Simple Summary
Insect vectors of plant diseases and insecticide resistance pose the greatest challenge to sustainable crop production in food security. In this study, we explored genetic mechanisms responsible for resistance to a common insecticide group, neonicotinoids, in a key insect vector of potato diseases—potato psyllids. These small insects with piercing-sucking mouthparts are common in potato and can transmit a bacterium that causes zebra chip disease. Zebra chip has had a devastating impact on potato producers and has contributed to a highly regimented and intense use of insecticides to suppress potato psyllids as soon as they are detected in a field, commonly using neonicotinoids. Widespread resistance to these insecticides is now evident in potato psyllid populations. Using susceptible and resistant psyllid populations, we sequenced portions of their genomes to elucidate genes involved in the evolution of resistance to neonicotinoid insecticides. We found several genes that are likely to be responsible for insecticide resistance, and these should be explored in further research. We also discovered that a method commonly used to separate potato psyllids into distinct groups based on their geographic origin does not adequately represent their genetic population structure and should be used in conjunction with other genetic techniques.
Abstract
(1) Background: Many hemipteran insects transmit plant pathogens that cause devastating crop diseases, while pest management frequently relies primarily on insecticide applications. These intense insecticide applications lead to the development of insecticide resistance, as was the case for potato psyllid, Bactericera cockerelli (Hemiptera: Triozidae), a vector of Candidatus Liberibacter solanacearum, which causes zebra chip disease in potato. (2) Methods: Here, we use double-digest restriction site-associated DNA (ddRAD) to genotype eight psyllid populations (one susceptible and seven resistant to neonicotinoid insecticides). (3) Results: Association tests identified over 400 loci that were strongly segregated between susceptible and resistant populations. Several loci were located within genes involved in insecticide resistance, gene regulation, fertility, and development. Moreover, we explored the genetic structure of these eight populations and discovered that routinely utilized haplotyping was not an accurate predictor of population structure. Pairwise comparisons of the fixation index (FST) of populations of the same haplotype were not different from pairwise FST of populations that belonged to different haplotypes. (4) Conclusions: Our findings suggest that neonicotinoid insecticide resistance has a genetic basis, most likely as a result of similar selection pressure. Furthermore, our results imply that using a single maternally inherited gene marker to designate genetic lineages for potato psyllids should be re-evaluated.
1. Introduction
Many insects in the order Hemiptera, such as aphids, psyllids, leafhoppers, and whiteflies, are highly injurious to crops due to their wide host range and rapid reproduction. Their injury is more severe when these insects transmit plant pathogens—bacteria, viruses, phytoplasmas, and fungi [1,2]. Management of these insect vectors is challenging, and chemical control aimed to suppress the insects remains the most widely implemented approach [1,3,4,5]. The widespread application of insecticides can disrupt ecosystem services that are dependent on natural enemies of pests, contribute to outbreaks of other primary arthropod pests, and cause outbreaks of secondary diseases transmitted by insects [6,7,8]. Further, insecticide-resistant populations have been reported in many crop systems globally [9,10,11,12,13,14], reducing the efficacy of pest management programs.
The potato psyllid, Bactericera cockerelli (Šulc) (Hemiptera: Triozidae), is a vector of pathogens that are casual agents of several diseases, including “psyllid yellows” (PY) [15], “zebra chip” (ZC) [16], and “vein greening disease” [17]. Among these, ZC is particularly injurious to potato, resulting in significant losses to producers and contributing to extremely intense pesticide applications to suppress potato psyllids. Zebra chip disease is caused by a bacterial pathogen, Candidatus Liberibacter solanacearum (Lso) [18,19], and has been responsible for extensive economic losses in most potato production regions of the US. Bactericera cockerelli is thought to be native to Central and North America and its current distribution ranges from southern Canada to Central America, New Zealand, and Australia [20,21]. Four different haplotypes of the potato psyllid have been identified based on the variation of Cytochrome c oxidase subunit I (COI) gene, Central, Western, Northwestern, and Southwestern, and the distribution of these haplotypes in the US roughly corresponds to geographical regions where the insects are prevalent [22,23]. Notably, the haplotype does not appear to affect the efficiency of Lso transmission or the likelihood of infection with the pathogen [24].
Owing to the lack of effective control measures for any of the pathogens transmitted by the potato psyllid, management is focused on the suppression of the psyllids and relies solely on pesticides [25,26]. The most frequently used systemic insecticides are neonicotinoids, which are commonly applied in-furrow during planting, as well as foliar sprays mid-season. Imidacloprid and thiamethoxam are the two most predominant neonicotinoids that are applied to soil or seed tubers [27,28]. Both insecticides are systemic and act as insect neurotoxins [29,30]. There is now widespread evidence that psyllids are resistant to these two commonly used neonicotinoid insecticides [14,31,32]. In fact, in our previous study, we demonstrated high resistance ratios in populations of the psyllids collected across potato-producing regions of Texas, New Mexico, and Colorado compared with a susceptible colony collected from the Lower Rio Grande Valley in Texas in 2006, prior to the intense use of neonicotinoids [14].
While the incidence and severity of the potato psyllid resistance to neonicotinoid insecticides have been documented in several separate studies, the mechanisms of this resistance are not known. Generally, hypotheses about molecular mechanisms of insecticide resistance are centered around targeted site mutations, which result in target insensitivity and increased expression of detoxification genes [9,11,12,13,33,34,35]. In previous RNA-seq or genome-wide association studies, a few genes associated with insecticide resistance have been identified [35,36,37,38]. However, genetic responses to insecticides are heterogeneous, as insect pests adapt to different and changing environments [38,39], and many other mechanisms that are outside these well-studied molecular mechanisms could contribute to insecticide resistance.
Here, we used genome-wide single nucleotide polymorphisms (SNPs) to investigate the genetic basis of resistance to neonicotinoid insecticides in resistant and susceptible potato psyllid populations collected across the potato-growing regions of the Southwestern US. We were interested in elucidating the genetic structure of the potato psyllid population and exploring the hypothesis that the selective pressure of the neonicotinoid insecticide on the potato psyllid population could result in heritable SNPs. In addition, we attempted to explore the genetic structure of the psyllid populations that are designated as Central and Western haplotypes. Given that the rapid evolution of insecticide resistance is a major challenge for sustainable agriculture, understanding the genetic resistance mechanisms is important for DNA-based resistance monitoring in field populations.
