Exploring the Effect of Methyl Jasmonate on the Expression of microRNAs Involved in Biosynthesis of Active Compounds of Rosemary Cell Suspension Cultures through RNA-Sequencing
Abstract
:1. Introduction
2. Results
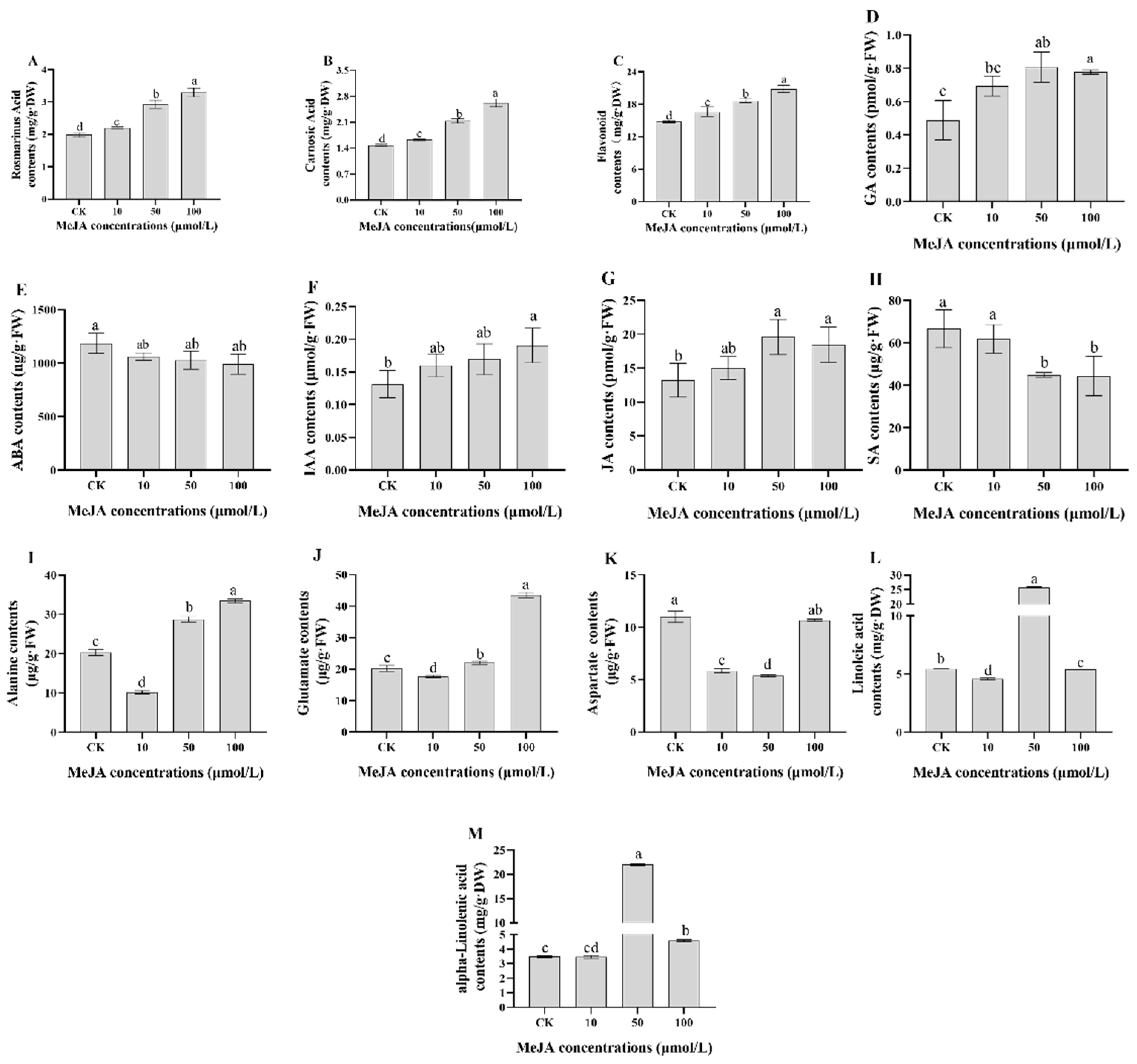
2.1. Physiological and Biochemical Indices of Rosemary Suspension Cells under Different Concentrations of MeJA
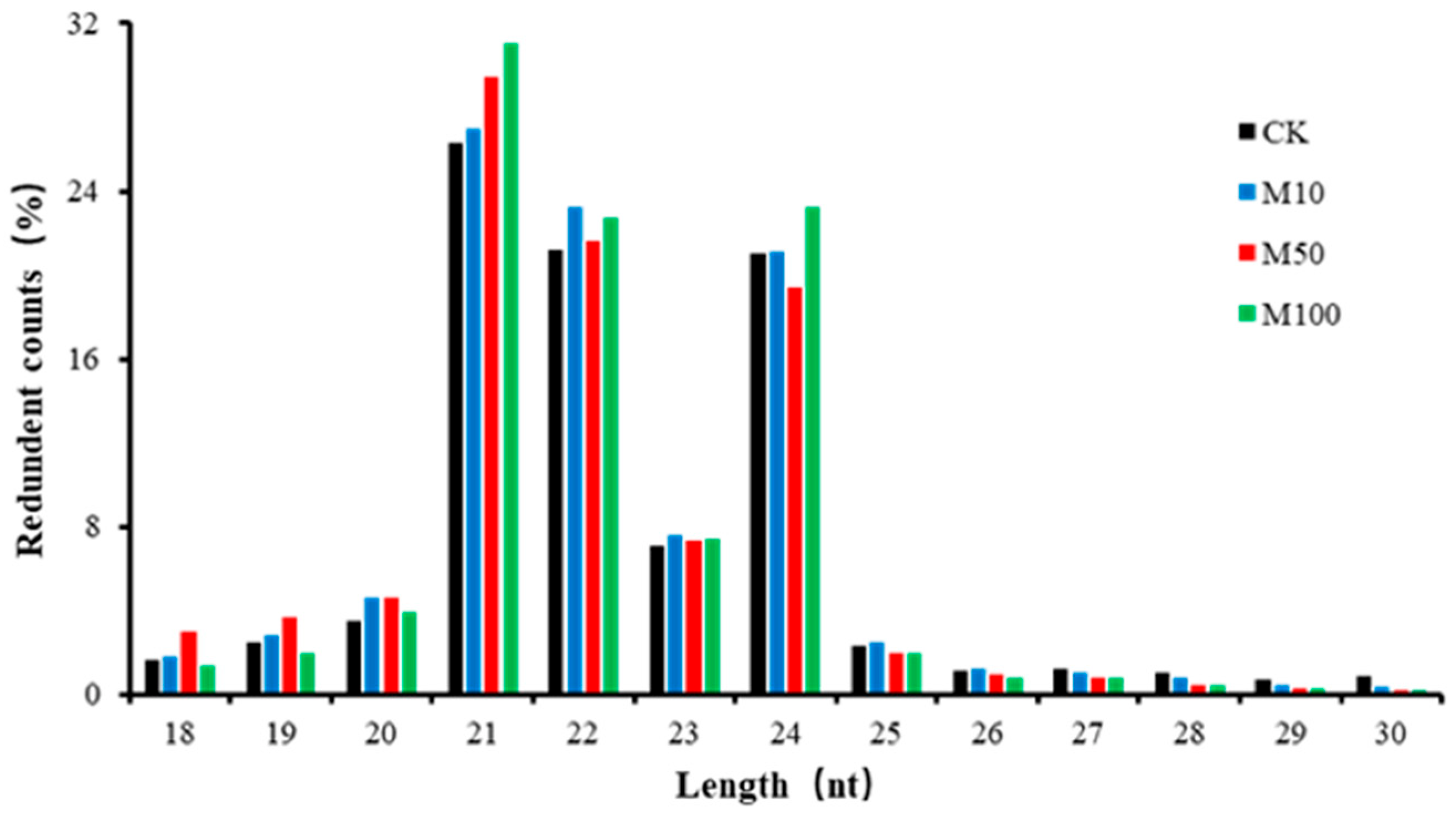
2.2. Small RNA Analysis of Rosemary Suspension Cells
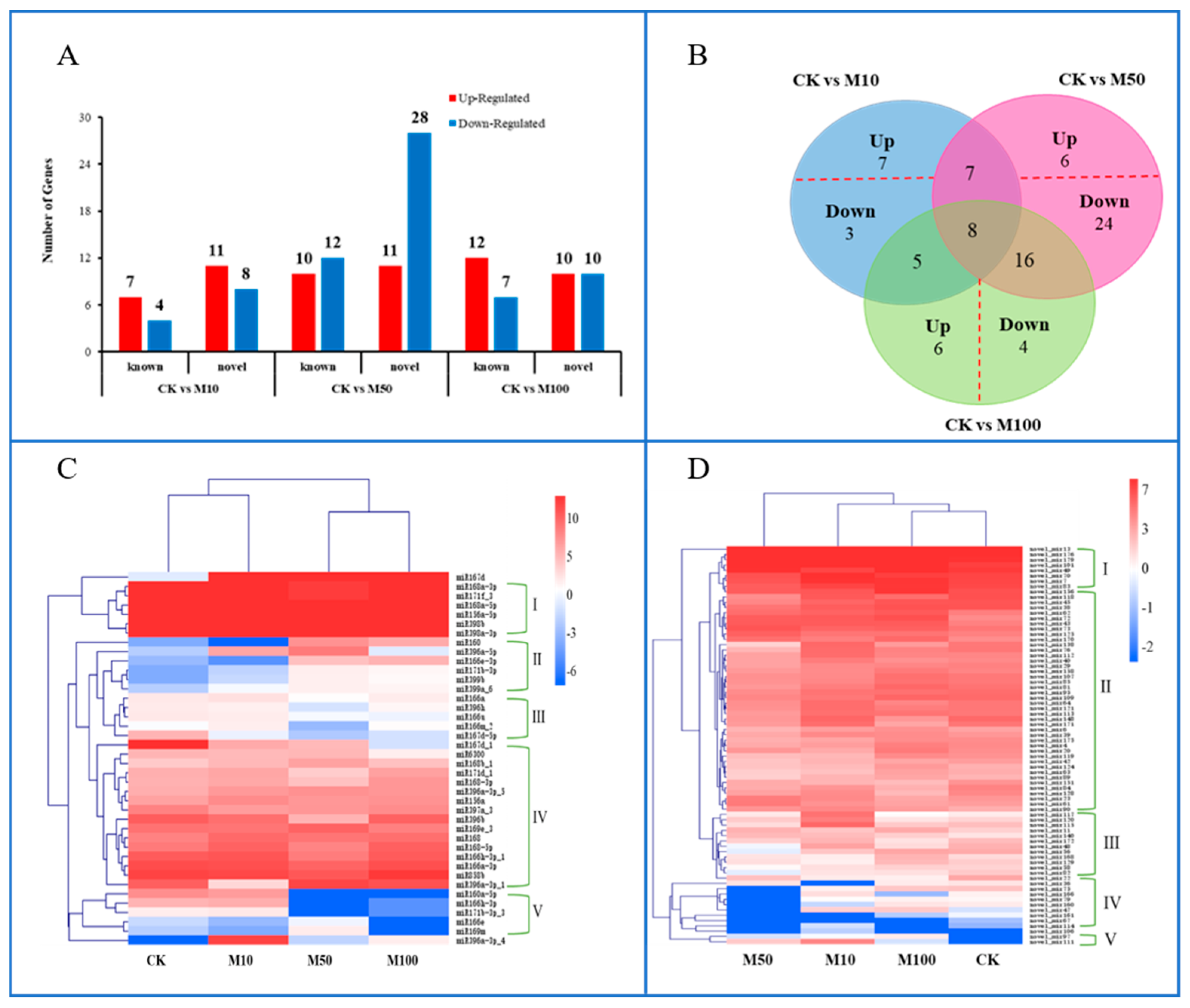
2.3. Identification of Known and Novel miRNAs of Rosemary Suspension Cells under Different Concentrations of MeJA
2.4. Differential Expression Analysis of Differentially Expressed miRNAs in Rosemary Suspension Cells under Different Concentrations of MeJA
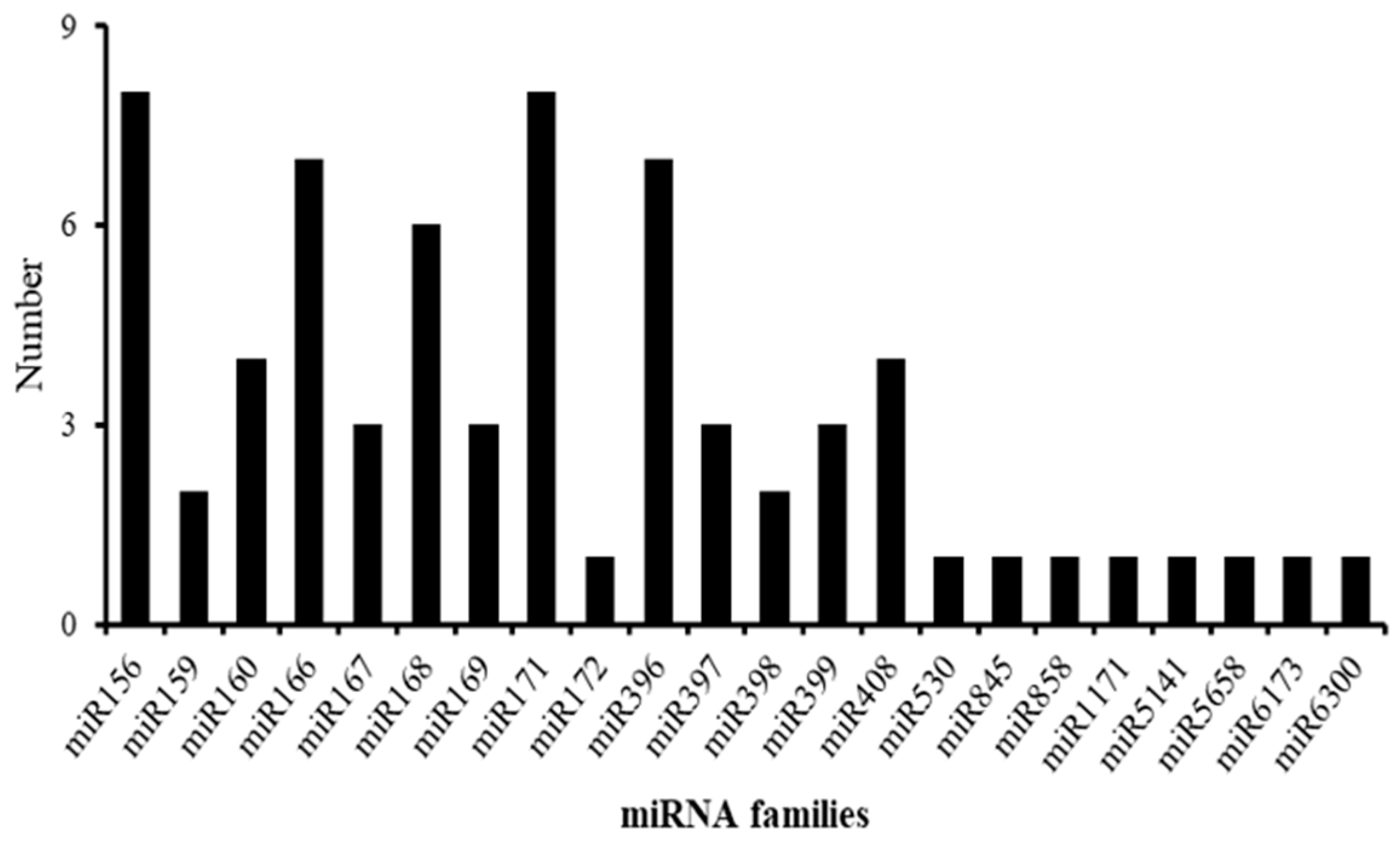
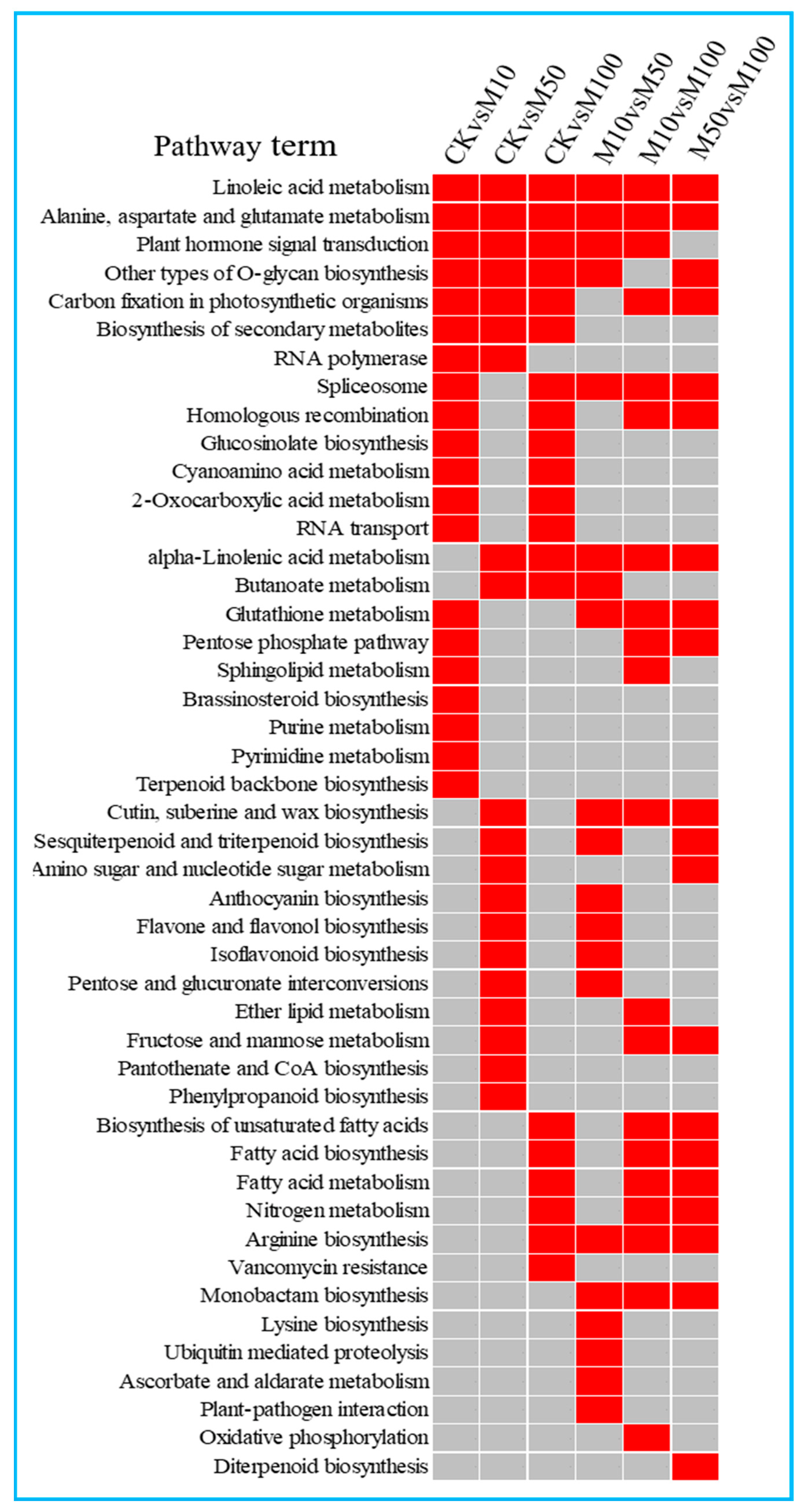
2.5. Functional Classification of Differentially Expressed miRNAs in Rosemary Suspension Cells under Different Concentrations of MeJA
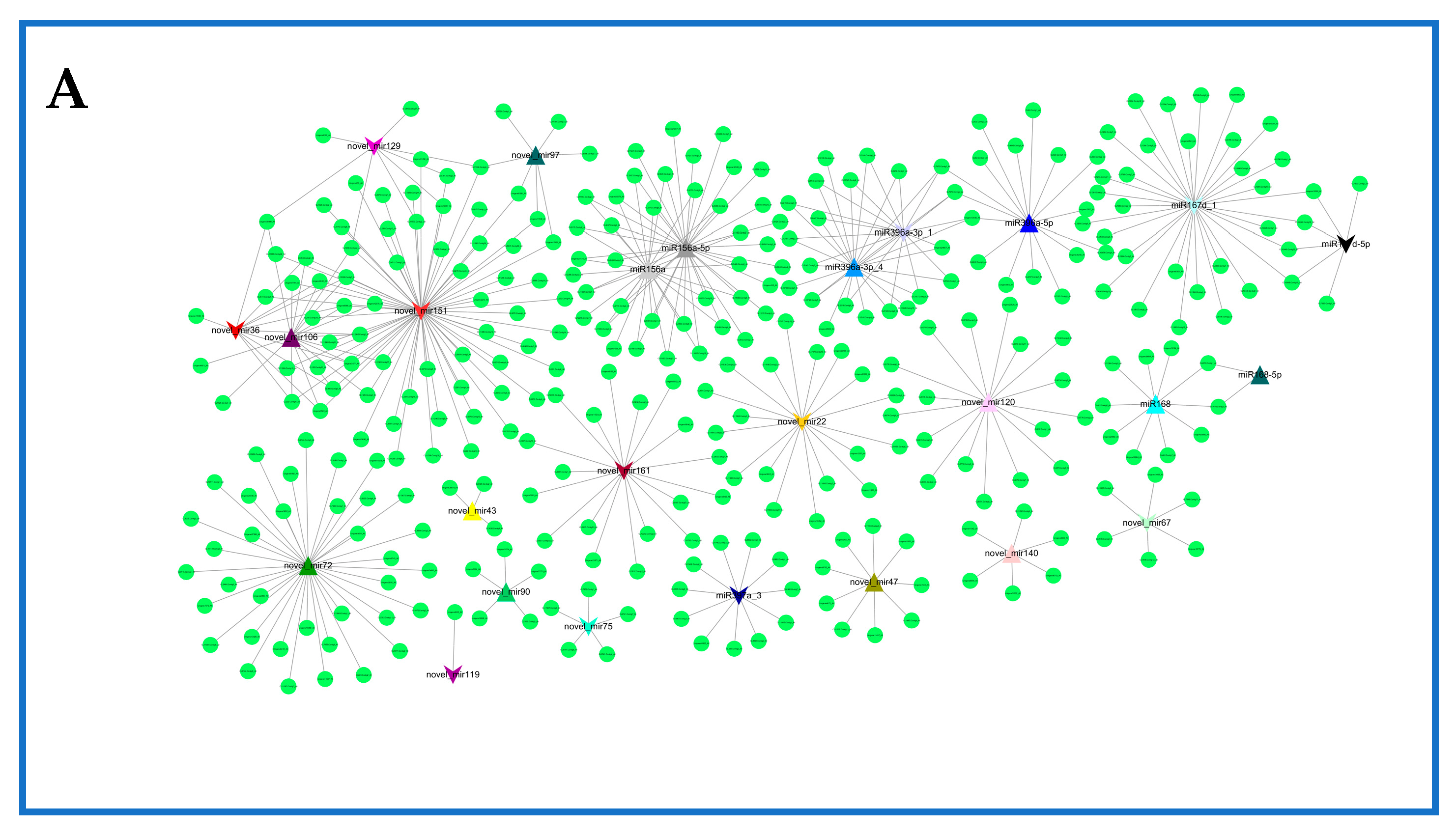
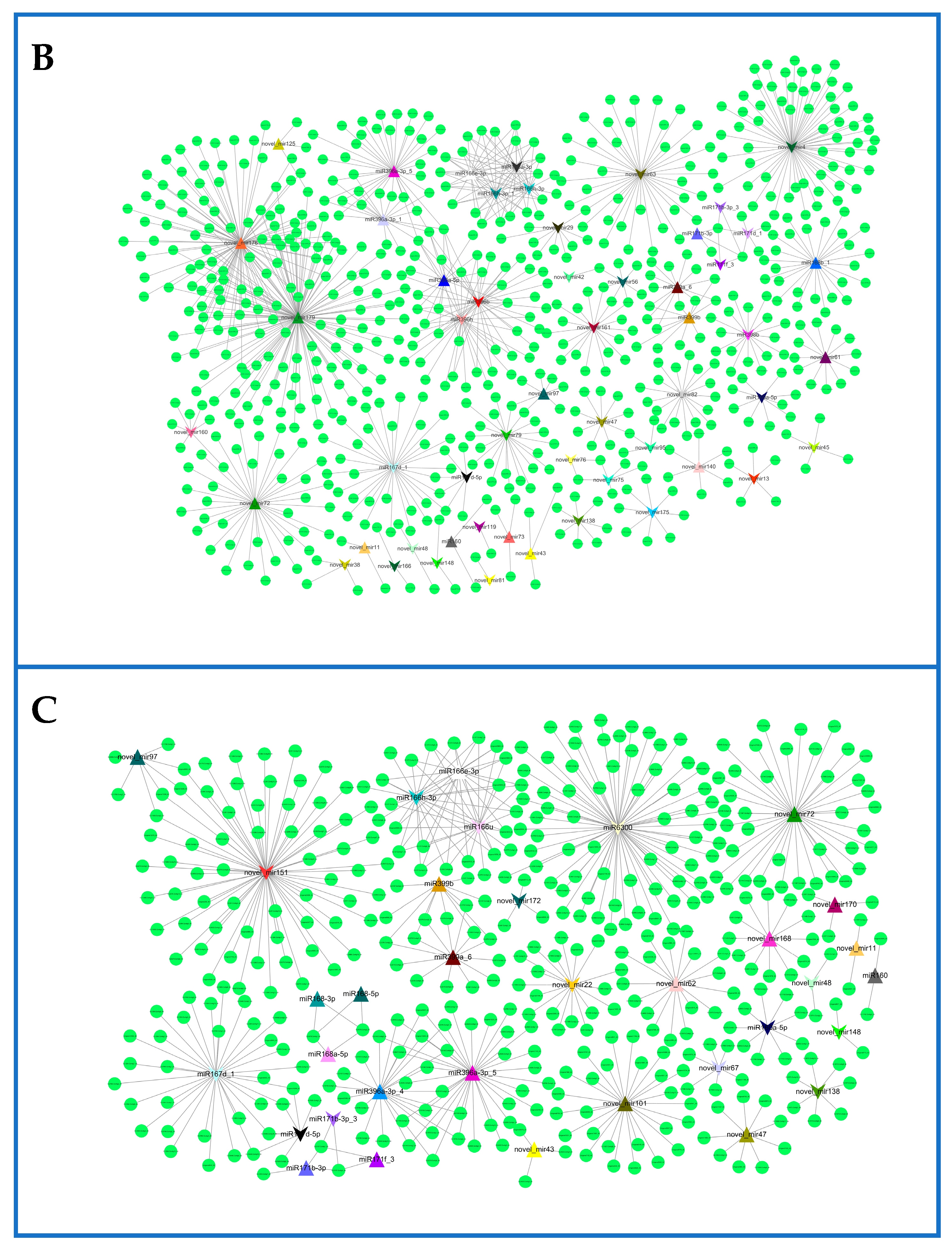
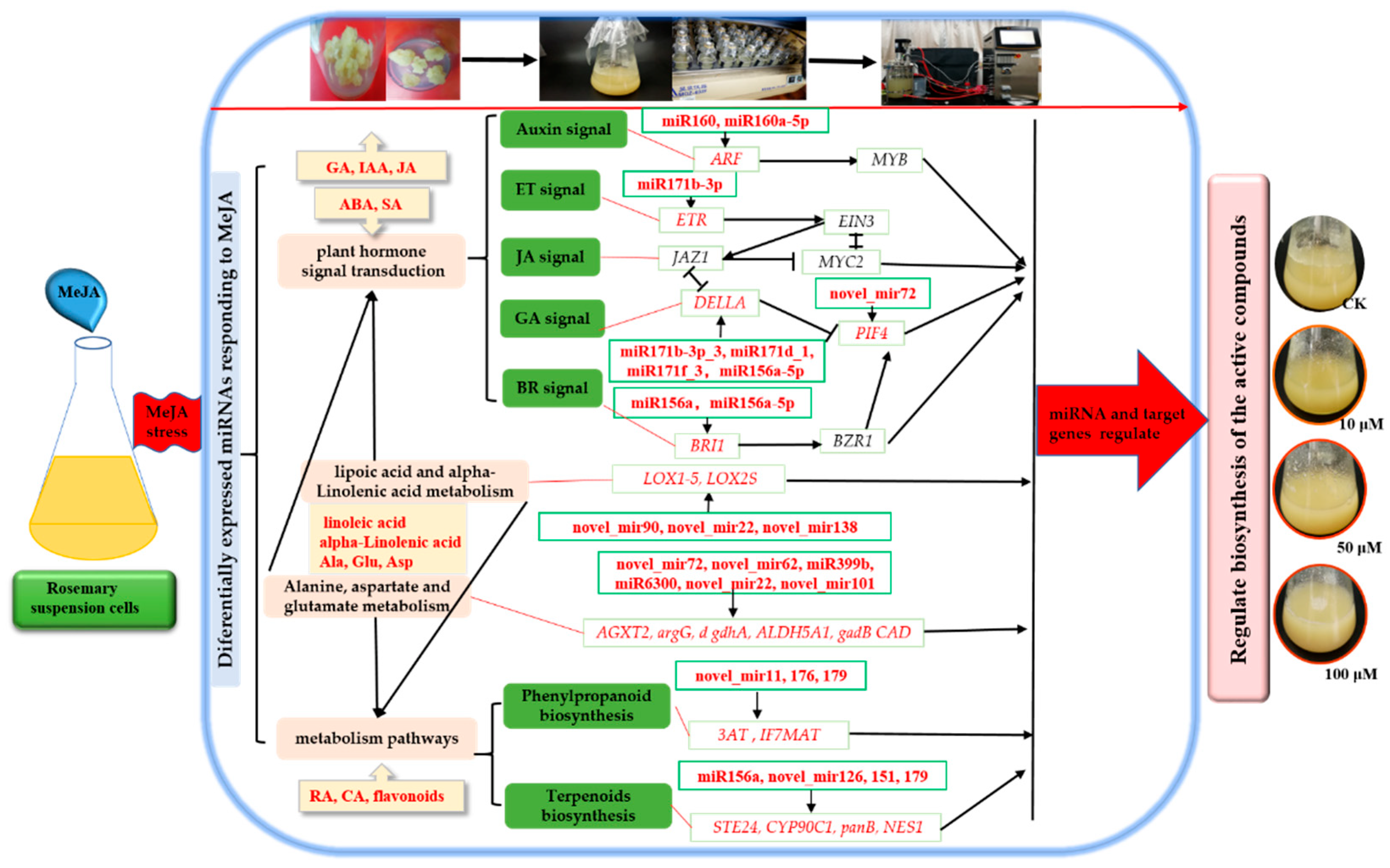
2.6. Network of Differentially Expressed miRNAs and Their Targets in Rosemary Suspension Cells under Different Concentrations of MeJA
2.7. miRNAs and Target Genes Related to the Plant Hormone Signal Transduction in Rosemary Suspension Cells under Different Concentrations of MeJA
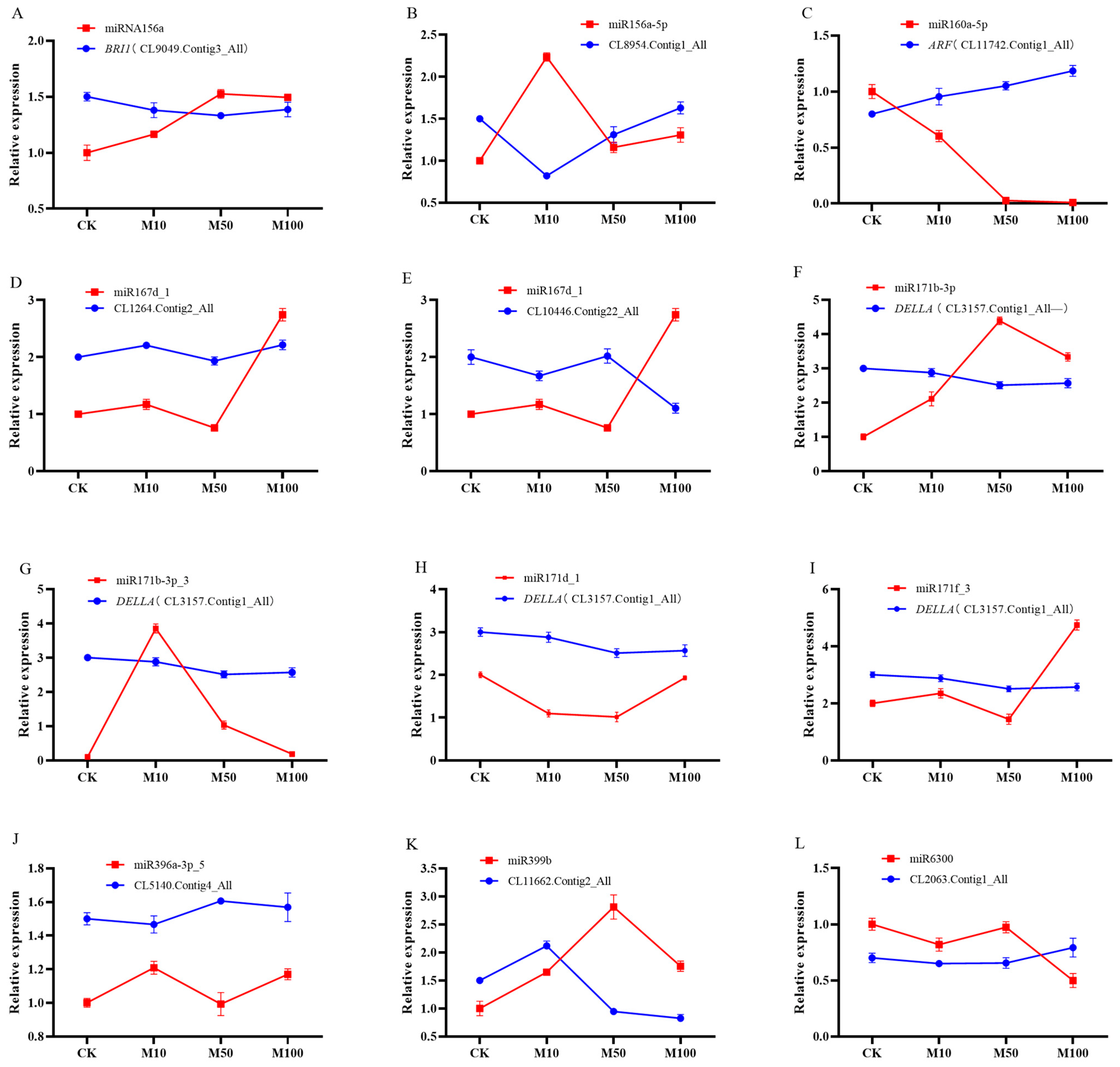
2.8. Identification of Differentially Expressed miRNAs and Their Targets in Rosemary Suspension Cells under Different Concentrations of MeJA by Quantitative qRT-PCR
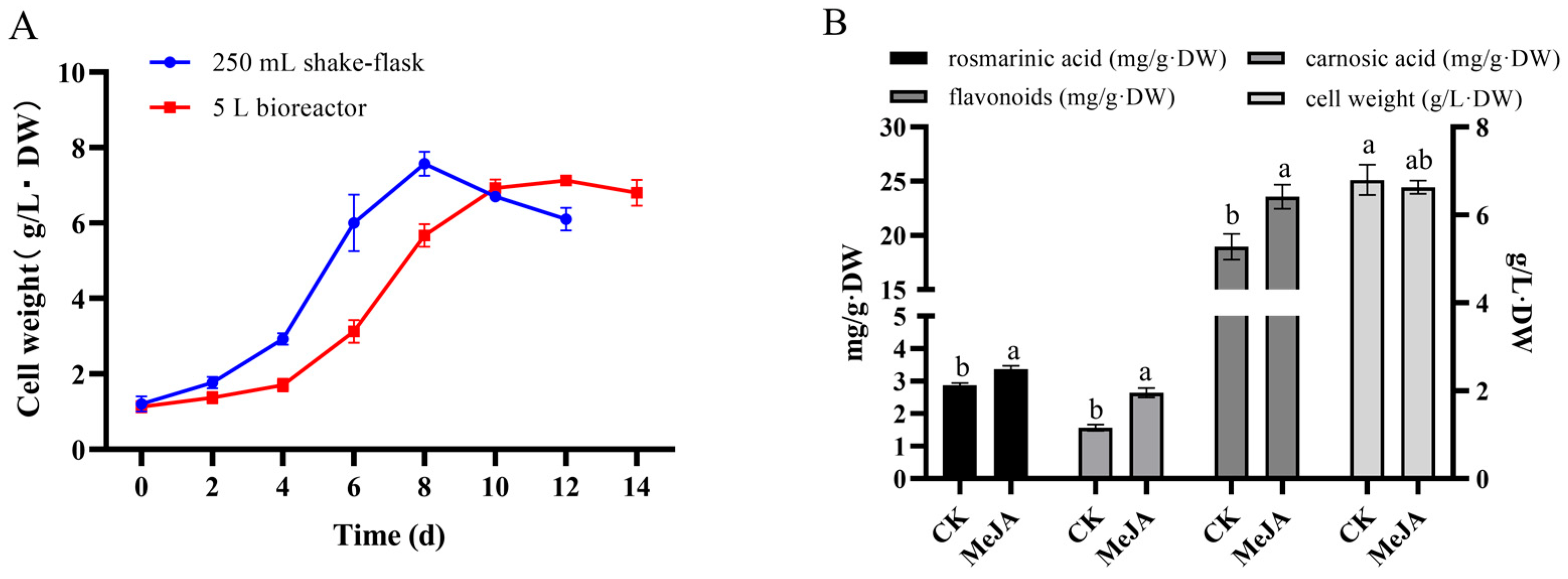
2.9. Analysis of Rosemary Cells Suspension Culture in 5 L Stirred Bioreactor
3. Discussion
3.1. miRNAs of Rosemary Suspension Cells Responding to MeJA
3.2. miRNAs Target Metabolic Pathway Genes to Regulat the Synthesis of Active Compounds in Rosemary Suspension Cells under Different Concentrations of MeJA
3.3. Differentially Expressed miRNAs Involved in Plant Hormone Signal Transduction Played a Key Role in Rosemary Suspension Cells Responding to MeJA
3.4. Amplification Culture of Rosemary Suspension Cells in 5 L Stirred Bioreactor
4. Materials and Methods
4.1. Plant Material and MeJA Treatments
4.2. Small RNA and RNA-Seq Library Construction
4.3. General Analysis of Small RNA and Prediction of miRNA Targets
4.4. Identification of Differentially Expressed miRNAs
4.5. qRT-PCR Validation of miRNAs and Their Targets
4.6. Determination of Flavonoid, Rosmarinic Acid, and Carnosic Acid
4.7. Determination of Physiological and Biochemical Indexes
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MeJA | methyl jasmonate |
| IAA | indoleacetic acid |
| ABA | abscisic acid |
| GA | gibberellin |
| SA | salicylic acid |
| JA | jasmonate |
| BR | brassinosteroids |
| ET | ethylene |
| 2,4-D | 2,4-dichlorophenoxyacetic acid |
| qRT-PCR | quantitative real-time PCR |
| DEMS | differentially expressed miRNAs |
| RA | rosmarinic acid |
| CA | carnosic acid |
| rpm | revolutions per minute |
| ccm | cubic centimeters per minute |
References
