miR-129-5p Participates in Hair Follicle Growth by Targeting HOXC13 in Rabbit
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture and Transfection Assay
2.3. Quantitative Real-Time Polymerase Chain Reaction
2.4. Construction of pcDNA3.1-HOXC13
2.5. Western Blot
2.6. Dual-Luciferase Reporter Gene Assay
2.7. Apoptosis Assay
2.8. Detection of Cell Proliferation
2.9. Statistical Analysis
3. Results
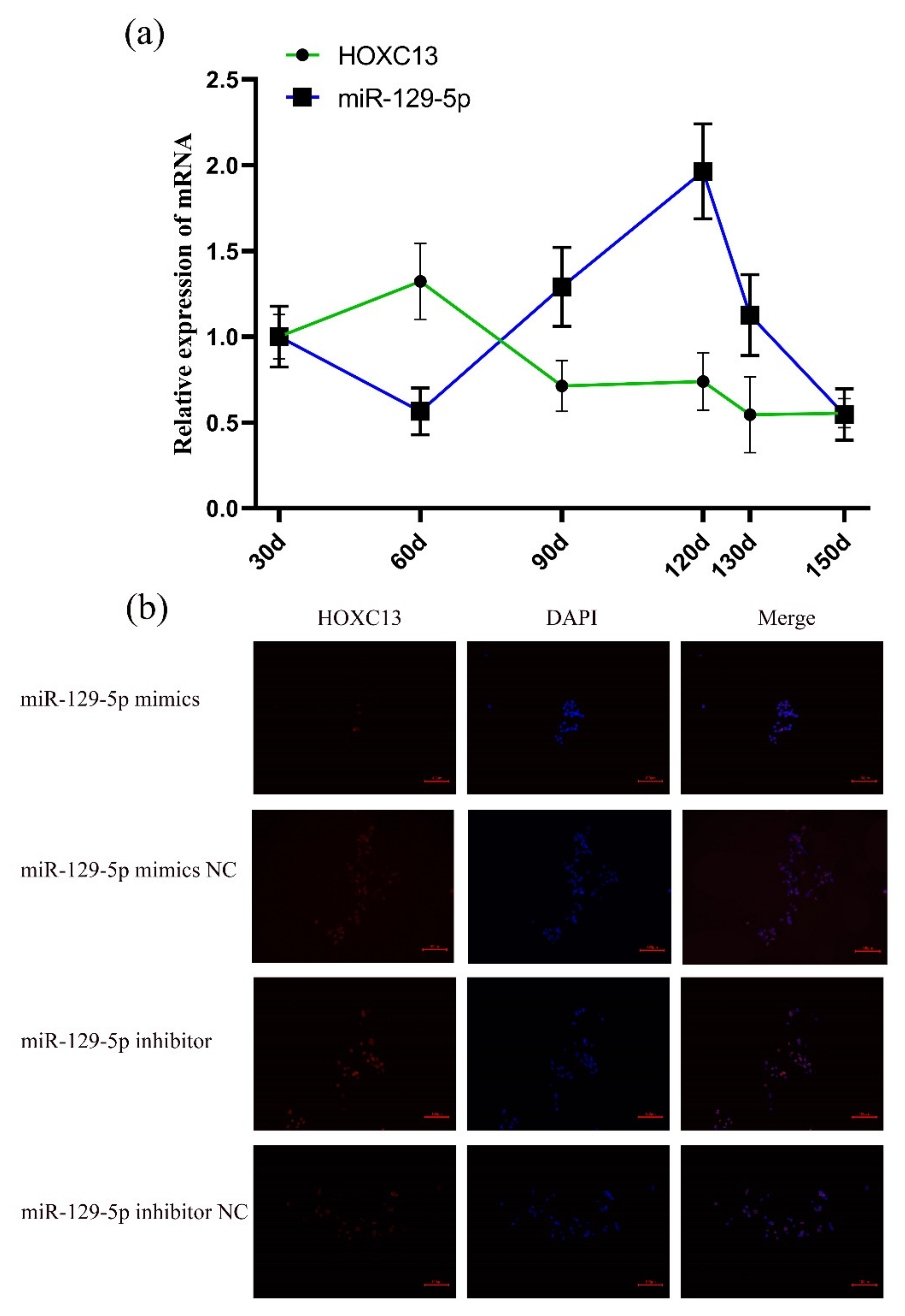
3.1. MiR-129-5p Regulates HOXC13 Expression
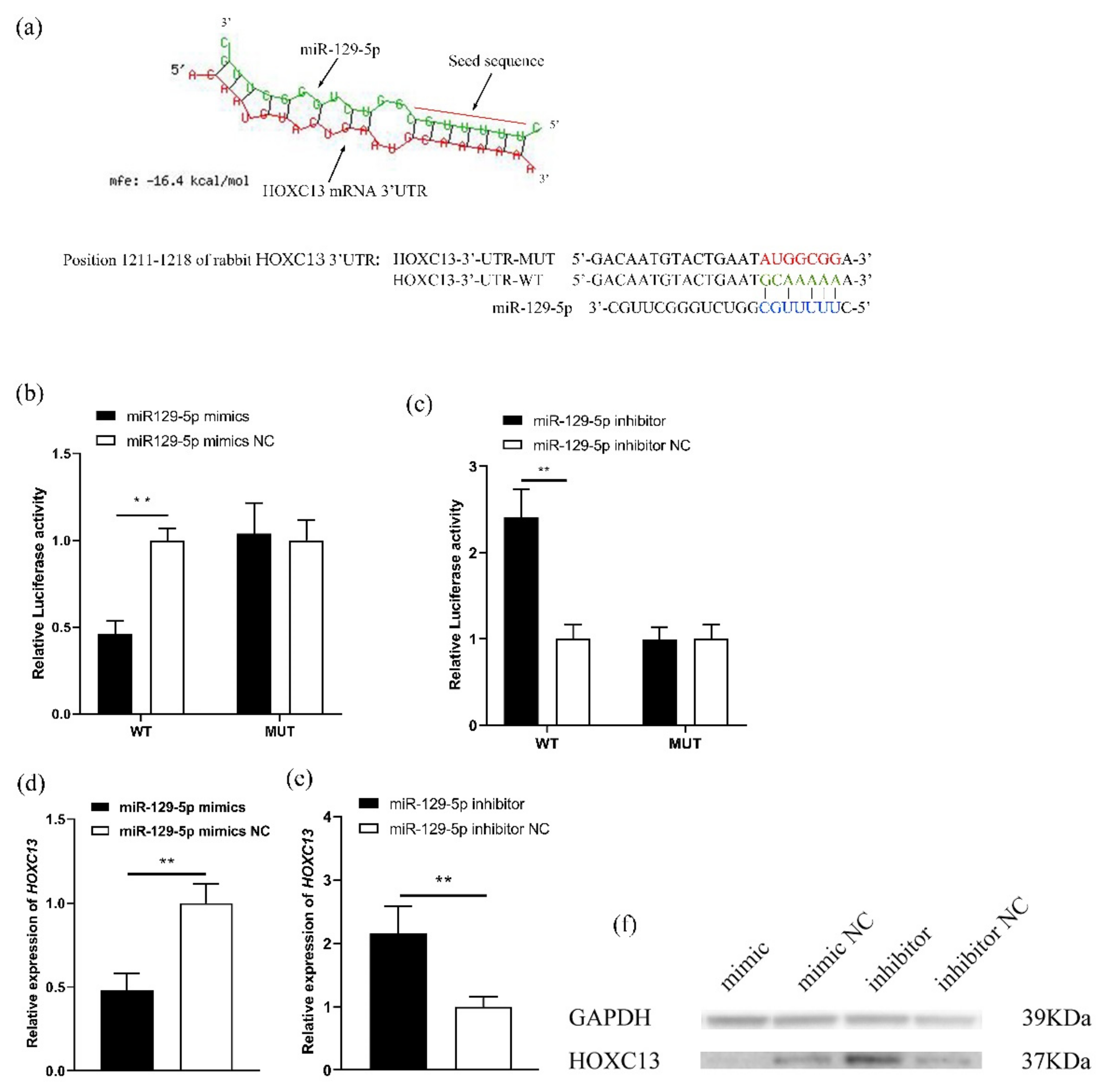
3.2. MiR-129-5p Directly Targets the Regulation of HOXC13
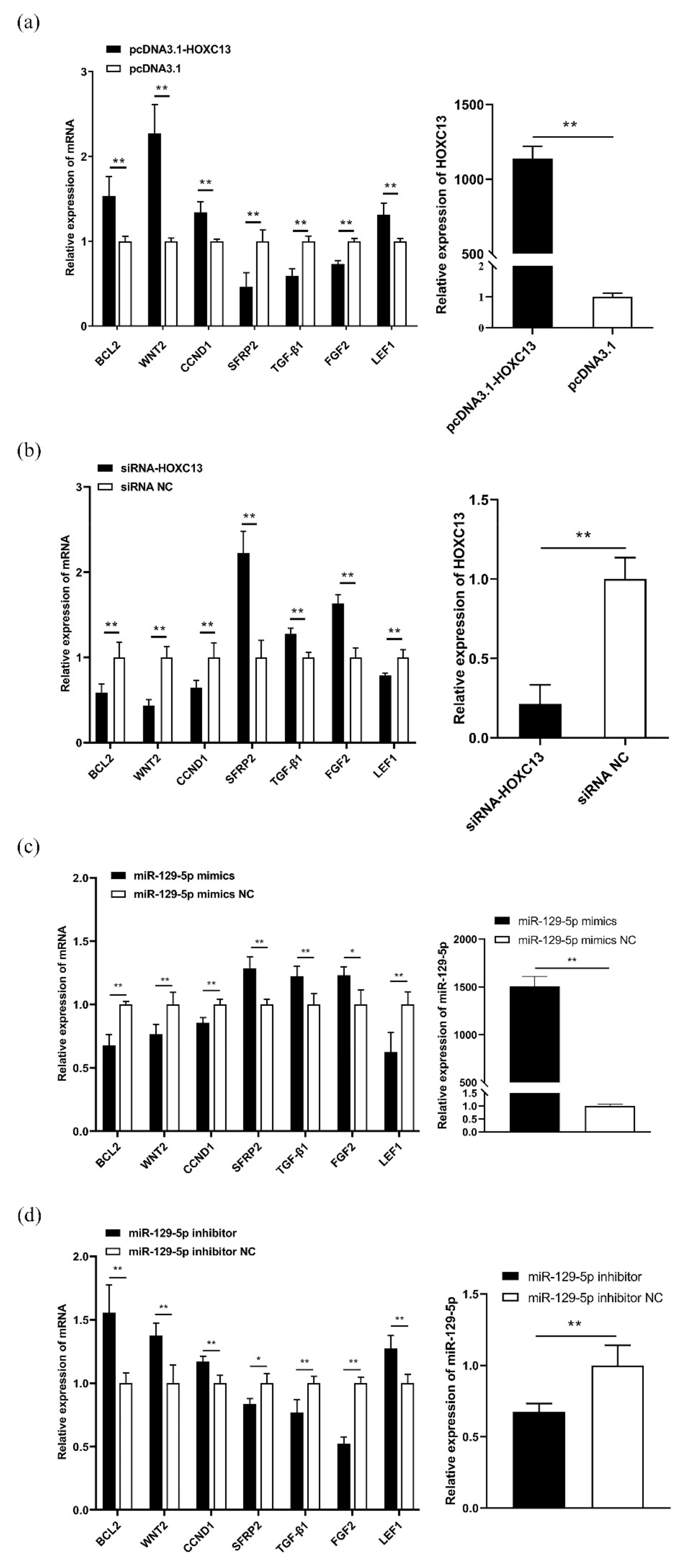
3.3. MiR-129-5p Targeted HOXC13 to Regulate the Expression of the HFDRGs
3.4. MiR-129-5p Enhances Apoptosis and Inhibits Proliferation of DPCs
3.5. HOXC13 Inhibits Apoptosis and Enhances the Proliferation of DPCs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ambros, V. The Functions of Animal MicroRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene Silencing by MicroRNAs: Contributions of Translational Repression and MRNA Decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Wang, H.; Xue, L.; Dong, Y.; Yang, L.; Fan, R.; Yu, X.; Tian, X.; Ma, S.; Smith, G.W. Coat Color Determination by MiR-137 Mediated down-Regulation of Microphthalmia-Associated Transcription Factor in a Mouse Model. RNA 2012, 18, 1679–1686. [Google Scholar] [CrossRef]
- Mardaryev, A.N.; Ahmed, M.I.; Vlahov, N.V.; Fessing, M.Y.; Gill, J.H.; Sharov, A.A.; Botchkareva, N.V. Micro-RNA-31 Controls Hair Cycle-Associated Changes in Gene Expression Programs of the Skin and Hair Follicle. FASEB J. 2010, 24, 3869–3881. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Z.; O’Loughlin, E.; Wang, L.; Fan, X.; Lai, E.C.; Yi, R. MicroRNA-205 Controls Neonatal Expansion of Skin Stem Cells by Modulating the PI(3)K Pathway. Nat. Cell Biol. 2013, 15, 1153–1163. [Google Scholar] [CrossRef]