2. Materials and Methods
2.1. Psyllid Collections and Colony Maintenance
Psyllid colonies were established from adult psyllids collected during 2013, 2015, and 2016, except for the Texas (TX) LRGV 2006 colony, which was originally collected in 2006 from the Lower Rio Grande Valley of Texas and was previously shown to be susceptible to neonicotinoids [14,28]. Potato psyllid colonies were maintained in the greenhouse complex at Texas A&M AgriLife Research Plant Stress Laboratory in Bushland, TX. Eight potato psyllid populations that were designated to two haplotypes (‘Western’ and ‘Central’) were included in this work (Table 1). All potato psyllids were maintained on tomato plants (Solanum lycopersicum L., variety Lance) in 60 × 60 × 60 cm insect-proof mesh cages (MegaView Science Education Services Co., Taipei, Taiwan) in the greenhouse with mean temperatures of 21 ± 3 °C (night) to 30 ± 3 °C (day) and 16:8 L:D photoperiod.

Table 1.
List of potato psyllid colonies included in this study.
2.2. Sample Collection and DNA Extraction
Six biological samples (replicates) consisting of five adult psyllids pooled together were used from each of the eight populations (Table 1). DNA was extracted using Puregene Core Kit A (Qiagen, Valencia, CA, USA). DNA quality and quantity were assessed using the NanoVue Plus spectrophotometer (GE, Healthcare, Piscataway, NJ, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) prior to ddRAD library preparation. The ddRAD-tag libraries were prepared from the genomic DNA of 48 samples using the restriction enzymes Spel and Mbol. The libraries were sequenced on one lane of the Illumina NovaSeq (Illumina, Inc., San Diego, CA, USA) with the 150 bp paired-end mode. The ddRAD library preparation, sequencing, and data demultiplexing were conducted by Texas AgriLife Research Genomics and Bioinformatics Center (College Station, TX, USA).
2.3. Data Analysis
2.3.1. SNP Calling
We took a de novo approach using Stacks v2.41 [40] because there was no reference genome available for potato psyllid. Quality filtering was performed with the process_radtags module in Stacks with default parameters. Putative orthologous tags (Stacks) per biological sample were assembled using ustacks with a minimum depth of coverage required to create a stack (m) of three and a maximum nucleotide mismatch (M) of three. Catalogs of loci were assembled using cstacks; the number of mismatches allowed between sample loci when generating the catalogs (n) was six. Matches of individual RAD loci to the catalog were searched using sstacks. Finally, we used the populations module of Stacks to filter and export the genotype calls and computed several common population statistics. The following filters were used for SNP calling: loci should be present in at least 85% of the samples and also in six populations with minimum allele frequency (MAF) > 0.05, minimum minor allele count (minmac) ten, maximum observed heterozygosity 0.7, and p-value cutoff 0.05 for keeping an FST measurement. To obtain a putatively neutral SNP data set for genetic structure analysis, an “unlinked” data set was generated with a single SNP per locus (the first SNP per locus) to reduce the effects of linkage. We then used BayeScan v2.01 [41], which estimates the posterior probability of SNPs under selection. We removed the outlier SNPs from the original vcf file using VCFtools [42] and used this filtered data set (neutral SNPs) for genetic structure analysis.
2.3.2. Genetic Basis of Insecticide Resistance Using Association Test and Annotation of De Novo Assembled Contigs
To identify loci that were strongly segregated between the insecticide susceptible (TX LRGV) and resistant populations (the remaining seven populations), we performed association analysis with the entire data set containing 12,681 variant sites using PLINK [43] (v1.9), and the p-value cutoff was set to 1 × 10−14.
Gene annotations were obtained for all specific RAD loci/contigs that contained significant SNPs in the association test using the Blast2GO annotation methodology embedded in OmicsBox (v 1.2.4; BioBam Bioinformatics S.L., Valencia, Spain). Sequences were queried against the Hemiptera database under “insects” (Taxid:6960), using OmicsBox with an E-value threshold of 1 × 10−4.
2.3.3. Genetic Structure and Variation
We deployed three complementary approaches to model population genetic structure without a priori population assignment. First, we conducted principal component analysis (PCA) implemented in the PLINK, using neutral SNPs marker set. Secondly, we used Structure software (v 2.3.4 [44]) to explore the population genetic structure based on our neutral SNP marker set. The optimal number of populations present within the 48 individuals was determined by running a continuous series of K = 1–10 with 5000 burn-in iterations and 50,000 MCMC repetitions in Structure. The calculation of the averaged likelihood at each K [In Pr (X|K) or In (Kn)] was performed in Structure Harvester [45]. For the final K analysis, a burn-in of 50,000 with a run length of 500,000 MCMC replications and 20 independent runs were conducted. Lastly, a neighbor-joining tree was constructed based on the Nei’s genetic distances using the “aboot” function in R package poppr [46] with the “nj” parameter and 500 bootstrapping replicates. The resulting tree was plotted using the R package ggtree [47].
3. Results and Discussion
3.1. Genetic Basis of Insecticide Resistance
Insecticide resistance is a growing challenge in potato production, and our research provides a foundation for understanding the genetics of insecticide resistance in potato psyllid. In this study, the potato psyllid population that was susceptible to imidacloprid and thiamethoxam [14] allowed us to characterize genotypes and alleles that were significantly different between this susceptible population and the neonicotinoid-resistant populations. We were able to identify a list of genes associated with insect gene regulation, fertility, and development, even though many of the loci containing the strongly segregated SNPs were not annotated (68.4%), as it is likely that they are located on non-coding or intergenic sections of the potato psyllid genome, or that they are unique to potato psyllids.
We generated 288 million pair-end reads with an average of 5.9 M reads per sample. As no reference genome was available for B. cockerelli, a de novo assembly was constructed using Stacks. The total ddRAD data set included 456,481 unique loci. We retained a total of 11,471 loci and 12,681 SNPs after filtering loci that did not pass sample/population constraints, and all these loci passed the HWE test (p-value < 0.01). Further filtering was also used to select only one SNP at each locus (the first SNP per locus) that generated 4294 SNPs.