- Holmes, P. Rosemary oil: The wisdom of the heart. Int. J. Aromather. 1999, 9, 62–66. [Google Scholar] [CrossRef]
- Neto, N.J.G.; Luz, I.D.S.; Tavares, A.G.; Honório, V.G.; Magnani, M.; De Souza, E.L. Rosmarinus officinalis L. Essential Oil and Its Majority Compound 1,8-Cineole at Sublethal Amounts Induce No Direct and Cross Protection in Staphylococcus aureus ATCC. Foodborne Pathog. Dis. 2012, 9, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Bomfim, N.D.S.; Nakassugi, L.P.; Oliveira, J.F.; Kohiyama, C.Y.; Mossini, S.A.; Grespan, R.; Nerilo, S.B.; Mallmann, C.; Abreu Filho, B.A.; Machinski, M., Jr. Antifungal activity and inhibition of fumonisin production by Rosmarinus officinalis L. essential oil in Fusarium verticillioides (Sacc.) Nirenberg. Food Chem. 2015, 166, 330–336. [Google Scholar] [CrossRef] [Green Version]
- Badreddine, B.S.; Olfa, E.; Samir, D.; Hnia, C.; Lahbib, B.J.M. Chemical composition of Rosmarinus and Lavandula essential oils and their insecticidal effects on Orgyia trigotephras (Lepidoptera, Lymantriidae). Asian Pac. J. Trop. Med. 2015, 8, 98–103. [Google Scholar] [CrossRef] [Green Version]
- Kiran, C.R.; Sasidharan, I.; Kumar, D.R.S.; Sundaresan, A. Influence of natural and synthetic antioxidants on the degradation of Soybean oil at frying temperature. J. Food Sci. Technol. 2015, 52, 5370–5375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, L.S.; Menis, M.E.C.; Jorge, N. Effect of rosemary (Rosmarinus officinalis) extracts on the oxidative stability and sensory acceptability of soybean oil. J. Sci. Food Agric. 2015, 95, 2021–2027. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, N.; Fan, Q.; Lin, M.; Zhang, C.; Fan, G.; Zhai, X.; Zhang, F.; Chen, Z.; Yao, J. Protective efficacy of carnosic acid against hydrogen peroxide induced oxidative injury in HepG2 cells through the SIRT1 pathway. Can. J. Physiol. Pharmacol. 2015, 93, 625–631. [Google Scholar] [CrossRef]
- Sedighi, R.; Zhao, Y.; Yerke, A.; Sang, S. Preventive and protective properties of rosemary (Rosmarinus officinalis L.) in obesity and diabetes mellitus of metabolic disorders: A brief review. Curr. Opin. Food Sci. 2015, 2, 58–70. [Google Scholar] [CrossRef]
- Yan, M.; Li, G.; Petiwala, S.M.; Householter, E.; Johnson, J.J. Standardized rosemary (Rosmarinus officinalis L.) extract induces Nrf2/sestrin-2 pathway in colon cancer cells. J. Funct. Foods 2015, 13, 137–147. [Google Scholar] [CrossRef]
- Bonfill, M.; Mangas, S.; Moyano, E.; Cusido, R.M.; Palazon, J. Production of centellosides and phytosterols in cell suspension cultures of Centella asiatica. Plant Cell Tissue Organ Cult. 2011, 104, 61–67. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Mai, V.C.; Drzewiecka, K.; Jeleń, H.; Narożna, D.; Rucińska-Sobkowiak, R.; Kęsy, J.; Floryszak-Wieczorek, J.; Gabryś, B.; Morkunas, I. Differential induction of Pisum sativum defense signaling molecules in response to pea aphid infestation. Plant Sci. 2014, 221, 1–12. [Google Scholar] [CrossRef] [PubMed]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; He, Y.; Li, G.; Liu, Y.; Gao, W.; Huang, L. Effects of Combined Elicitors on Tanshinone Metabolic Profiling and SmCPS Expression in Salvia miltiorrhiza Hairy Root Cultures. Molecules 2013, 18, 7473–7485. [Google Scholar] [CrossRef]