- Yuan, S.; Li, F.; Meng, Q.; Zhao, Y.; Chen, L.; Zhang, H.; Xue, L.; Zhang, X.; Lengner, C.; Yu, Z. Post-Transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by MiR-22. PLoS Genet. 2015, 11, 1005253. [Google Scholar] [CrossRef]
- Li, J.L.; Liu, L.B.; Zhang, J.P.; Cheng, L.; Ren, L.T.; Zhao, Y.J. The expression of miR-129-5p and its target genes in the skin of goats. Anim. Biotechnol. 2020, 32, 573–579. [Google Scholar] [CrossRef]
- Greco, V.; Chen, T.; Rendl, M.; Schober, M.; Pasolli, H.A.; Stokes, N.; dela Cruz-Racelis, J.; Fuchs, E. A Two-Step Mechanism for Stem Cell Activation during Hair Regeneration. Cell Stem Cell 2009, 4, 464. [Google Scholar] [CrossRef][Green Version]
- Rendl, M.; Lewis, L.; Fuchs, E. Molecular Dissection of Mesenchymal-Epithelial Interactions in the Hair Follicle. PLoS Biol. 2005, 3, 1910–1924. [Google Scholar] [CrossRef]
- Andl, T.; Reddy, S.T.; Gaddapara, T.; Millar, S.E. WNT Signals Are Required for the Initiation of Hair Follicle Development. Dev. Cell. 2002, 2, 643–653. [Google Scholar] [CrossRef]
- Niimori, D.; Kawano, R.; Felemban, A.; Niimori-Kita, K.; Tanaka, H.; Ihn, H.; Ohta, K. Tsukushi Controls the Hair Cycle by Regulating TGF-Β1 Signaling. Dev. Biol. 2012, 372, 81–87. [Google Scholar] [CrossRef] [PubMed]
- St-Jacques, B.; Dassule, H.R.; Karavanova, I.; Botchkarev, V.A.; Li, J.; Danielian, P.S.; Mcmahon, J.A.; Lewis, P.M.; Paus, R.; Mcmahon, A.P. Sonic Hedgehog Signaling Is Essential for Hair Development. Curr. Biol. 1998, 8, 1058–1069. [Google Scholar] [CrossRef]