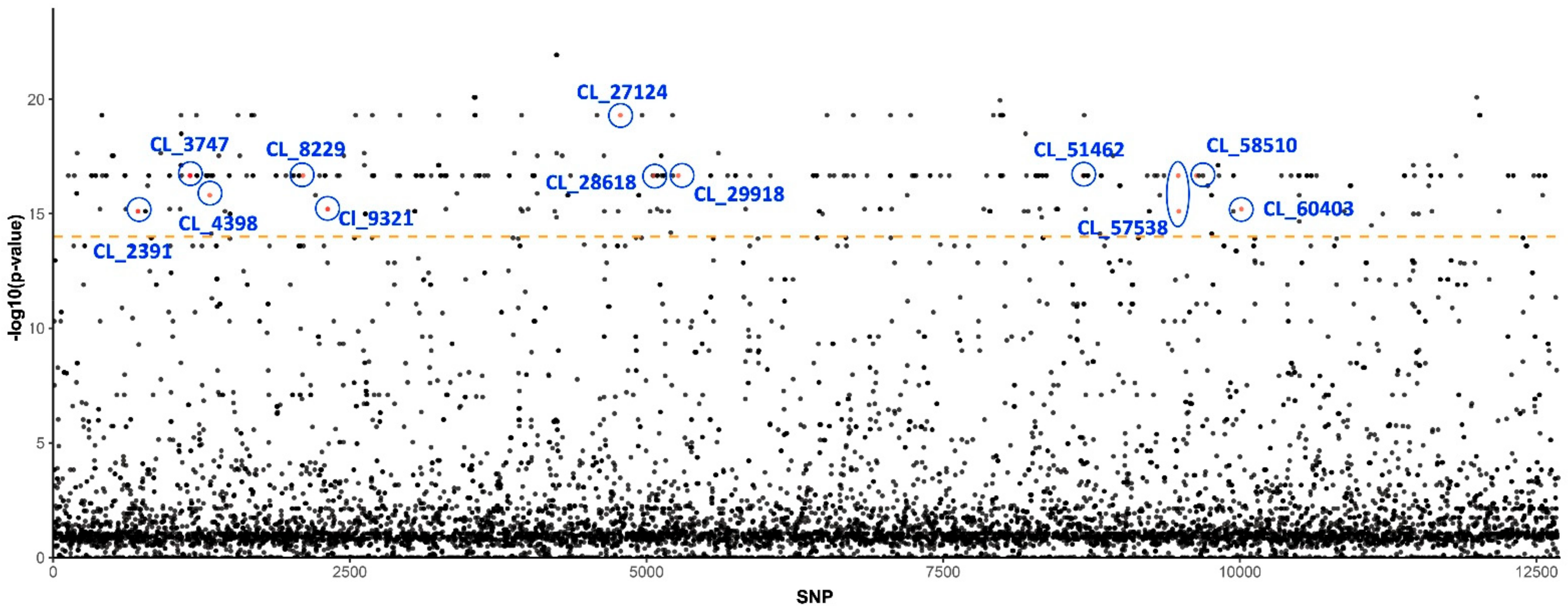
Based on association test results, we detected 429 SNPs out of 12,681 on 215 unique loci that were strongly segregated between the susceptible and insecticide-resistant populations (Supplementary Table S1, Figure 1). Approximately one-third (68/215) of the unique loci were found to be homologous to known genes or sequences in the annotation database when the E-value cutoff was set as 1 × 10−4 (Supplementary Table S2). Notably, a number of these genes have been identified as being associated with insecticide resistance, insect gene control, fertility, and development. Furthermore, a few loci were found to be homologous to genes from microbes that are endosymbiont of or transmitted by potato psyllids, such as genes from Wolbachia and Lso (Table 2 and Figure 1).
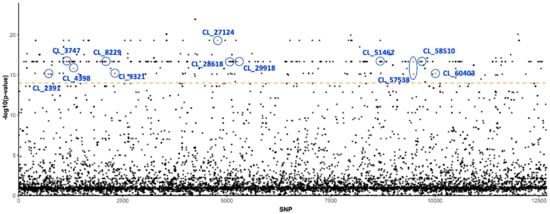
Figure 1.
SNPs and p-values from association test with insecticide resistance. The SNP index on y-axis is sorted by locus names and where they are located. Highlighted loci that host strongly segregated SNPs and their annotation are also presented in Table 2. Orange dashed line indicates the p-value cutoff (1 × 10−14) for association test.

Table 2.
A subset of loci that host strongly segregated SNPs between insecticide susceptible and resistant psyllid populations.
Within insecticide resistance loci, we found the locus CL_8229 to be homologous of the Paramoysin (prm) gene, which was reported to be one of the two key loci involved in imidacloprid resistance in populations of Drosophila melanogaster based on a large-scale genome-wide association study (GWAS) [38]. The prm encodes a protein that constitutes the structure of muscle filament, and mutation of the prm would impact the indirect flight muscle and power generation in D. melanogaster [48]. In addition, locus CL_28618 was found to be homologous to wheat stem sawfly, Cephus cinctus inositol 1,4,5-trisphosphate receptor (IP3R), a family of the Ca2+ release channel protein involved in increasing the cytoplasmic free calcium concentration. Guo et al. [37] reported that silencing of ip3r in a silverleaf whitefly, Bemisia tabaci, decreased the susceptibility of adult B. tabaci to cyantraniliprole, a diamide pesticide that is widely used to manage sucking insect pests. The last locus within this group, CL_51462, is homologous to the non-lysosomal glucosylceramidase-like gene. Although the function of glucosylceramidase in insects is not well characterized, it was also reported to be differentially expressed in the abdomen of insecticide-resistant strains of mosquito, Anopheles gambiae, a vector of malaria [36].
Within loci related to gene regulation, DNA repair, and innate immunity, we found interesting homologous, including the locus CL_3747, which is homologous to DDB1- and CUL4-associated factor 5 in Asian citrus psyllid, Diaphorina citri (Table 2). This locus is involved in gene regulation, DNA repair, and innate immunity, and it is reported to be involved in DNA damage response [49]. Another notable locus, CL_4398, is homologous to D. citri histone deacetylase 11. This gene has been proven to be a crucial regulator in insect larval development [50].
Moreover, several loci that differed between the susceptible and resistant populations were related to the innate immunity of insects. For example, CL_2391 is homologous to spaetzle 4-like in D. citri, a ligand that responds to the bacterial or fungal infection by binding Toll receptors to induce the secretion of antimicrobial peptides [51,52]. Another locus in this class, CL_9321, is homologous to a pea aphid (Acyrthosiphon pisum) CD109 antigen, which is induced by ecdysone signaling and involved in the cellular immunity of insects [53]. It is noteworthy that multiple studies have linked insect immunity pathways to insecticide resistance [54,55,56]. For example, in a recent study characterizing gene expression of Bt-resistant and Bt-susceptible bollworm, Helicoverpa zea, several immunity genes were found to be differentially expressed between resistant and susceptible populations, including two CD109 antigen genes [55].