- Zhang, S.; Yan, Y.; Wang, B.; Liang, Z.; Liu, Y.; Liu, F.; Qi, Z. Selective responses of enzymes in the two parallel pathways of rosmarinic acid biosynthetic pathway to elicitors in Salvia miltiorrhiza hairy root cultures. J. Biosci. Bioeng. 2014, 117, 645–651. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.F.; Deng, K.; He, X.Y.; Zhan, R.T.; Tang, L. Methyl Jasmonate affects metabolism and gene transcription of volatile terpenoids from Amomum villosum Lour. World Sci. Technol. Mod. Tradit. Chin. Med. Mater. Med. 2014, 16, 1528–1536. [Google Scholar] [CrossRef]
- Zhang, W.J.; Cao, X.Y.; Jiang, J.H. Triterpene biosynthesis in Euphorbia pekinensis induced by methyl jasmonate. Guihaia 2015, 35, 590–596. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, B.; Lu, B.; Kai, G.; Wang, Z.; Xia, Y.; Ding, R.; Zhang, H.; Sun, X.; Chen, W. Tropane alkaloids production in transgenic Hyoscyamus niger hairy root cultures over-expressing putrescinen-methyltransferase is methyl jasmonate-dependent. Planta 2007, 225, 887–896. [Google Scholar] [CrossRef]
- Zhang, C.H.; Mei, X.G.; Liu, L.; Yu, L.J. Enhanced paclitaxel production induced by the combination of elicitors in cell sus-pension cultures of Taxus chinensis. Biotechnol. Lett. 2000, 22, 1561–1564. [Google Scholar] [CrossRef]
- Rischer, H.; Orešič, M.; Seppänen-Laakso, T.; Katajamaa, M.; Lammertyn, F.; Diaz, W.A.; Montagu, M.V.; Inzé, D.; Oksman-Caldentey, K.M.; Goossens, A. Gene-to-metabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc. Natl. Acad. Sci. USA 2006, 103, 5614–5619. [Google Scholar] [CrossRef] [Green Version]
- Sutter, V.; Vanderhaeghen, R.; Tilleman, S.; Lammertyn, F.; Vanhoutte, I.; Karimi, M.; Inzé, D.; Goossens, A.; Hilson, P. Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J. 2005, 44, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Gális, I.; Šimek, P.; Narisawa, T.; Sasaki, M.; Horiguchi, T.; Fukuda, H.; Matsuoka, K. A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J. 2010, 46, 573–592. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Zhang, Y.; Hua, W.P.; Wu, Y.C.; Jin, X.X.; Song, S.-H.; Wang, Z.-Z. Combination of transcriptomic and metabolomic analyses reveals a JAZ repressor in the jasmonate signaling pathway of Salvia miltiorrhiza. Sci. Rep. 2015, 5, 14048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hedhili, S.; Montiel, G.; Zhang, Y.; Chatel, G.; Pré, M.; Gantet, P.; Memelink, J. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 2011, 67, 61–71. [Google Scholar] [CrossRef]
- De Boer, K.; Tilleman, S.; Pauwels, L.; Bossche, R.V.; De Sutter, V.; Vanderhaeghen, R.; Hilson, P.; Hamill, J.D.; Goossens, A. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 2011, 66, 1053–1065. [Google Scholar] [CrossRef]
- Shoji, T.; Kajikawa, M.; Hashimoto, T. Clustered Transcription Factor Genes Regulate Nicotine Biosynthesis in Tobacco. Plant Cell 2010, 22, 3390–3409. [Google Scholar] [CrossRef] [Green Version]