- Stenn, K.S.; Paus, R. Controls of Hair Follicle Cycling. Physiol. Rev. 2001, 81, 449–494. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Chen, Q.; Shi, L.; Lee, M.; Giehl, K.A.; Tang, Z.; Wang, H.; Zhang, J.; Yin, J.; Wu, L.; et al. Loss-of-Function Mutations in HOXC13 Cause Pure Hair and Nail Ectodermal Dysplasia. Am. J. Hum. Genet. 2012, 91, 906–911. [Google Scholar] [CrossRef]
- Farooq, M.; Kurban, M.; Fujimoto, A.; Fujikawa, H.; Abbas, O.; Nemer, G.; Saliba, J.; Sleiman, R.; Tofaili, M.; Kibbi, A.G.; et al. A Homozygous Frameshift Mutation in the HOXC13 Gene Underlies Pure Hair and Nail Ectodermal Dysplasia in a Syrian Family. Hum. Mutat. 2013, 34, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Jiang-hong, W.; Yan-jun, Z.; Jia-xin, Z.; Zi-li, C.; Jin-quan, L.; Zu-wei, Y.; Wen-guang, Z. Hoxc13/β-Catenin Correlation with Hair Follicle Activity in Cashmere Goat. J. Integr. Agric. 2012, 2012, 11. [Google Scholar]
- Qiu, W.; Lei, M.; Tang, H.; Yan, H.; Wen, X.; Zhang, W.; Tan, R.; Wang, D.; Wu, J. Hoxc13 Is a Crucial Regulator of Murine Hair Cycle. Cell Tissue Res. 2016, 364, 149–158. [Google Scholar] [CrossRef]
- Kahata, K.; Dadras, M.S.; Moustakas, A. TGF-β Family Signaling in Epithelial Differentiation and Epithelial-Mesenchymal Transition. Cold Spring Harb. Perspect. Biol. 2018, 10, 022194. [Google Scholar] [CrossRef]
- Potter, C.S.; Kern, M.J.; Baybo, M.A.; Pruett, N.D.; Godwin, A.R.; Sundberg, J.P.; Awgulewitsch, A. Dysregulated Expression of Sterol O-Acyltransferase 1 (Soat1) in the Hair Shaft of Hoxc13 Null Mice. Exp. Mol. Pathol. 2015, 99, 441–444. [Google Scholar] [CrossRef]