In addition to insect DNA, we discovered three contigs (including the previously mentioned Wolbachia homolog) that presumably originated from the microbiome of potato psyllids (Table 2). Two contigs, CL_27124 and CL_57510, are homologous to Lso, the bacterial pathogen transmitted by potato psyllids. It is unknown whether the plant pathogen that psyllids transmit has any impact on insecticide resistance.
Insecticide resistance has been a prevailing challenge to sustainable pest management and has been the focus of research for decades. Two major mechanisms are thought to result in insecticide resistance: (1) insecticide targeted site gene mutations, which result in target insensitivity [9,12,13,33,34], and (2) increases in detoxification gene products, also known as metabolic resistance [11,13,35]. In the case of target site insensitivity, mutations occur at the active sites of genes that encode proteins that are the targets of insecticides due to long-term insecticide selection. Insecticide resistance can also originate from the target organism’s increased ability to detoxify the pesticide’s active ingredient. Usually, detoxifying enzymes comprise three main superstructures: cytochrome P450 (CYP), glutathione S-transferases (GST), and carboxylesterases (COE). Among these, P450s tend to play a key role in interactions between four plants and insects and have been studied intensively [57,58,59,60]. Several studies employed bioassays that target these genes, and they are informative in detecting the prevalence of insecticide resistance genes [61,62,63].
Beyond these few well-known targets, different insect species might have evolved distinct genes and molecular mechanisms of insecticide resistance [64], and insecticide resistance is often a complex and polygenetic phenotype. Therefore, examining genotypes of insecticide-resistant populations, lines, and individuals offers a different perspective on the origin, expansion, and dynamics of insecticide resistance-related genes or alleles. In a study that used the ddRAD approach, Yang et al. [65] identified several candidate loci strongly associated with population-level resistance to synthetic pyrethroids and organophosphates in a phytophagous mite, Halotydeus destructor, a major pest of pastures and crops in Australia. These candidate loci were located in the genomic regions that code for transmembrane transport and signaling proteins, with a few other genes related to pyrethroid resistance. In another study in which whole genome sequencing was conducted on multiple field-collected fruit fly, D. melanogaster, two major loci appeared to be related to imidacloprid resistance [38]. One locus was in the coding region of prm gene, and another locus was 60 k upstream of the coding region of the Nicotinic-Acetylcholine Receptor Alpha 3 gene. These studies illustrate the distinct genetic mechanisms of insecticide resistance in different arthropod species.
In our study, we found a strongly segregated SNP located on contigs that is homologous to the prm gene. However, we did not identity SNPs that are on contigs that are homologous to previously reported genes involved in insecticide resistance, such as P450, sodium channel gene, gst, or coe. We speculate that this may be due to the fact that in ddRAD library preparation, only DNA fragments flanking the restriction enzyme cutting sites are included in the DNA library, resulting in just a portion of the genome being sequenced. Furthermore, the lack of a reference genome makes annotating these short de novo assembled tags/loci challenging. We anticipate that the annotation of SNPs will improve with the availability of the reference genome. In the future, examining the transcriptomics will be informative to test whether these SNPs impact the gene expression level of detoxification-related gene products.