- Caretto, S.; Quarta, A.; Durante, M.; Nisi, R.; De Paolis, A.; Blando, F.; Mita, G. Methyl jasmonate and miconazole differently affect arteminisin production and gene expression in Artemisia annua suspension cultures. Plant Biol. 2015, 13, 51–58. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Q. MicroRNA-Based Biotechnology for Plant Improvement. J. Cell. Physiol. 2015, 230, 1–15. [Google Scholar] [CrossRef]
- Sunkar, R.; Li, Y.-F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Pareek, M.; Yogindran, S.; Mukherjee, S.K.; Rajam, M.V. Plant MicroRNAs: Biogenesis, Functions and Applications. In Plant Biology and Biotechnology; Springer: New Delhi, India, 2015; pp. 639–661. [Google Scholar] [CrossRef]
- Budak, H.; Kabtara, M.; Bulut, R.; Akpinar, B.A. Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 2015, 235, 1–13. [Google Scholar] [CrossRef]
- Shen, E.M.; Singh, S.K.; Ghosh, J.S.; Patra, B.; Paul, P.; Yuan, L.; Pattanaik, S. The miRNAome of Catharanthus roseus: Identification, expression analysis, and potential roles of microRNAs in regulation of terpenoid indole alkaloid biosynthesis. Sci. Rep. 2017, 7, 43027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, D.; Pan, X.; Wilson, I.W.; Li, F.; Liu, M.; Teng, W.; Zhang, B. High throughput sequencing technology reveals that the taxoid elicitor methyl jasmonate regulates microRNA expression in Chinese yew (Taxus chinensis). Gene 2009, 436, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Jiang, Y.M.; Wang, N.; Xia, B.; Jiang, Y.L.; Li, X.D.; Zhang, Z.Z.; Li, Y.K.; Wang, R. Identification and differential regulation of microRNAs in response to methyl jasmonate treatment in Lycoris aurea by deep sequencing. BMC Genom. 2016, 17, 789–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulcheski, F.R.; De Oliveira, L.F.; Molina, L.G.; Almerão, M.P.; Rodrigues, F.A.; Marcolino, J.; Barbosa, J.F.; Stolf-Moreira, R.; Nepomuceno, A.L.; Marcelino-Guimarães, F.C.; et al. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 2011, 12, 307. [Google Scholar] [CrossRef] [Green Version]
- Pantaleo, V.; Szittya, G.; Moxon, S.; Miozzi, L.; Moulton, V.; Dalmay, T.; Burgyan, J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010, 62, 960–976. [Google Scholar] [CrossRef]
- Yao, Y.; Guo, G.; Ni, Z.; Sunkar, R.; Du, J.; Zhu, J.K.; Sun, Q.X. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol. 2007, 8, R96. [Google Scholar] [CrossRef] [Green Version]
- Thiebaut, F.; Grativol, C.; Tanurdzic, M.; Carnavalebottino, M.; Vieira, T.; Motta, M.R.; Cristian, R.; Renato, V.; Moutinho, C.S.; Silva, H.A.; et al. Differential sRNA Regulation in Leaves and Roots of Sugarcane under Water Depletion. PLoS ONE 2014, 9, e93822. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.W.; Luo, L.X.; Liang, C.Q.; Jiang, N.; Liu, P.F.; Li, J.Q. High-Throughput Sequencing Identifies Novel and Conserved Cucumber (Cucumis sativus L.) microRNAs in Response to Cucumber Green Mottle Mosaic Virus Infection. PLoS ONE 2015, 10, e0129002. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.L.; Lai, Z.X. Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant Physiol. Biochem. 2013, 66, 20–25. [Google Scholar] [CrossRef]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [Green Version]
