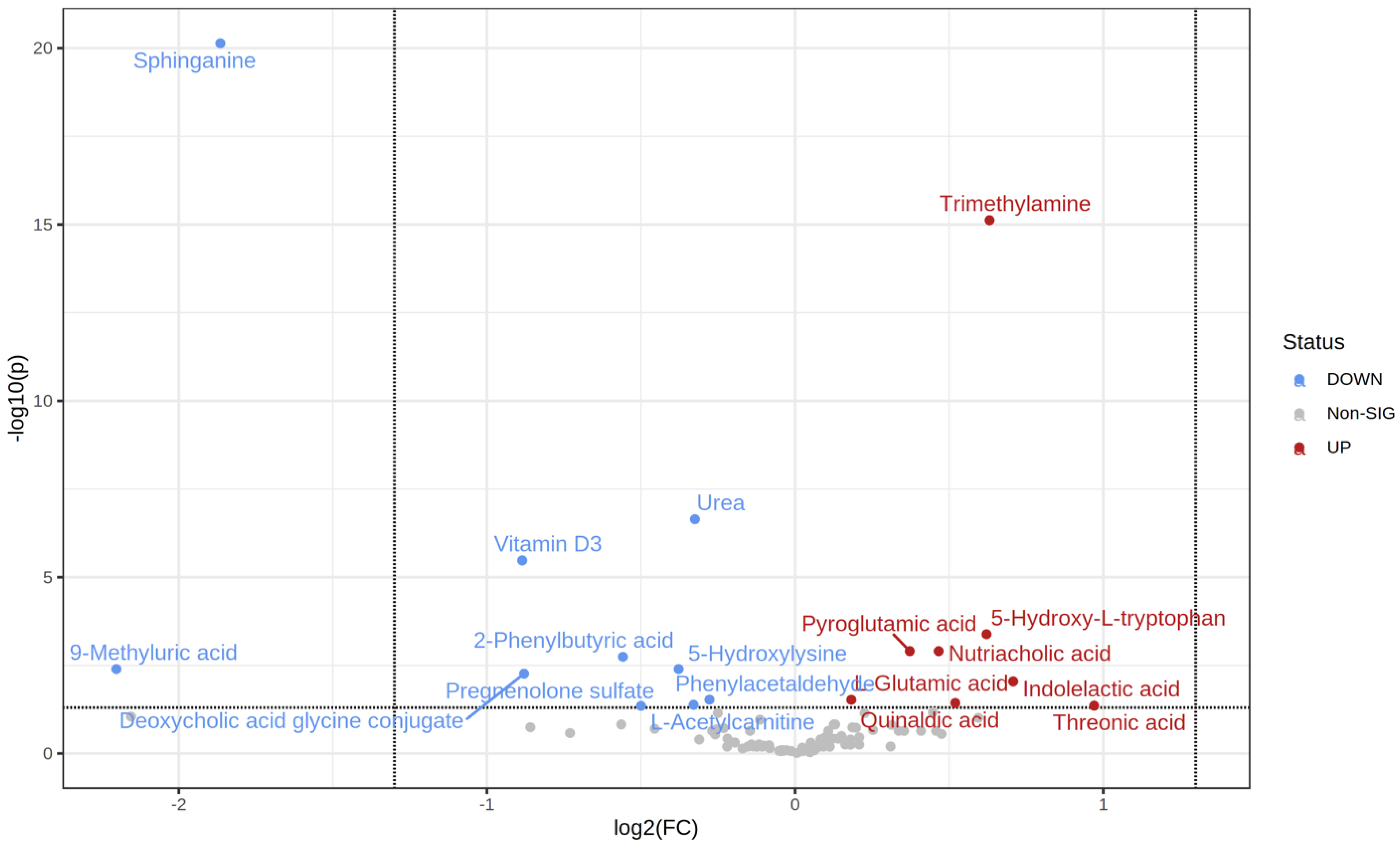
2.2. Metabolic Changes
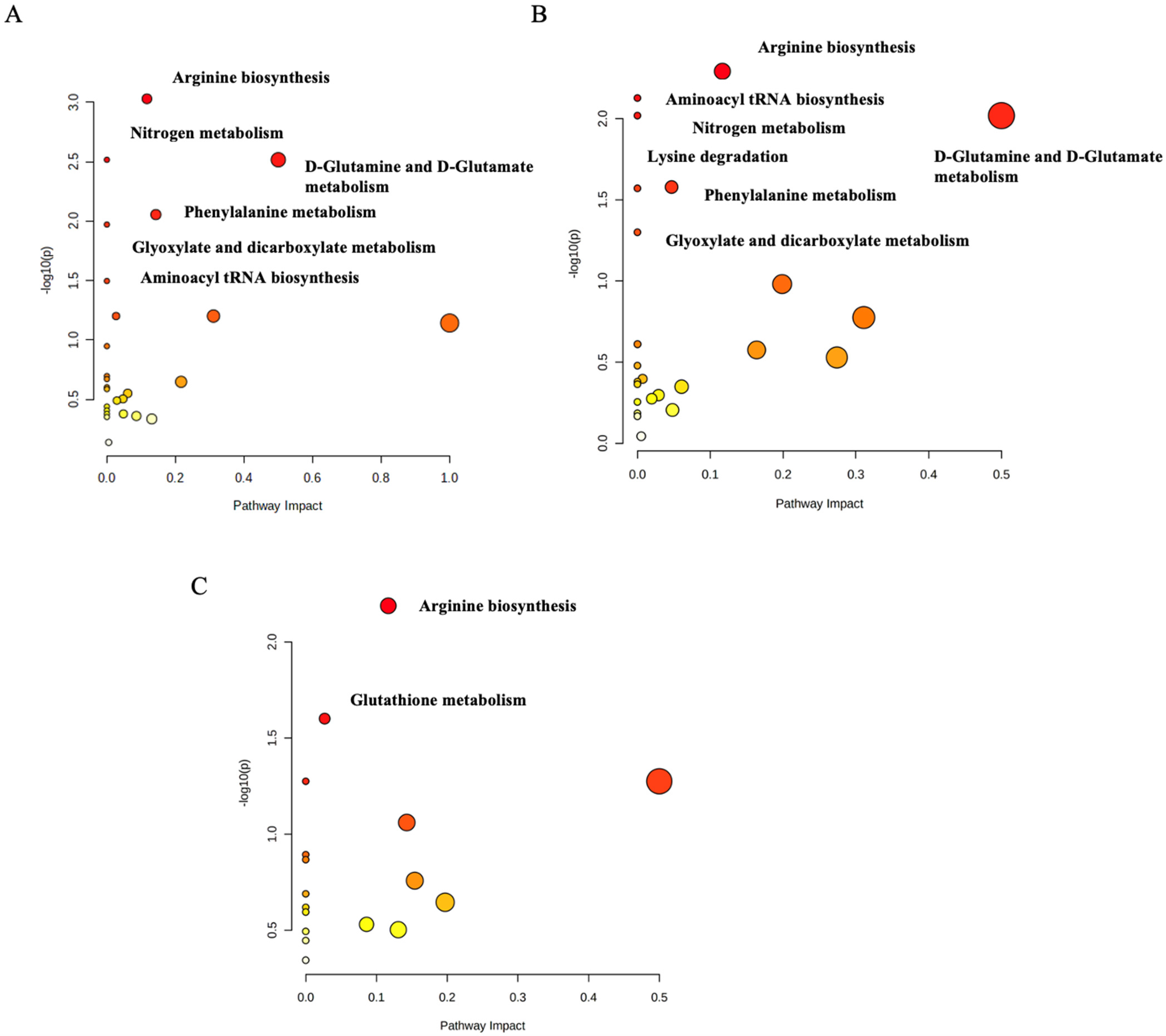
2.3. Functional Analysis Pathway Changes
3. Discussion
3.1. The Arginine Biosynthesis Pathway
3.2. The D-Glutamine and D-Glutamate Metabolism Pathway
3.3. The Phenylalanine Metabolism Pathway
3.4. The Lysine Degradation Pathway
3.5. The Glutathione Metabolism Pathway
3.6. Aminoacyl tRNA Biosynthesis
3.7. Glyoxylate and Dicarboxylate Metabolism
3.8. Nitrogen Metabolism
4. Materials and Methods
4.1. Population and Study Design
4.2. Collection of Samples
4.3. Preparation of the Samples for Metabolomics Extraction
4.4. Ultra-High-Performance Liquid Chromatography Coupled to Electrospray Ionization and Quadrupole Time-of-Flight Mass Spectrometry (UHPLC-ESI-QTOF-MS)
4.5. Data Processing and Analysis
4.6. Metabolic Pathway and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fahed, G.; Aoun, L.; Zerdan, M.B.; Allam, S.; Zerdan, M.B.; Bouferraa, Y.; Assi, H.I. Metabolic Syndrome: Updates on Pathophysiology and Management in 2021. Int. J. Mol. Sci. 2022, 23, 786. [Google Scholar] [CrossRef] [PubMed]
- Belete, R.; Ataro, Z.; Abdu, A.; Sheleme, M. Global Prevalence of Metabolic Syndrome among Patients with Type I Diabetes Mellitus: A Systematic Review and Meta-Analysis. Diabetol. Metab. Syndr. 