We have found multiple variants that are strongly segregated between insecticide susceptible and resistant populations; these variants and related genes could provide a candidate list for further testing. Insecticide resistance is recognized as a complex, polygenic, and quantitative trait, according to growing knowledge and studies on the mechanism and genetic basis of insecticide resistance [9,38]. Thus, the outcomes of our work suggest taking interdisciplinary approaches that include the sampling of natural populations that exhibit a varying degree of resistance, whole genome sequencing, and functional tests to understand this complex trait.
3.2. Population Genetic Structure
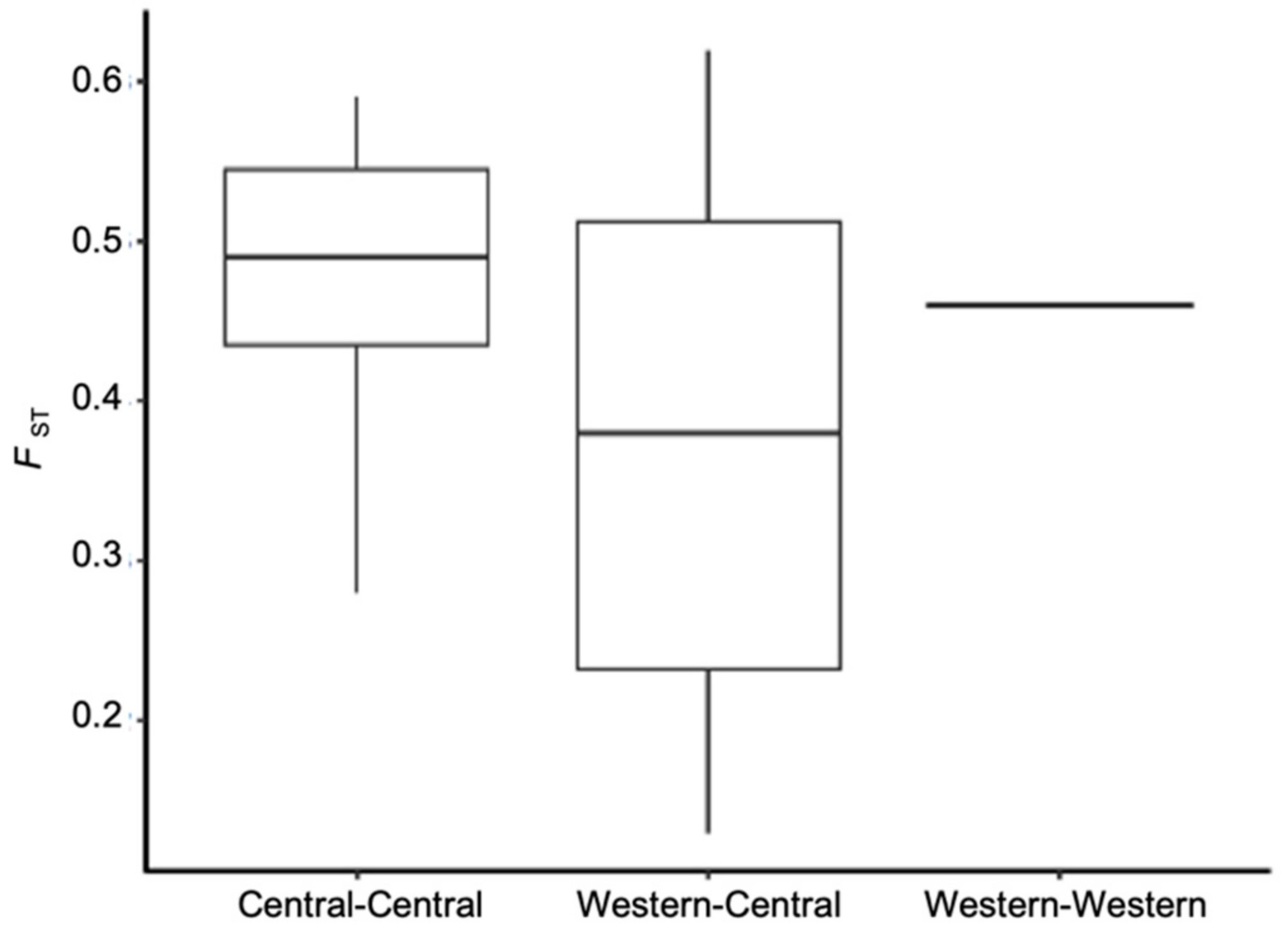
The percentage of polymorphic loci and observed and expected heterozygosity (Ho and He respectively) for all loci were estimated (Table 3). The TX Weslaco potato psyllid population was polymorphic at the highest percentage of loci (10%), while all other populations were polymorphic at 6 to 9% of loci. The average observed heterozygosity (Ho) among all populations ranged from 1.6 × 10−4 to 2.5 × 10−4, and the highest was observed in the TX Weslaco population. To examine variance in allele frequencies among populations and their degree of differentiation, we also measured the fixation index (FST) using the data set containing 4294 independent SNPs. The mean FST for all pairwise comparisons was 0.44. The TX Panhandle was, on average, the most differentiated from all other populations (mean FST = 0.52), and Western-2 was the least differentiated population (mean FST = 0.31) (Table S1). Surprisingly, pairwise FST of populations of Central haplotype were not significantly different from pairwise FST of populations that belonged to different haplotypes, e.g., Central and Western (t-test, p-value = 0.095, Figure 2, Table S1).

Table 3.
Summary of genetics statistics calculated by the Stacks. Private refers to number of variable sites unique to each population; %Polymorphic_Loci refers to percentage of polymorphic loci, Obs_He and Exp_Het refer to average observed and expected heterozygosity per locus, respectively.
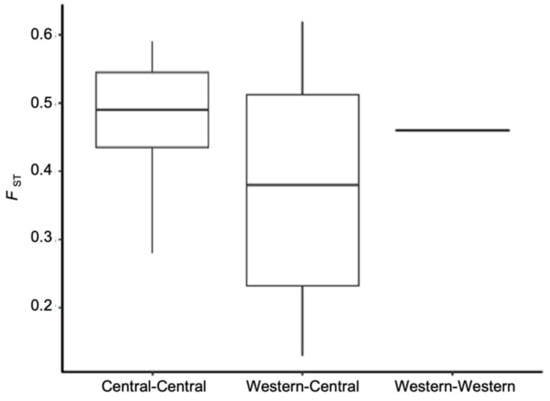
Figure 2.
Pairwise FST of psyllid populations separated by whether the pair were different haplotypes or the same haplotype. Note, only two populations were designated as Western haplotype in our study.
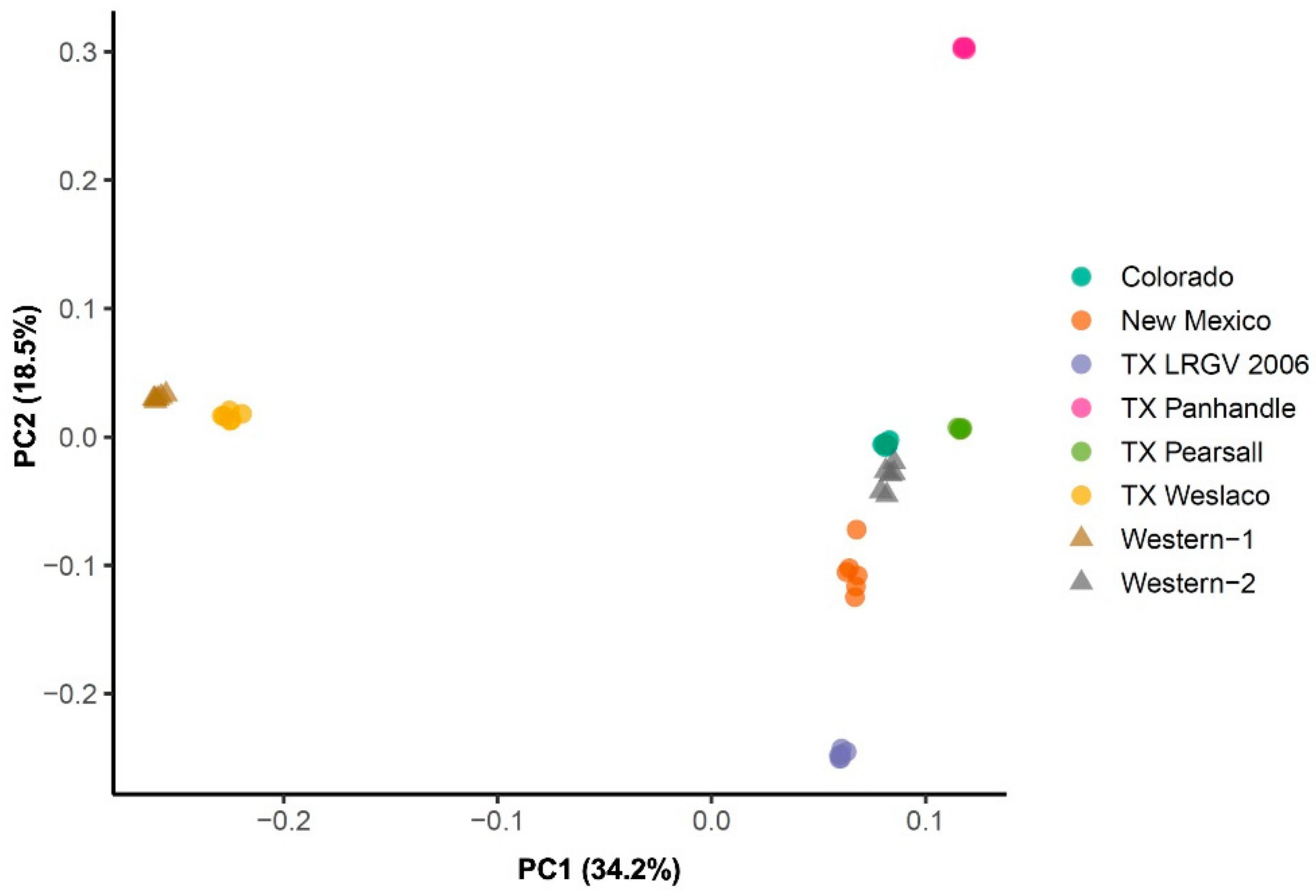
Further, the principal component 1 (PC1), which explained over 30% of the overall variation (Figure 3), separated the TX Weslaco and Western-1 cluster from the rest of the six populations. At PC2, TX LRGV 2006 and TX Panhandle diverged from the rest of the four populations, i.e., TX Pearsall, Colorado, Western-2, and New Mexico. The populations from New Mexico, Colorado, Western-2, and TX Pearsall grouped together. Similar to the FST results, we did not find that populations from the same haplotypes clustered together, e.g., Western-1 and Western-2 did not cluster together, and TX Weslaco was separated from the other Central haplotype populations.