- Lakhotia, N.; Joshi, G.; Bhardwaj, A.R.; Katiyar-Agarwal, S.; Agarwal, M.; Jagannath, A.; Goel, S.; Kumar, A. Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC Plant Biol. 2014, 14, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moxon, S.; Jing, R.; Szittya, G.; Schwach, F.; Pilcher, R.L.R.; Moulton, V.; Dalmay, T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008, 18, 1602–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.-L.; Wen, Z.; Yang, K.; Wen, X.-P. Conserved miR396b-GRF Regulation Is Involved in Abiotic Stress Responses in Pitaya (Hylocereus polyrhizus). Int. J. Mol. Sci. 2019, 20, 2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoratti, L.; Karppinen, K.; Escobar, A.L.; Häggman, H.; Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 2014, 5, 534. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Tian, Z.; Zhang, Q.; Zhang, Y.; Wang, M.; Fang, X.A.; Shi, W.J.; Cai, X. MicroRNAome Profile of Euphorbia kansui in Response to Methyl Jasmonate. Int. J. Mol. Sci. 2019, 20, 1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.; Cheng, C.; Lin, Y.; Zhu, Q.; Lin, J.; Lai, Z. Combined small RNA and degradome sequencing reveals complex microRNA regulation of catechin biosynthesis in tea (Camellia sinensis). PLoS ONE 2017, 12, e0171173. [Google Scholar] [CrossRef]
- Zhou, F.M.; Bai, Z.C.; a Lu, S.F. MicroRNA in medicinal plants. Chin. Tradit Herb. Drugs. 2013, 44, 232–237. [Google Scholar]
- Kushalappa, A.C.; Gunnaiah, R. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 2013, 18, 522–531. [Google Scholar] [CrossRef]
- Andreou, A.; Brodhun, F.; Feussner, I. Biosynthesis of oxylipins in non-mammals. Prog. Lipid Res. 2009, 48, 148–170. [Google Scholar] [CrossRef]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.-R.; Chen, G.Q.; Kim, H.U. Current progress towards the metabolic engineering of plant seed oil for hydroxy fatty acids production. Plant Cell Rep. 2015, 34, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 Gene in Plants: Occurrence, Regulation, and Role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Chen, B.; Win, A.N.; Fu, C.; Lian, J.; Liu, X.; Wang, R.; Zhang, X.; Chai, Y. Omega-3 fatty acid desaturase gene family from two ω-3 sources, Salvia hispanica and Perilla frutescens: Cloning, characterization and expression. PLoS ONE 2018, 13, e0191432. [Google Scholar] [CrossRef]
- Wang, H.Z.; Xu, Q.J.; Yan, H.F. Summary of regulation of anthocyanin synthesis by light, sugar and hormone. Acta Agric. Jiangxi 2016, 28, 35–41. [Google Scholar] [CrossRef]
- Oudin, A.; Mahroug, S.; Courdavault, V.; Hervouet, N.; Zelwer, C.; Rodriguez-Concepcion, M.; St-Pierre, B.; Burlat, V. Spatial distribution and hormonal regulation of gene products from methyl erythritol phosphate and monoterpene-secoiridoid pathways in Catharanthus roseus. Plant Mol. Biol. 2007, 65, 13–30. [Google Scholar] [CrossRef]
- Kerchev, P.I.; Fenton, B.; Foyer, C.; Hancock, R.D. Plant responses to insect herbivory: Interactions between photosynthesis, reactive oxygen species and hormonal signalling pathways. Plant Cell Environ. 2012, 35, 441–453. [Google Scholar] [CrossRef]