2021, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Azarpazhooh, M.R.; Najafi, F.; Darbandi, M.; Kiarasi, S.; Oduyemi, T.; Spence, J.D. Triglyceride/High-Density Lipoprotein Cholesterol Ratio: A Clue to Metabolic Syndrome, Insulin Resistance, and Severe Atherosclerosis. Lipids 2021, 56, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.C.; Ng, O.C.; Wong, T.W.; Joseph, A.; Chan, Y.M.; Hejar, A.R.; Tan, M.C.; Ng, O.C.; Wong, T.W.; Joseph, A.; et al. Prevalence of Metabolic Syndrome in Type 2 Diabetic Patients: A Comparative Study Using WHO, NCEP ATP III, IDF and Harmonized Definitions. Health 2013, 5, 1689–1696. [Google Scholar] [CrossRef] [Green Version]
- Alberti, K.G.M.M.; Zimmet, P.; Shaw, J. Metabolic Syndrome—A New World-Wide Definition. A Consensus Statement from the International Diabetes Federation. Diabet. Med. 2006, 23, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Eizirik, D.L.; Pasquali, L.; Cnop, M. Pancreatic β-Cells in Type 1 and Type 2 Diabetes Mellitus: Different Pathways to Failure. Nat. Rev. Endocrinol. 2020, 16, 349–362. [Google Scholar] [CrossRef]
- Husein, R.; Depkes, R.I.; Soelistijo, S.A.; Novida, H.; Rudijanto, A.; BPJS; Of, S.; Carediabetes, M.; Ceriello, A.; Gavin, J.R.; et al. Classification of Diabetes Mellitus. Dep. Manag. NCD Disabil. Violence Inj. Prev. 2018, 138, 271–281. [Google Scholar]
- Association, A.D. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care 2021, 44, S15–S33. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative Omics for Health and Disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- TimsTOF | Bruker. Available online: https://www.bruker.com/en/products-and-solutions/mass-spectrometry/timstof.html (accessed on 20 April 2022).