- Wang, X.; Xu, H.R.; Li, T.; Qu, L.; Zhao, Z.D.; Zhang, Z.Y. Expression Analysis of KAP9.2 and Hoxc13 Genes during Different Cashmere Growth Stages by QRT-PCR Method. Mol. Biol. Rep. 2014, 41, 5665–5668. [Google Scholar] [CrossRef]
- Wang, S.; Luo, Z.; Zhang, Y.; Yuan, D.; Ge, W.; Wang, X. The Inconsistent Regulation of HOXC13 on Different Keratins and the Regulation Mechanism on HOXC13 in Cashmere Goat (Capra Hircus). BMC Genom. 2018, 19, 630. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Zhao, B.H.; Zhang, C.; Zhang, X.Y.; Dai, Y.Y.; Hu, S.S.; Yang, N.S.; Chen, Y.; Wu, X.S. Establishment and Functional Characterization of Immortalized Rabbit Dermal Papilla Cell Lines. 2021; preprint. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Liu, Z.; Xiao, H.; Li, H.; Zhao, Y.; Lai, S.; Yu, X.; Cai, T.; Du, C.; Zhang, W.; Li, J. Identification of Conserved and Novel MicroRNAs in Cashmere Goat Skin by Deep Sequencing. PLoS ONE 2012, 7, e50001. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive Analysis of MicroRNA Expression Patterns in Hepatocellular Carcinoma and Non-Tumorous Tissues. Oncogene 2006, 25, 2537–2545. [Google Scholar] [CrossRef]
- Polytarchou, C.; Hommes, D.W.; Palumbo, T.; Hatziapostolou, M.; Koutsioumpa, M.; Koukos, G.; Van Der Meulen-De Jong, A.E.; Oikonomopoulos, A.; Van Deen, W.K.; Vorvis, C.; et al. MicroRNA214 Is Associated With Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology 2015, 149, 981–992.e11. [Google Scholar] [CrossRef]
- Ma, H.; Pan, J.S.; Jin, L.X.; Wu, J.; Ren, Y.D.; Chen, P.; Xiao, C.; Han, J. MicroRNA-17~92 Inhibits Colorectal Cancer Progression by Targeting Angiogenesis. Cancer Lett. 2016, 376, 293–302. [Google Scholar] [CrossRef]