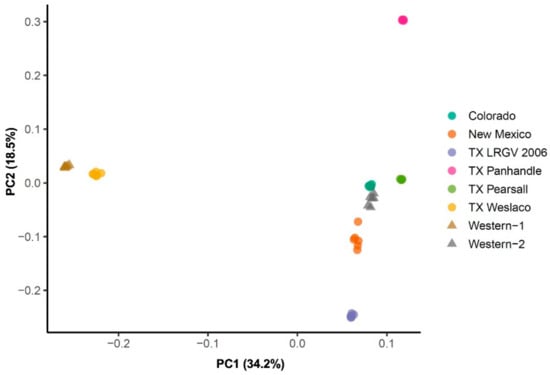
Figure 3.
Principal component analysis of eight psyllid populations. PCA implemented in the PLINK using 4294 neutral SNPs marker set.
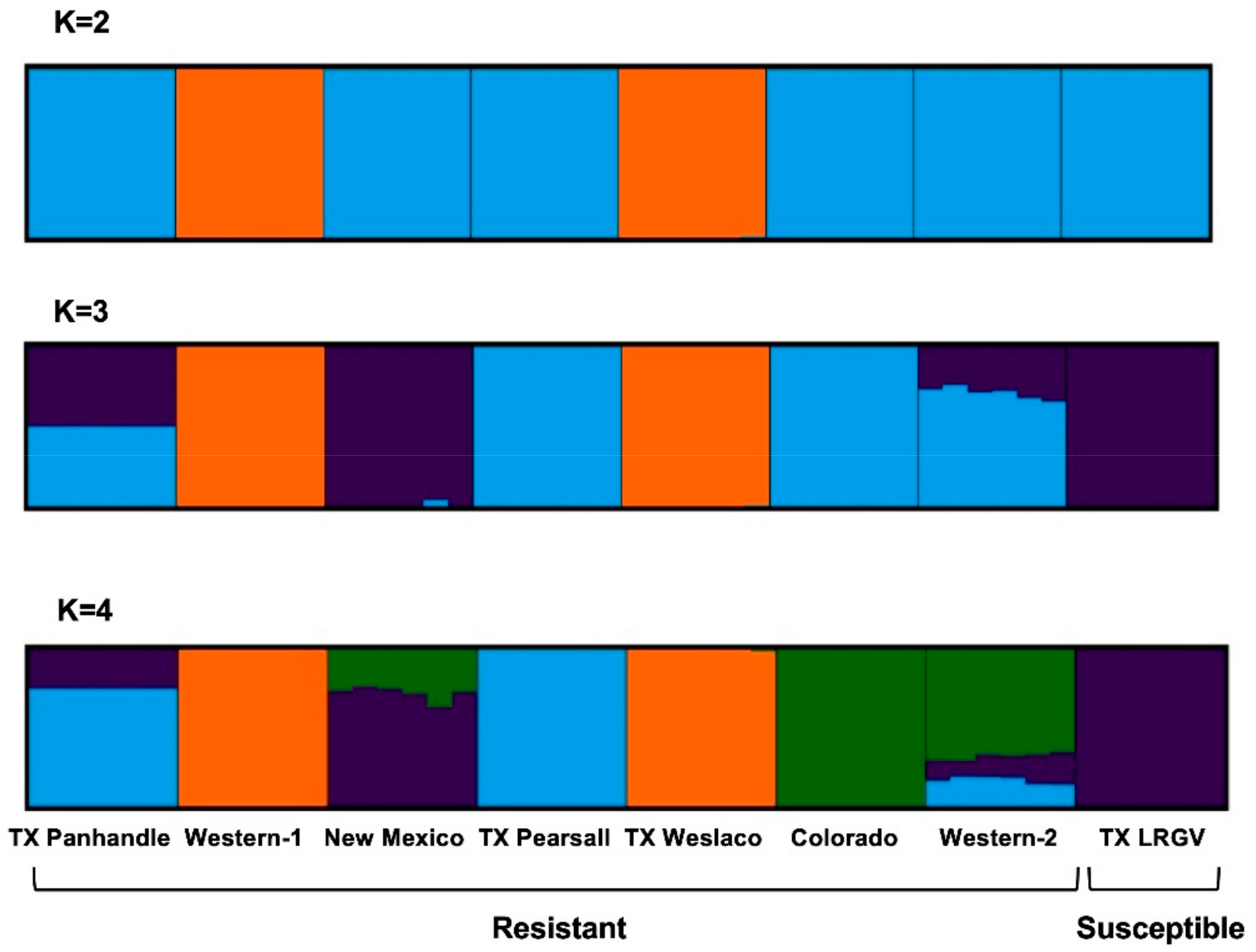
Moreover, Structure analysis indicated the most likely number of genetic clusters to be four (K = 4), based on the agreement between the ΔK method and estimators of the structure selector. At K = 2, Western-1 and TX Weslaco were placed in one cluster (Figure 4), and the rest of the six populations were placed in a different cluster. At K = 3, New Mexico and TX LRGV formed a new cluster that separated them from Western-1 and TX Weslaco. TX Panhandle and Western-2 seemed to be admixed between the TX Pearsall‒Colorado cluster and the New Mexico‒TX LRGV cluster. At K = 4, the Colorado samples separated and formed their own cluster, and Western-2 seemed to be most similar to Colorado while sharing some genetic similarities to TX Pearsall and TX LRGV. This pattern was consistent with the PCA outcomes. The overall pattern of the genetic structure indicated that populations of the same haplotype did not cluster together.