- Zhong, F.; Xu, M.; Bruno, R.S.; Ballard, K.D.; Zhu, J. Targeted High Performance Liquid Chromatography Tandem Mass Spectrometry-Based Metabolomics Differentiates Metabolic Syndrome from Obesity. Exp. Biol. Med. (Maywood) 2017, 242, 773–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azab, S.M.; de Souza, R.J.; Lamri, A.; Shanmuganathan, M.; Kroezen, Z.; Schulze, K.M.; Desai, D.; Williams, N.C.; Morrison, K.M.; Atkinson, S.A.; et al. Metabolite Profiles and the Risk of Metabolic Syndrome in Early Childhood: A Case-Control Study. BMC Med. 2021, 19, 292. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Kwak, S.Y.; Jo, G.; Song, T.J.; Shin, M.J. Serum Metabolite Profile Associated with Incident Type 2 Diabetes in Koreans: Findings from the Korean Genome and Epidemiology Study. Sci. Rep. 2018, 8, 8207. [Google Scholar] [CrossRef] [Green Version]
- Schrauben, S.J.; Jepson, C.; Hsu, J.Y.; Wilson, F.P.; Zhang, X.; Lash, J.P.; Robinson, B.M.; Townsend, R.R.; Chen, J.; Fogelfeld, L.; et al. Insulin Resistance and Chronic Kidney Disease Progression, Cardiovascular Events, and Death: Findings from the Chronic Renal Insufficiency Cohort Study. BMC Nephrol. 2019, 20, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumari, S.; Sharma, P. Targeted Metabolomics Reveals Hippurate as a Urinary Potential Marker for Diabetic Nephropathy. Curr. Metab. 2015, 4, 121–128. [Google Scholar] [CrossRef]
- Metabolomics Studies of Obesity, Prediabetes and Type Two Diabetes: Potential Biomarkers Identified in Human and Animal Models—UCL Discovery. Available online: https://discovery.ucl.ac.uk/id/eprint/10133649/ (accessed on 26 April 2022).
- Perez-Pozo, S.E.; Schold, J.; Nakagawa, T.; Sánchez-Lozada, L.G.; Johnson, R.J.; Lillo, J.L. Excessive Fructose Intake Induces the Features of Metabolic Syndrome in Healthy Adult Men: Role of Uric Acid in the Hypertensive Response. Int. J. Obes. (Lond.) 2010, 34, 454–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.S.; Wu, Q.J.; Xia, Y.; Zhang, J.Y.; Jiang, Y.T.; Chang, Q.; Zhao, Y.H. Carbohydrate Intake and Risk of Metabolic Syndrome: A Dose-Response Meta-Analysis of Observational Studies. Nutr. Metab. Cardiovasc. Dis. 2019, 29, 1288–1298. [Google Scholar] [CrossRef]
- Taskinen, M.R.; Packard, C.J.; Borén, J. Dietary Fructose and the Metabolic Syndrome. Nutrients 2019, 11, 1987. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.K.; Chang, Y.; Kim, I.; Ryu, S. U-Shaped Association Between Serum Uric Acid Level and Risk of Mortality: A Cohort Study. Arthritis Rheumatol. 2018, 70, 1122–1132. [Google Scholar] [CrossRef] [Green Version]
- Human Metabolome Database: Showing Metabocard for 9-Methyluric Acid (HMDB0001973). Available online: https://hmdb.ca/metabolites/HMDB0001973 (accessed on 21 April 2022).
- Chen, C.J.; Liao, W.L.; Chang, C.T.; Lin, Y.N.; Tsai, F.J. Identification of Urinary Metabolite Biomarkers of Type 2 Diabetes Nephropathy Using an Untargeted Metabolomic Approach. J. Proteome Res. 2018, 17, 3997–4007. [Google Scholar] [CrossRef]
- Wang, H.; Fang, J.; Chen, F.; Sun, Q.; Xu, X.; Lin, S.H.; Liu, K. Metabolomic Profile of Diabetic Retinopathy: A GC-TOFMS-Based Approach Using Vitreous and Aqueous Humor. Acta Diabetol. 2020, 57, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Shimizu, F.; Takao, T.; Masuda, J.; Takada, A.; Shimizu, F.; Takao, T.; Masuda, J. Measurement of Tryptophan Metabolites in Healthy Old Men and Patients of Type 2 Diabetes Mellitus (T2DM). Food Nutr. Sci. 2018, 9, 1206–1220. [Google Scholar] [CrossRef] [Green Version]
- Roff, C.F.; Goldin, E.; Comly, M.E.; Cooney, A.; Brown, A.; Vanier, M.T.; Miller, S.P.F.; Brady, R.O.; Pentchev, P.G. Type C Niemann-Pick Disease: Use of Hydrophobic Amines to Study Defective Cholesterol Transport. Dev. Neurosci. 1991, 13, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Human Metabolome Database: Showing Metabocard for Sphinganine (HMDB0000269). Available online: https://hmdb.ca/metabolites/HMDB0000269 (accessed on 21 April 2022).