- Zhang, P.; Li, J.; Song, Y.; Wang, X. MiR-129-5p Inhibits Proliferation and Invasion of Chondrosarcoma Cells by Regulating SOX4/Wnt/β-Catenin Signaling Pathway. Cell. Physiol. Biochem. 2017, 42, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Jin, B.; Huang, L.; Zhou, W. MiR-129-5p Inhibits Glioma Cell Progression in Vitro and in Vivo by Targeting TGIF2. J. Cell. Mol. Med. 2018, 22, 2357–2367. [Google Scholar] [CrossRef]
- Wu, J.H.; Yan, Z.W.; Zhang, W.G.; Li, J.Q. Hoxc13 and the Development of Hair Follicle. Yi Chuan = Hereditas 2010, 32, 656–662. [Google Scholar] [PubMed]
- Jave-Suarez, L.F.; Winter, H.; Langbein, L.; Rogers, M.A.; Schweizer, J. HOXC13 Is Involved in the Regulation of Human Hair Keratin Gene Expression. J. Biol. Chem. 2002, 277, 3718–3726. [Google Scholar] [CrossRef]
- Pruett, N.D.; Tkatchenko, T.V.; Jave-Suarez, L.; Jacobs, D.F.; Potter, C.S.; Tkatchenko, A.V.; Schweizer, J.; Awgulewitsch, A. Krtap16, Characterization of a New Hair Keratin-Associated Protein (KAP) Gene Complex on Mouse Chromosome 16 and Evidence for Regulation by Hoxc13. J. Biol. Chem. 2004, 279, 51524–51533. [Google Scholar] [CrossRef] [PubMed]
- Ahlawat, S.; Arora, R.; Sharma, R.; Sharma, U.; Kaur, M.; Kumar, A.; Singh, K.V.; Singh, M.K.; Vijh, R.K. Skin Transcriptome Profiling of Changthangi Goats Highlights the Relevance of Genes Involved in Pashmina Production. Sci. Rep. 2020, 10, 6050. [Google Scholar] [CrossRef]
- Yao, Y.; Luo, J.; Sun, Q.; Xu, T.; Sun, S.; Chen, M.; Lin, X.; Qian, Q.; Zhang, Y.; Cao, L.; et al. Original Article HOXC13 Promotes Proliferation of Lung Adenocarcinoma via Modulation of CCND1 and CCNE1. Am. J. Cancer Res. 2017, 7, 1820. [Google Scholar]