- Fang, R.; Zhao, H.; Liao, Y.; Tang, C.; Wu, F.; Zhu, Y.; Pang, Y.; Lu, G.; Wang, X.; Yang, R.; et al. Dual Regulating Effects of Ethylene on the Formation of Plant Secondary Metabolites. Chin. Bull. Bot. 2014, 49, 626. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; MacLean, D.; Jikumaru, Y.; Hill, L.; Yamaguchi, S.; Kamiya, Y.; Jones, J.D. The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 2011, 67, 218–231. [Google Scholar] [CrossRef]
- Khusnutdinov, E.; Sukhareva, A.; Panfilova, M.; Mikhaylova, E. Anthocyanin Biosynthesis Genes as Model Genes for Genome Editing in Plants. Int. J. Mol. Sci. 2021, 22, 8752. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, W.; Wang, X.; Yang, R.; Wu, Z.; Wang, H.; Wang, L.; Hu, Z.; Guo, S.; Zhang, H.; et al. MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic. Res. 2020, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, L.; Bari, R.; Achard, P.; Lisón, P.; Nemri, A.; Harberd, N.P.; Jones, J.D.G. DELLAs Control Plant Immune Responses by Modulating the Balance of Jasmonic Acid and Salicylic Acid Signaling. Curr. Biol. 2008, 18, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Deng, X.W. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, L.; Li, Y.; Hu, Q.; Zhu, L.; Gao, W.; Wu, Y.; Ding, Y.; Liu, S.; Yang, X.; Zhang, X. Sugar and Auxin Signaling Pathways Respond to High-Temperature Stress during Anther Development as Revealed by Transcript Profiling Analysis in Cotton. Plant Physiol. 2014, 164, 1293–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephenson, P.G.; Fankhauser, C.; Terry, M.J. PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 2009, 106, 7654–7659. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.; Man, C.; Xie, Y.; Yan, J.; Chu, J.; Huang, J. A Crucial Role of GA-Regulated Flavonol Biosynthesis in Root Growth of Arabidopsis. Mol. Plant 2019, 12, 521–537. [Google Scholar] [CrossRef] [Green Version]
- Li, W.R.; Xing, B.C.; Mao, R.J.; Bai, Z.Q.; Yang, D.F.; Xu, J.Y.; Liang, Z.S. SmGRAS3 negatively responds to GA signaling while promotes tanshinones biosynthesis in Salvia miltiorrhiza. Ind. Crop. Prod. 2020, 144, 112004. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 Modulates Antagonism between Jasmonate and Ethylene Signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, B.W.; Liu, J.; Zhong, P.; Guo, X.R. Effects of Exogenous Ethylene on Physiology and Alkaloid Accumulations in Catharanthus roseus. Bull. Bot. Res. 2018, 38, 284–2918. [Google Scholar]
- Zhang, H. Effect on Phosphorus Metabolism Disorders of latex by Ethylene Stimulus on Hevea brasiliensis; Hainan University: Hainan, China, 2017. [Google Scholar]
- Pan, Y.T. The Role of mi R171b in the Regulation of Growth and Development in Tomato; Zhejiang University: Zhejiang, China, 2020. [Google Scholar]