- Rhee, E.J.; Han, K.; Ko, S.H.; Ko, K.S.; Lee, W.Y. Increased Risk for Diabetes Development in Subjects with Large Variation in Total Cholesterol Levels in 2,827,950 Koreans: A Nationwide Population-Based Study. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Li, Z.; Liu, H.; Wang, X.; Lv, C.; Wang, R.; Zhang, D.; Li, Y.; Du, X.; Li, Y.; et al. Metabolomics-Based Clinical Efficacy and Effect on the Endogenous Metabolites of Tangzhiqing Tablet, a Chinese Patent Medicine for Type 2 Diabetes Mellitus with Hypertriglyceridemia. Evid.-Based Complementary Altern. Med. 2018, 2018, 5490491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Han, M.; Rezaei, A.; Li, D.; Wu, G.; Ma, X. L-Arginine Modulates Glucose and Lipid Metabolism in Obesity and Diabetes. Curr. Protein. Pept. Sci. 2017, 18, 599–608. [Google Scholar] [CrossRef]
- Ginguay, A.; Cynober, L.A. Amino Acids: Amino Acid Metabolism. In Encyclopedia of Biological Chemistry, 3rd ed.; Willey: Hoboken, NJ, USA, 2021; Volume 1, pp. 2–9. [Google Scholar] [CrossRef]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite Profiles and the Risk of Developing Diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Zhuang, Y.; Zhang, P.; Chen, S.; Asakawa, T.; Gao, B. Serum Metabolomic Profiles Associated with Untreated Metabolic Syndrome Patients in the Chinese Population. Clin. Transl. Sci. 2020, 13, 1271–1278. [Google Scholar] [CrossRef]
- Yelamanchi, S.D.; Jayaram, S.; Thomas, J.K.; Gundimeda, S.; Khan, A.A.; Singhal, A.; Keshava Prasad, T.S.; Pandey, A.; Somani, B.L.; Gowda, H. A Pathway Map of Glutamate Metabolism. J. Cell Commun. Signal. 2016, 10, 69. [Google Scholar] [CrossRef] [Green Version]
- Adams, S.H. Emerging Perspectives on Essential Amino Acid Metabolism in Obesity and the Insulin-Resistant State. Adv. Nutr. 2011, 2, 445–456. [Google Scholar] [CrossRef] [Green Version]
- Bala, C.G.; Rusu, A.; Ciobanu, D.; Bucsa, C.; Roman, G. Amino Acid Signature of Oxidative Stress in Patients with Type 2 Diabetes: Targeted Exploratory Metabolomic Research. Antioxidants 2021, 10, 610. [Google Scholar] [CrossRef] [PubMed]
- Wurtz, P.; Soininen, P.; Kangas, A.J.; Rönnemaa, T.; Lehtimäki, T.; Kähönen, M.; Viikari, J.S.; Raitakari, O.T.; Ala-Korpela, M. Branched-Chain and Aromatic Amino Acids Are Predictors of Insulin Resistance in Young Adults. Diabetes Care 2013, 36, 648–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichter-Konecki, U.; Hipke, C.M.; Konecki, D.S. Human Phenylalanine Hydroxylase Gene Expression in Kidney and Other Nonhepatic Tissues. Mol. Genet. Metab. 1999, 67, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Mirmiranpour, H.; Bathaie, S.Z.; Khaghani, S.; Nakhjavani, M.; Kebriaeezadeh, A. Investigation of the Mechanism(s) Involved in Decreasing Increased Fibrinogen Activity in Hyperglycemic Conditions Using L-Lysine Supplementation. Thromb. Res. 2012, 130, e13–e19. [Google Scholar] [CrossRef] [PubMed]
- del Coco, L.; Vergara, D.; de Matteis, S.; Mensà, E.; Sabbatinelli, J.; Prattichizzo, F.; Bonfigli, A.R.; Storci, G.; Bravaccini, S.; Pirini, F.; et al. NMR-Based Metabolomic Approach Tracks Potential Serum Biomarkers of Disease Progression in Patients with Type 2 Diabetes Mellitus. J. Clin. Med. 2019, 8, 720. [Google Scholar] [CrossRef] [Green Version]
- Dall’Olio, F.; Vanhooren, V.; Chen, C.C.; Slagboom, P.E.; Wuhrer, M.; Franceschi, C. N-Glycomic Biomarkers of Biological Aging and Longevity: A Link with Inflammaging. Ageing Res. Rev. 2013, 12, 685–698. [Google Scholar] [CrossRef] [Green Version]
- Lutchmansingh, F.K.; Hsu, J.W.; Bennett, F.I.; Badaloo, A.V.; Norma, M.A.; Georgiana, M.G.S.; Rosemarie, A.W.P.; Jahoor, F.; Boyne, M.S. Glutathione Metabolism in Type 2 Diabetes and Its Relationship with Microvascular Complications and Glycemia. PLoS ONE 2018, 13, e0198626. [Google Scholar] [CrossRef] [Green Version]
- Excessive Excretion of 5-Oxoproline and Decreased Levels of Blood Glutathione in Type II Diabetes Mellitus. Semantic Scholar. Available online: https://www.semanticscholar.org/paper/Excessive-excretion-of-5-oxoproline-and-decreased-Forrester-Badaloo/ad79ab69c5124de27af3daa5018fd0a6690b9bda (accessed on 21 April 2022).