- Kasiri, S.; Ansari, K.I.; Hussain, I.; Bhan, A.; Mandal, S.S. Antisense Oligonucleotide Mediated Knockdown of HOXC13 Affects Cell Growth and Induces Apoptosis in Tumor Cells and over Expression of HOXC13 Induces 3D-Colony Formation. RSC Adv. 2013, 3, 3260–3269. [Google Scholar] [CrossRef]
- Zhao, B.H.; Chen, Y.; Yang, N.S.; Chen, Q.R.; Bao, Z.Y.; Liu, M.; Hu, S.S.; Li, J.L.; Wu, X.S. miR-218-5p regulates skin and hair follicle development through Wnt/β-catenin signaling pathway by targeting SFRP2. J. Cell. Physiol. 2019, 234, 20329–20341. [Google Scholar] [CrossRef]
- Fuchs, E.; Raghavan, S. Getting under the Skin of Epidermal Morphogenesis. Nat. Rev. Genet. 2002, 3, 199–209. [Google Scholar]
- Liu, Y.; Zhou, Q.; Zhou, D.; Huang, C.; Meng, X.; Li, J. Secreted Frizzled-Related Protein 2-Mediated Cancer Events: Friend or Foe? Pharmacol. Rep. 2017, 69, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Byrne, C.; Jacobs, J.; Fuchs, E. Lymphoid Enhancer Factor 1 Directs Hair Folhcle Patterning and Epithelial Cell Fate. Genes Dev. 1995, 9, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Foitzik, K.; Lindner, G.; Mueller-Roever, S.; Maurer, M.; Botchkareva, N.; Botchkarev, V.; Handjiski, B.; Metz, M.; Hibino, T.; Soma, T.; et al. Control of Murine Hair Follicle Regression (Catagen) by TGF-β1 in Vivo. FASEB J. 2000, 14, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.K.; Yoon, S.K. Expression of Sfrp2 Is Increased in Catagen of Hair Follicles and Inhibits Keratinocyte Proliferation. Ann. Dermatol. 2014, 26, 79–87. [Google Scholar] [CrossRef]