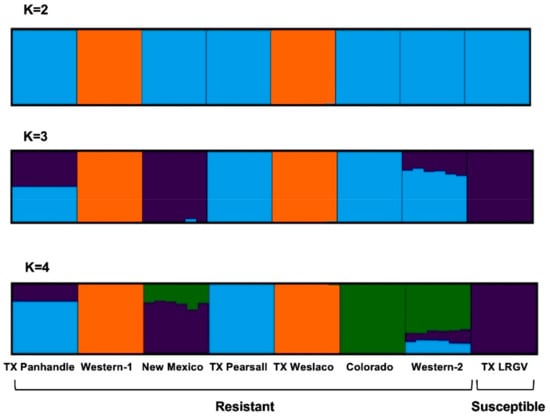
Figure 4.
Cluster assignment of the individuals revealed by Structure. Each individual is represented by a single column divided into K genetic clusters. K indicates the number of clusters that maximized the probability of the model. The color proportions of each bar correspond to individuals’ estimated membership fractions of each of the clusters.
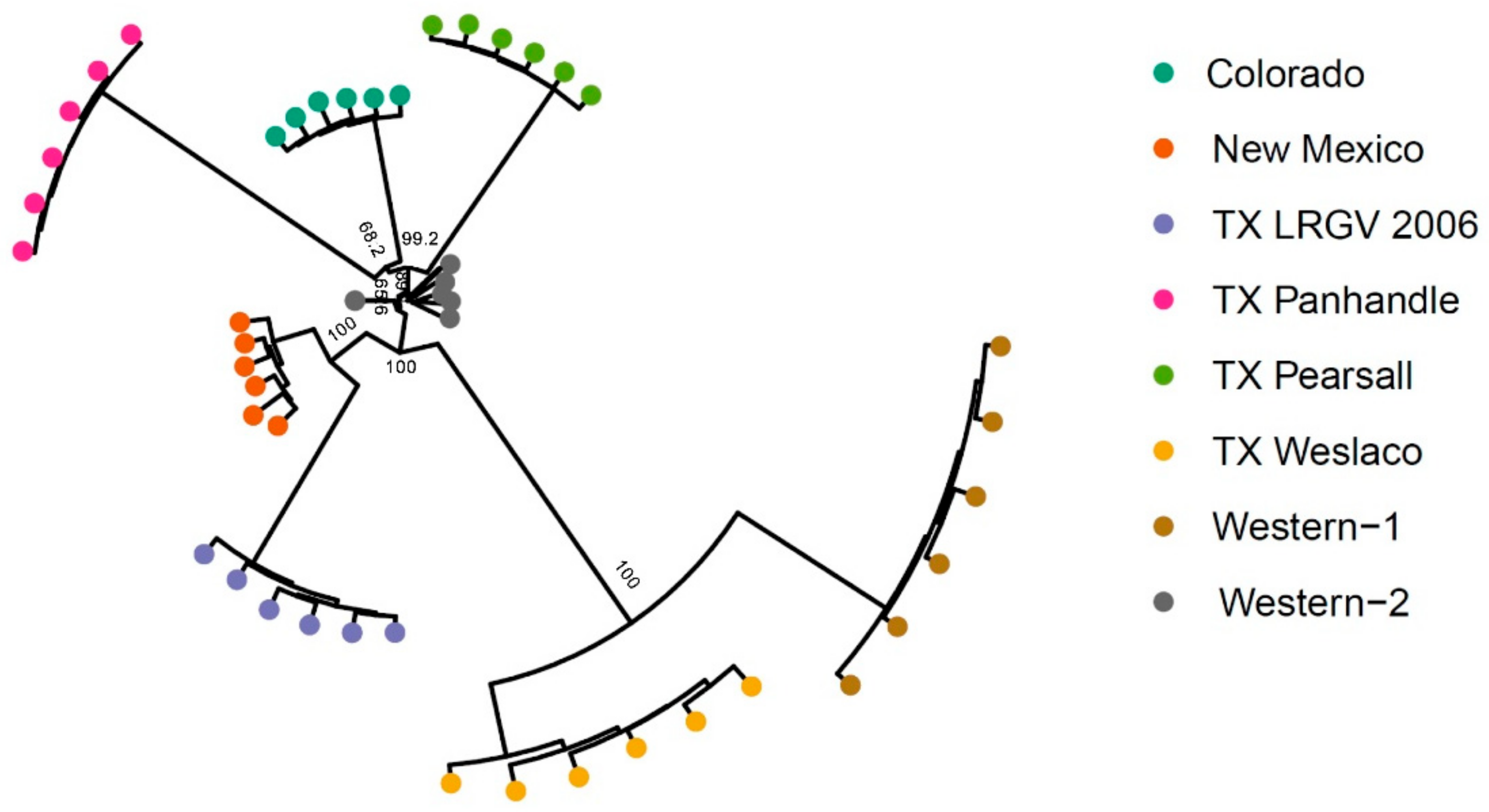
To further examine the genetic structure of psyllid populations, we used the 4294 neutral SNPs data set to conduct a phylogenetic analysis based on Nei’s distance. Similar to results in PCA and Structure, TX Weslaco and Western-1 were found to be closely related. The New Mexico and TX LRGV 2006 populations clustered together, and three TX Panhandle, Colorado, and TX Pearsall populations were found to be closely related (Figure 5). All Western-2 samples seemed to form a tight cluster with the exception of one sample. The neighbor-joining tree revealed a pattern similar to PCA, Structure, and FST, suggesting that haplotype does not appear to be a good indication of genetic structure.
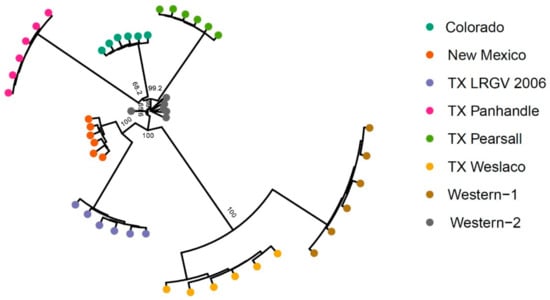
Figure 5.