- Gomez, M.A.R.; Ibba, M. Aminoacyl-TRNA Synthetases. RNA 2020, 26, 910–936. [Google Scholar] [CrossRef]
- Sang, G.P.; Schimmel, P.; Kim, S. Aminoacyl TRNA Synthetases and Their Connections to Disease. Proc. Natl. Acad. Sci. USA 2008, 105, 11043–11049. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, H.; Rong, S.; Bian, C.; Yang, Y.; Pan, H. NMR Spectroscopy Based Metabolomics Confirms the Aggravation of Metabolic Disorder in Metabolic Syndrome Combined with Hyperuricemia. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 2449–2457. [Google Scholar] [CrossRef]
- OuYang, Y.N.; Jin, Y.X.; Zhao, X.R.; Chen, M.; Yang, P.F.; Zheng, X.W.; Zeng, L.; Chen, L.; Tian, Z. Revealing Metabolic Pathways Relevant to Prediabetes Based on Metabolomics Profiling Analysis. Biochem. Biophys. Res. Commun. 2020, 533, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Amin Pharmacy, A.M.; Pharm, M.; Arwa Amin, C.M. The Metabolic Signatures of Cardiometabolic Diseases: Does the Shared Metabotype Offer New Therapeutic Targets? Lifestyle Med. 2021, 2, e25. [Google Scholar] [CrossRef]
- Cano, K.E.; Li, L.; Bhatia, S.; Bhatia, R.; Forman, S.J.; Chen, Y. NMR-Based Metabolomic Analysis of the Molecular Pathogenesis of Therapy-Related Myelodysplasia/Acute Myeloid Leukemia. J. Proteome Res. 2011, 10, 2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, C.K.; Sindhu, K.K. Oxidative Stress and Metabolic Syndrome. Life Sci. 2009, 84, 705–712. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Liang, Y.; Hu, R.; Xu, W.; Liu, Y. Plasma Targeted Metabolomics Analysis for Amino Acids and Acylcarnitines in Patients with Prediabetes, Type 2 Diabetes Mellitus, and Diabetic Vascular Complications. Diabetes Metab. J. 2021, 45, 195. [Google Scholar] [CrossRef]
- Merino, J.; Leong, A.; Liu, C.T.; Porneala, B.; Walford, G.A.; von Grotthuss, M.; Wang, T.J.; Flannick, J.; Dupuis, J.; Levy, D.; et al. Metabolomics Insights into Early Type 2 Diabetes Pathogenesis and Detection in Individuals with Normal Fasting Glucose. Diabetologia 2018, 61, 1315–1324. [Google Scholar] [CrossRef] [Green Version]
- Sharaf, B.M.; Giddey, A.D.; Alniss, H.; Al-Hroub, H.M.; El-Awady, R.; Mousa, M.; Almehdi, A.; Soares, N.C.; Semreen, M.H. Untargeted Metabolomics of Breast Cancer Cells MCF-7 and SkBr3 Treated With Tamoxifen/Trastuzumab. Cancer Genom. Proteom. 2022, 19, 79–93. [Google Scholar] [CrossRef]
- MetaboScape | Bruker. Available online: https://www.bruker.com/en/products-and-solutions/mass-spectrometry/ms-software/metaboscape.html (accessed on 19 May 2022).
- Human Metabolome Database. Available online: https://hmdb.ca/ (accessed on 20 April 2022).
- MetaboAnalyst. Available online: https://www.metaboanalyst.ca/ (accessed on 20 April 2022).