- Sun, Y.; Fan, X.-Y.; Cao, D.-M.; Tang, W.; He, K.; Zhu, J.-Y.; He, J.-X.; Bai, M.-Y.; Zhu, S.; Oh, E.; et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.-W.; Wang, Z.-Y. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu. Rev. Plant Biol. 2010, 61, 681–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Z.; Han, C.; Yuan, L.; Zhang, K.; Huang, H.; Ren, C. Brassinosteroid Enhances Jasmonate-Induced Anthocyanin Accu-mulation in Arabidopsis Seedlings. J. Integr. Plant Biol. 2011, 53, 632–640. [Google Scholar] [CrossRef]
- Sun, Z.; He, Y.; Li, J.; Wang, X.; Chen, J. Genome-Wide Characterization of Rice Black Streaked Dwarf Virus-Responsive MicroRNAs in Rice Leaves and Roots by Small RNA and Degradome Sequencing. Plant Cell Physiol. 2014, 56, 688–699. [Google Scholar] [CrossRef] [Green Version]
- Hettenhausen, C.; Schuman, M.C.; Wu, J. MAPK signaling: A key element in plant defense response to insects. Insect Sci. 2015, 22, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Arnaud, D.; Hwang, I. A Sophisticated Network of Signaling Pathways Regulates Stomatal Defenses to Bacterial Pathogens. Mol. Plant 2015, 8, 566–581. [Google Scholar] [CrossRef] [Green Version]
- He, H.J.; Huang, N.; Cao, R.J.; Meng, L.J. Structures, Antioxidation Mechanism, and Antioxidation Test of the Common Natural Antioxidants in Plants. Biophysics 2015, 3, 25–47. [Google Scholar] [CrossRef]
- Li, H.; Chen, X.; Wang, Y.; Yao, D.; Lin, Y.; Lai, Z. Exploration of the effect of blue light on microRNAs involved in the accumulation of functional metabolites of longan embryonic calli through RNA-sequencing. J. Sci. Food Agric. 2019, 99, 1533–1547. [Google Scholar] [CrossRef]
- Banton, M.C.; Tunnacliffe, A. MAPK phosphorylation is implicated in the adaptation to desiccation stress in nematodes. J. Exp. Biol. 2012, 215, 4288–4298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, S.; Cusido, R.M.; Mirjalili, M.H.; Moyano, E.; Palazon, J.; Bonfill, M. Production of the anticancer drug taxol in Taxus baccata suspension cultures: A review. Process Biochem. 2011, 46, 23–34. [Google Scholar] [CrossRef]
- Eibl, R.; Eibl, D. Design of bioreactors suitable for plant cell and tissue cultures. Phytochem. Rev. 2008, 7, 593–598. [Google Scholar] [CrossRef]
- Bentebibel, S.; Moyano, E.; Palazón, J.; Cusidó, R.; Bonfill, M.; Eibl, R.; Piñol, M. Effects of immobilization by entrapment in alginate and scale-up on paclitaxel and baccatin III production in cell suspension cultures of Taxus baccata. Biotechnol. Bioeng. 2005, 89, 647–655. [Google Scholar] [CrossRef]
- Martin, E.K.; Vishal, G.; Susan, C.R. Pharmaceutically active natural product synthesis and supply via plant cell culture technology. Mol. Pharm. 2008, 5, 243–256. [Google Scholar]
- Li, Y.L.; Meng, T.T.; Wang, M.M.; Li, X.X.; Li, R.R.; Guo, X.Q. Amplification Culture of Glycyrrhiza uralensis Cell in Stirring Bioreactor. Plant Physiol. J. 2015, 51, 302–306. [Google Scholar]
- Tang, C.; Xie, Y.; Guo, M.; Yan, W. AASRA: An anchor alignment-based small RNA annotation pipeline. bioRxiv 2017, 105, 267–277. [Google Scholar] [CrossRef]
- Evers, M.; Huttner, M.; Dueck, A.; Meister, G.; Engelmann, J.C. miRA: Adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinform. 2015, 16, 370. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.-J.; Ma, Y.-K.; Chen, T.; Wang, M.; Wang, X.-J. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012, 40, W22–W28. [Google Scholar] [CrossRef]