| Genes | Primers Sequence(5′→3′) |
|---|---|
| GAPDH | F: TCACCATCTTCCAGGAGCGA R: CACAATGCCGAAGTGGTCGT |
| BCL2 | F: ACATCGCCCTGTGGATGACTG R: CGAGGGTGATGCAAGCTCCTAT |
| WNT2 | F: AGCCATCCAGGTCGTCATGAACCAG R: TGCACACACGACCTGCTGTACCC |
| CCND1 | F: GAACGCTACCTTCCCCAGTGCTC R: CCTCACAGACCTCCAGCATCCAG |
| SFRP2 | F: CCAGCCCGACTTCTCCTACAAGC R: TCCAGCACCTCTTTCATGGTCT |
| TGF-β1 | F: CAGGTCCTTGCGGAAGTCAA R: CTGGAACGGGCTCAACATCTA |
| FGF2 | F: GTGTGTGCAAACCGTTACCTT R: TCGTTTCAGTGCCACATACCAG |
| LEF1 | F: CATCTCGGGTGGATTCAGG R: ATGAGGGATGCCAGTTGTG |
| HOXC13 | F: TGGAGAAAGAGTACGCAGCC R: TCTTCTCTTTGACTCGGCGG |
| miR-129-5p | CTTTTTGCGGTCTGGGCTTGC |
| U6 | CAAGGATGACACGCAAATTCG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, F.; Zhao, B.; Hu, S.; Bai, S.; Jin, R.; Zhang, C.; Chen, Y.; Wu, X. miR-129-5p Participates in Hair Follicle Growth by Targeting HOXC13 in Rabbit. Genes 2022, 13, 679. https://doi.org/10.3390/genes13040679
Yao F, Zhao B, Hu S, Bai S, Jin R, Zhang C, Chen Y, Wu X. miR-129-5p Participates in Hair Follicle Growth by Targeting HOXC13 in Rabbit. Genes. 2022; 13(4):679. https://doi.org/10.3390/genes13040679
Chicago/Turabian StyleYao, Fan, Bohao Zhao, Shuaishuai Hu, Shaocheng Bai, Rongshuai Jin, Chen Zhang, Yang Chen, and Xinsheng Wu. 2022. "miR-129-5p Participates in Hair Follicle Growth by Targeting HOXC13 in Rabbit" Genes 13, no. 4: 679. https://doi.org/10.3390/genes13040679
APA StyleYao, F., Zhao, B., Hu, S., Bai, S., Jin, R., Zhang, C., Chen, Y., & Wu, X. (2022). miR-129-5p Participates in Hair Follicle Growth by Targeting HOXC13 in Rabbit. Genes, 13(4), 679. https://doi.org/10.3390/genes13040679