| (A) Patients’ Demographics | ||||||
| Total Sample Mean ± SD (n = 86) | Controls Group Mean ± SD (n = 30) | Normoglycemic (MetS) Mean ± SD (n = 30) | Prediabetic MetS Mean ± SD (n = 26) | p-Value | ||
| Female, n (%) | 46 (53.5%) | 16 (53.3%) | 19 (63.3%) | 11 (42.3%) | 0.113 | |
| Age (years) | 48.75 ± 12.87 | 37.83 ± 11.1 | 54.13 ± 10.72 | 53.73 ± 9.55 | <0.001 | |
| (B) Patients’ clinical characteristics | ||||||
| Control Group Mean ± SD (n = 30) | Normoglycemic MetS Mean ± SD (n = 30) | Prediabetic MetS Mean ± SD (n = 26) | p1-Value | p2-Value | p3-Value | |
| BMI (kg/m2) | 22.91 ± 2 | 32.74 ± 4.3 | 32.87 ± 3.8 | <0.001 | <0.001 | 1 |
| SBP (mmHg) | 111.23 ± 9.05 | 132.20 ± 11.61 | 138.17 ± 11.55 | <0.001 | <0.001 | 0.106 |
| DBP (mmHg) | 73.77 ± 6.82 | 81.43 ± 9.22 | 87.50 ± 8.93 | 0.002 | <0.001 | 0.019 |
| HbA1C% | 5.13 ± 0.3 | 5.43 ± 0.243 | 6.65 ± 1.32 | 0.471 | <0.001 | <0.001 |
| FPG (mg/dL) | 88.18 ± 8.68 | 101.07 ± 15.95 | 124.31 ± 47.38 | 0.275 | <0.001 | 0.009 |
| TG (mg/dL) | 76.33 ± 23.61 | 168.74 ± 47.14 | 221.84 ± 102.7 | <0.001 | <0.001 | 0.008 |
| LDL-C (mg/dL) | 117.1 ± 36.57 | 129.7 ± 36.57 | 141.03 ± 43.12 | 0.493 | 0.028 | 0.632 |
| HDL-C (mg/dL) | 58.4 ± 10.21 | 45.87 ± 8.68 | 45.40 ± 13.03 | <0.001 | <0.001 | 1 |
| LDL-C/HDL C ratio | 2.06 ± 0.53 | 2.86 ± 0.73 | 3.28 ± 1.15 | <0.001 | 0.01 | 0.188 |
| WC (cm) | 77.37 ± 7.74 | 111.03 ± 10.5 | 115.2 ± 10.26 | <0.001 | <0.001 | 0.287 |
| Surrogate insulin resistance (sIR) indices | ||||||
| MetS-IR | 13.08 ± 1.67 | 22.68 ± 3.018 | 24.55 ± 3.85 | <0.001 | <0.001 | 0.06 |
| TyG Index | 8.06 ± 0.39 | 9.0 ± 0.24 | 9.38 ± 0.62 | <0.001 | <0.001 | 0.005 |
| TyG-BMI | 185.14 ± 21.01 | 294.66 ± 37.93 | 308.38 ± 39.81 | <0.001 | <0.001 | 0.365 |
| TyG-WC | 625.3 ± 82.47 | 999.07 ± 87.59 | 1081.6 ± 127.91 | <0.001 | <0.001 | 0.007 |
| t.Stat | p.Value | FDR | Fold Change | |
|---|---|---|---|---|
| Hippuric acid | −5.0653 | 4.29 × 10−6 | 2.36 × 10−5 | 144.62 |
| L-Sorbose | −7.0091 | 2.61 × 10−9 | 3.23 × 10−8 | 80.759 |
| Homoveratric acid | −10.342 | 2.30 × 10−15 | 2.28 × 10−13 | 5.6759 |
| 5-Hydroxy-L-tryptophan | −9.366 | 1.10 × 10−13 | 5.42 × 10−12 | 5.0849 |
| 1,3-Dimethyluric acid | −6.4478 | 1.21 × 10−8 | 1.33 × 10−7 | 4.3219 |
| Alpha-Tocopherol | −5.0201 | 3.74 × 10−6 | 2.18 × 10−5 | 3.748 |
| Indolelactic acid | −8.028 | 1.86 × 10−11 | 4.61 × 10−10 | 3.547 |
| L-Glutamine | −4.5216 | 2.79 × 10−5 | 0.00013146 | 3.4823 |
| L-Aspartyl-L-phenylalanine | −4.5417 | 1.69 × 10−5 | 8.78 × 10−5 | 3.1962 |
| L-Threonine | −2.6831 | 0.0091206 | 0.027362 | 1.8144 |
| Glycerophosphocholine | −5.2978 | 1.30 × 10−6 | 9.19 × 10−6 | 1.6951 |
| Medroxyprogesterone | −3.4628 | 0.00076961 | 0.0030477 | 1.5083 |
| Nonadecanoic acid | −4.1575 | 7.15 × 10−5 | 0.00030394 | 1.4801 |
| Indole-3-carbinol | −2.4999 | 0.013962 | 0.040654 | 1.4384 |
| 3-Hexenedioic acid | −2.681 | 0.0083979 | 0.025981 | 1.3861 |
| 2,4-Diaminobutyric acid | −3.2445 | 0.001658 | 0.0060793 | 1.3345 |
| All-trans-retinoic acid | −4.2232 | 6.02 × 10−5 | 0.00027072 | 1.3167 |
| 3-Hydroxymethylglutaric acid | −3.2301 | 0.0016254 | 0.0060793 | 1.2938 |
| Elaidic acid | −5.0285 | 1.79 × 10−6 | 1.10 × 10−5 | 1.2811 |
| Phenylacetaldehyde | −4.1249 | 7.37 × 10−5 | 0.00030394 | 1.2643 |
| 2-Furoylglycine | −3.0855 | 0.002555 | 0.0090336 | 1.1606 |
| L-Glutamic acid | −2.4676 | 0.015037 | 0.042534 | 1.1302 |
| Pyroglutamic acid | 5.0629 | 1.71 × 10−6 | 1.10 × 10−5 | 0.77785 |