- Bonnet, E.; He, Y.; Billiau, K.; Van De Peer, Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 2010, 26, 1566–1568. [Google Scholar] [CrossRef]
- Fahlgren, N.; Carrington, J.C. miRNA Target Prediction in Plants. Plants. Methods Mol. Biol. 2009, 592, 51–57. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.-M. The Significance of Digital Gene Expression Profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Abdi, H. The Bonferroni and Sidak corrections for multiple comparisons. In Encyclopedia of Measurement and Statistics; Salkind, N.J., Ed.; Sage: Thousand Oaks, CA, USA, 2007. [Google Scholar]
| Combination | Target Genes | miRNAs | Target Finder Score | Corresponding Hormone |
|---|---|---|---|---|
| CKvsM50 CKvsM100 | CL11742.Contig5_All | miR160 | 1 | Auxin (ARF) |
| CL7275.Contig1_All | miR160a-5p | 1 | ||
| CL7275.Contig2_All | miR160a-5p | 1 | ||
| CL8053.Contig1_All | miR160a-5p | 0.5 | ||
| CL8053.Contig2_All | miR160a-5p | 0.5 | ||
| CL8053.Contig3_All | miR160a-5p | 0.5 | ||
| CL11742.Contig1_All | miR160a-5p | 1 | ||
| CL11742.Contig3_All | miR160a-5p | 1 | ||
| CL11742.Contig4_All | miR160a-5p | 1 | ||
| CKvsM10 | CL4553.Contig1_All | miR156a-5p | 1 | Gibberellin (DELLA) |
| CKvsM50 CKvsM100 | CL3157.Contig1_All CL3157.Contig4_All | miR171b-3p | 2 | |
| CL3157.Contig1_All CL3157.Contig4_All | miR171b-3p_3 | 3 | ||
| CL3157.Contig1_All CL3157.Contig4_All | miR171d_1 | 1 | ||
| CL3157.Contig1_All CL3157.Contig4_All | miR171f_3 | 1 | ||
| CKvsM50 CKvsM100 | CL1203.Contig7_All | miR171b-3p | 3.5 | Ethylene (ETR) |
| CKvsM10 | CL9049.Contig3_All | miR156a | 2 | Brassinosteroid (BRI1) |
| CL9049.Contig3_All | miR156a-5p | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, D.; Chen, Y.; Xu, X.; Lin, Y.; Lai, Z. Exploring the Effect of Methyl Jasmonate on the Expression of microRNAs Involved in Biosynthesis of Active Compounds of Rosemary Cell Suspension Cultures through RNA-Sequencing. Int. J. Mol. Sci. 2022, 23, 3704. https://doi.org/10.3390/ijms23073704
Yao D, Chen Y, Xu X, Lin Y, Lai Z. Exploring the Effect of Methyl Jasmonate on the Expression of microRNAs Involved in Biosynthesis of Active Compounds of Rosemary Cell Suspension Cultures through RNA-Sequencing. International Journal of Molecular Sciences. 2022; 23(7):3704. https://doi.org/10.3390/ijms23073704
Chicago/Turabian StyleYao, Deheng, Yukun Chen, Xiaoping Xu, Yuling Lin, and Zhongxiong Lai. 2022. "Exploring the Effect of Methyl Jasmonate on the Expression of microRNAs Involved in Biosynthesis of Active Compounds of Rosemary Cell Suspension Cultures through RNA-Sequencing" International Journal of Molecular Sciences 23, no. 7: 3704. https://doi.org/10.3390/ijms23073704
APA StyleYao, D., Chen, Y., Xu, X., Lin, Y., & Lai, Z. (2022). Exploring the Effect of Methyl Jasmonate on the Expression of microRNAs Involved in Biosynthesis of Active Compounds of Rosemary Cell Suspension Cultures through RNA-Sequencing. International Journal of Molecular Sciences, 23(7), 3704. https://doi.org/10.3390/ijms23073704