| Saccharopine | 2.7334 | 0.0072891 | 0.023278 | 0.76389 |
| Androsterone | 2.9115 | 0.0045829 | 0.015124 | 0.75177 |
| Linoleic acid | 4.5263 | 1.77 × 10−5 | 8.78 × 10−5 | 0.749 |
| 4-Aminohippuric acid | 5.4352 | 3.43 × 10−7 | 2.83 × 10−6 | 0.55491 |
| Glycyl-L-leucine | 6.6067 | 1.28 × 10−9 | 2.11 × 10−8 | 0.54098 |
| Urea | 8.2841 | 9.31 × 10−12 | 3.07 × 10−10 | 0.52527 |
| Nutriacholic acid | 6.4667 | 1.34 × 10−8 | 1.33 × 10−7 | 0.4331 |
| 3-Indolepropionic acid | 3.0702 | 0.0030686 | 0.010476 | 0.30739 |
| Acetic acid | 5.8209 | 1.40 × 10−7 | 1.26 × 10−6 | 0.2955 |
| 9-Methyluric acid | 6.6234 | 2.53 × 10−9 | 3.23 × 10−8 | 0.2591 |
| 3,4,5-Trimethoxycinnamic acid | 5.4243 | 7.53 × 10−7 | 5.73 × 10−6 | 0.20262 |
| 3-Methylxanthine | 7.1403 | 6.53 × 10−10 | 1.29 × 10−8 | 0.11684 |
| t.Stat | p.Value | FDR | Fold Change | |
|---|---|---|---|---|
| Hippuric acid | −4.997 | 7.23 × 10−6 | 3.41 × 10−5 | 168.99 |
| L-Sorbose | −6.5991 | 2.34 × 10−8 | 1.54 × 10−7 | 112.29 |
| 5-Hydroxy-L-tryptophan | −14.973 | 3.26 × 10−21 | 3.23 × 10−19 | 7.8145 |
| Homoveratric acid | −13.381 | 3.46 × 10−19 | 1.14 × 10−17 | 6.7333 |
| Indolelactic acid | −8.0659 | 8.53 × 10−11 | 9.39 × 10−10 | 5.7961 |
| 1,3-Dimethyluric acid | −7.1566 | 1.76 × 10−9 | 1.58 × 10−8 | 5.7356 |
| L-Aspartyl-L-phenylalanine | −6.2668 | 2.84 × 10−8 | 1.76 × 10−7 | 4.8287 |
| L-Glutamine | −7.0562 | 3.24 × 10−9 | 2.47 × 10−8 | 4.7812 |
| Alpha-Tocopherol | −4.1864 | 9.14 × 10−5 | 0.00037703 | 3.2137 |
| Indole-3-carbinol | −4.082 | 0.00010219 | 0.00038912 | 1.8178 |
| Glycerophosphocholine | −11.298 | 2.17 × 10−19 | 1.07 × 10−17 | 1.7227 |
| Nonadecanoic acid | −3.5226 | 0.00079411 | 0.002536 | 1.5918 |
| 3-Hexenedioic acid | −2.9064 | 0.0047055 | 0.010587 | 1.5583 |
| Medroxyprogesterone | −3.3856 | 0.0010441 | 0.0031729 | 1.5147 |
| Heptadecanoic acid | −3.8583 | 0.00025945 | 0.00095133 | 1.4928 |
| Trimethylamine | −8.3473 | 2.96 × 10−13 | 4.19 × 10−12 | 1.4759 |
| Quinaldic acid | −3.17 | 0.0024236 | 0.0063142 | 1.4694 |
| 3-Hydroxymethylglutaric acid | −2.8211 | 0.0063286 | 0.013923 | 1.4489 |
| L-Norleucine | −2.6868 | 0.0085954 | 0.017728 | 1.355 |
| L-Glutamic acid | −5.3425 | 5.02 × 10−7 | 2.62 × 10−6 | 1.2829 |
| 2-Furoylglycine | −4.6727 | 8.55 × 10−6 | 3.85 × 10−5 | 1.2707 |
| Elaidic acid | −4.7976 | 5.15 × 10−6 | 2.55 × 10−5 | 1.2702 |
| Benzoic acid | −3.2383 | 0.001675 | 0.0047047 | 1.2563 |
| o-Tyrosine | −2.9137 | 0.0045066 | 0.010587 | 1.2511 |
| m-Coumaric acid | −3.0439 | 0.0029627 | 0.0075208 | 1.2423 |
| Pantothenic acid | −3.3059 | 0.0013618 | 0.0039652 | 1.238 |
| 2,4-Diaminobutyric acid | −3.1673 | 0.0020146 | 0.0053904 | 1.2297 |
| L-Proline | −2.7595 | 0.0070984 | 0.014952 | 1.2077 |
| Allantoic acid | −3.2264 | 0.0017108 | 0.0047047 | 1.2058 |
| Pipecolic acid | −2.3124 | 0.022748 | 0.041705 | 1.2013 |
| L-Carnitine | −2.4289 | 0.017903 | 0.034085 | 1.1728 |
| Creatinine | −3.0407 | 0.0031613 | 0.0076335 | 1.1642 |
| m-Aminobenzoic acid | −2.5326 | 0.012834 | 0.025931 | 1.1603 |
| Indole | −2.894 | 0.0046922 | 0.010587 | 1.1492 |
| L-Histidine | −2.3403 | 0.021428 | 0.040027 | 1.1351 |
| 3-Methylindole | −3.587 | 0.00051176 | 0.0016888 | 1.1249 |
| Cinnamic acid | −2.4588 | 0.015506 | 0.030701 | 1.1234 |
| Phenylacetic acid | −2.756 | 0.006908 | 0.014867 | 1.0948 |
| L-Tryptophan | −2.2723 | 0.025179 | 0.044513 | 1.0784 |
| Benzamide | 3.0269 | 0.0030779 | 0.0076178 | 0.89678 |
| 5-Hydroxylysine | 2.3014 | 0.023624 | 0.042523 | 0.84997 |
| Pregnenolone sulfate | 2.4517 | 0.015934 | 0.030931 | 0.79827 |
| Sphingosine | 2.2348 | 0.027463 | 0.047699 | 0.79163 |
| Androsterone | 3.384 | 0.0010576 | 0.0031729 | 0.70978 |
| Saccharopine | 3.7427 | 0.00029123 | 0.0010297 | 0.65817 |
| Nutriacholic acid | 4.4071 | 3.36 × 10−5 | 0.00014474 | 0.59816 |
| Thyroxine | 4.0722 | 9.57 × 10−5 | 0.00037908 | 0.59602 |
| 4-Aminohippuric acid | 6.6679 | 2.82 × 10−9 | 2.33 × 10−8 | 0.51509 |
| Glycyl-L-leucine | 6.9782 | 2.37 × 10−10 | 2.34 × 10−9 | 0.49622 |
| Vitamin D3 | 5.7192 | 1.93 × 10−7 | 1.06 × 10−6 | 0.47169 |
| Urea | 10.172 | 5.50 × 10−15 | 1.09 × 10−13 | 0.41933 |
| Sphinganine | 10.711 | 1.67 × 10−18 | 4.12 × 10−17 | 0.31225 |
| Acetic acid | 6.5618 | 1.11 × 10−8 | 7.85 × 10−8 | 0.2382 |
| 3-Indolepropionic acid | 3.6696 | 0.00049507 | 0.0016888 | 0.18522 |
| 3,4,5-Trimethoxycinnamic acid | 6.3521 | 3.30 × 10−8 | 1.92 × 10−7 | 0.1117 |
| 9-Methyluric acid | 9.4247 | 1.56 × 10−13 | 2.58 × 10−12 | 0.059596 |
| 3-Methylxanthine | 8.2614 | 1.96 × 10−11 | 2.42 × 10−10 | 0.026165 |
| t.Stat | p.Value | FDR | Fold Change | |
|---|---|---|---|---|
| Threonic acid | 2.7699 | 0.0075509 | 0.043973 | 1.9592 |
| Indolelactic acid | 3.393 | 0.0010926 | 0.0090136 | 1.6339 |
| Trimethylamine | 10.182 | 1.53 × 10−17 | 7.55 × 10−16 | 1.5493 |
| 5-Hydroxy-L-tryptophan | 4.4509 | 2.08 × 10−5 | 0.00041226 | 1.5388 |
| Quinaldic acid | 2.8608 | 0.0055763 | 0.036804 | 1.434 |
| Nutriacholic acid | 4.105 | 8.81 × 10−5 | 0.001246 | 1.3812 |
| Pyroglutamic acid | 4.1094 | 7.98 × 10−5 | 0.001246 | 1.2941 |
| L-Glutamic acid | 2.9216 | 0.0042316 | 0.029923 | 1.1351 |
| Phenylacetaldehyde | −2.9494 | 0.0038999 | 0.029699 | 0.82477 |
| Urea | −6.2806 | 6.91 × 10−9 | 2.28 × 10−7 | 0.79831 |
| L-Acetylcarnitine | −2.7699 | 0.0067091 | 0.041513 | 0.79586 |
| 5-Hydroxylysine | −3.6543 | 0.00040633 | 0.0040226 | 0.7697 |
| Pregnenolone sulfate | −2.7043 | 0.0080906 | 0.044498 | 0.70729 |
| 2-Phenylbutyric acid | −3.9444 | 0.00014566 | 0.0018025 | 0.67892 |
| Deoxycholic acid glycine conjugate | −3.5347 | 0.00060533 | 0.005448 | 0.54356 |
| Vitamin D3 | −5.7136 | 1.36 × 10−7 | 3.36 × 10−6 | 0.54128 |
| Sphinganine | −12.621 | 7.37 × 10−23 | 7.29 × 10−21 | 0.27442 |
| 9-Methyluric acid | −3.7467 | 0.00037414 | 0.0040226 | 0.2172 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsoud, L.O.; Soares, N.C.; Al-Hroub, H.M.; Mousa, M.; Kasabri, V.; Bulatova, N.; Suyagh, M.; Alzoubi, K.H.; El-Huneidi, W.; Abu-Irmaileh, B.; et al. Identification of Insulin Resistance Biomarkers in Metabolic Syndrome Detected by UHPLC-ESI-QTOF-MS. Metabolites 2022, 12, 508. https://doi.org/10.3390/metabo12060508
Alsoud LO, Soares NC, Al-Hroub HM, Mousa M, Kasabri V, Bulatova N, Suyagh M, Alzoubi KH, El-Huneidi W, Abu-Irmaileh B, et al. Identification of Insulin Resistance Biomarkers in Metabolic Syndrome Detected by UHPLC-ESI-QTOF-MS. Metabolites. 2022; 12(6):508. https://doi.org/10.3390/metabo12060508
Chicago/Turabian StyleAlsoud, Leen Oyoun, Nelson C. Soares, Hamza M. Al-Hroub, Muath Mousa, Violet Kasabri, Nailya Bulatova, Maysa Suyagh, Karem H. Alzoubi, Waseem El-Huneidi, Bashaer Abu-Irmaileh, and et al. 2022. "Identification of Insulin Resistance Biomarkers in Metabolic Syndrome Detected by UHPLC-ESI-QTOF-MS" Metabolites 12, no. 6: 508. https://doi.org/10.3390/metabo12060508