Neighbor-joining tree of 48 samples from eight populations. Five hundred bootstrapping resampling was conducted to obtain the support values indicated at the branch. Support values are not labeled for the end branch because the eight samples within a population are rather similar, given they were pooled samples.
Based on genetic structure results, the susceptible population (TX LRGV) was found to be somewhat related to the population of New Mexico; however, the FST between the two is moderate (Table S1). We found no genetic structure linkage to haplotypes, as the two Western haplotype psyllid populations did not cluster together using any of the three methods tested, including PCA, Structure, and neighbor-joining tree. Similarly, the pairwise FST value of the populations that originated from different haplotypes was not different from the FST from the same haplotype population pairs. This suggests that haplotyping is not an informative marker to determine the genetic differentiation of potato psyllids. Similar results were shown in other studies that examined both haplotypes and genotypes of potato psyllids [66,67]. Specifically, Fu et al. [67] employed genome-wide SNP to analyze the genetic structure of potato psyllids and found that a handful of Western haplotype psyllids were genetically more similar to psyllids of the Northwestern haplotype. In potato psyllids, haplotyping is based on nucleotide variation within a 500 bp- stretch of the potato psyllid COI gene [20], which is located in the mitochondrial genome and is inherited maternally. This means that if there is any hybridization among psyllids from different haplotypes, the haplotype of the offspring will only reflect the maternal lineage [67]. More broadly, in the animal kingdom, conflicting phylogenetics and population genetics patterns are common between mitochondrial and nuclear DNA markers [68]. We suggest that future research should focus on including a wider range of molecular markers rather than relying solely on COI haplotyping to provide more thorough information on potato psyllid ancestry and lineage.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects13030257/s1, Table S1: pairwise comparison of genetic (FST) among eight populations of potato psyllid based on 4294 independent loci, Table S2: significant SNPs for association test.
Author Contributions
Conceptualization, A.S. and M.K.; methodology, A.S., M.K. and Z.F.; software, M.K. and Z.F.; validation, M.K. and Z.F.; formal analysis, M.K. and Z.F.; investigation, M.K., Z.F. and A.S.; resources, M.K. and A.S.; data curation, M.K. and Z.F.; writing—original draft preparation, M.K., Z.F. and A.S.; writing—review and editing, M.K., Z.F. and A.S.; visualization, M.K., Z.F. and A.S.; supervision, A.S.; project administration, A.S.; funding acquisition, A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the USDA NIFA (TEX09638), Texas Department of Agriculture Zebra Chip Initiative, and Texas Department of Agriculture Specialty Crop Block Grants Program to A.S.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All raw sequencing reads have been submitted to the NCBI Sequence Read Archive and are available under BioProject ID: PRJNA773566: https://www.ncbi.nlm.nih.gov/sra/PRJNA773566 (accessed on 19 October 2021).
Acknowledgments
The authors would like to thank K. Varela and E. Huff for assistance in psyllid colony maintenance, L. Paetzold for her support in the laboratory, and J. Trumble, A. Silva, I. Badillo-Vargas, and C. Higgins for providing psyllids from their locations. We also thank two anonymous reviewers, whose comments and suggestions helped improve this manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Perilla-Henao, L.M.; Casteel, C.L. Vector-borne bacterial plant pathogens: Interactions with hemipteran insects and plants. Front. Plant Sci. 2016, 7, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Eigenbrode, S.D.; Bosque-Pérez, N.A.; Davis, T.S. Insect-borne plant pathogens and their vectors: Ecology, evolution, and complex interactions. Annu. Rev. Entomol. 2018, 63, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Perring, T.M.; Gruenhagen, N.M.; Farrar, C.A. Management of plant viral diseases through chemical control of insect vectors. Annu. Rev. Entomol. 1999, 44, 457–481. [Google Scholar] [CrossRef] [PubMed]
- Mota-Sanchez, D.; Whalon, M.E.; Holingworth, R.M.; Xue, Q. Documentation of pesticide resistance in arthropods. In Global Pesticide Resistance in Arthropods, 1st ed.; Whalon, M.E., Mota-Sanchez, D., Hollingworth, R.M., Eds.; CABI: Wallingford, UK, 2008; pp. 32–39. [Google Scholar]
- Stillson, P.T.; Bloom, E.H.; Illán, J.G.; Szendrei, Z. A novel plant pathogen management tool for vector management. Pest Manag. Sci. 2020, 76, 3729–3737. [Google Scholar] [CrossRef] [PubMed]
- Biondi, A.; Desneux, N.; Siscaro, G.; Zappalà, L. Using organic-certified rather than synthetic pesticides may not be safer for biological control agents: Selectivity and side effects of 14 pesticides on the predator Orius laevigatus. Chemosphere 2012, 87, 803–881. [Google Scholar] [CrossRef]
- Ndakidemi, B.; Mtei, K.; Ndakidemi, P.A. Impacts of synthetic and botanical pesticides on beneficial insects. Agric. Sci. 2016, 7, 364–372. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Pierson, E.A.; Setubal, J.C.; Xu, J.; Levy, J.G.; Zhang, Y.; Li, J.; Rangel, L.T.; Martins, J., Jr. The Candidatus Liberibacter—Host interface: Insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 2017, 55, 451–482. [Google Scholar] [CrossRef]
- ffrench-Constant, R.H.; Anthony, N.; Aronstein, K.; Rocheleau, T.; Stilwell, G. Cyclodiene insecticide resistance: From molecular to population genetics. Annu. Rev. Entomol. 2000, 45, 449–466. [Google Scholar] [CrossRef]
- Lietti, M.M.; Botto, E.; Alzogaray, R.A. Insecticide resistance in argentine populations of Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae). Neotrop. Entomol. 2005, 34, 113–119. [Google Scholar] [CrossRef]
- Bass, C.; Field, L.M. Gene amplification and insecticide resistance. Pest Manag. Sci. 2011, 67, 886–890. [Google Scholar] [CrossRef]
- Casida, J.E.; Durkin, K.A. Neuroactive insecticides: Targets, selectivity, resistance, and secondary effects. Annu. Rev. Entomol. 2013, 58, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Liu, N. Insecticide resistance in mosquitoes: Impact, mechanisms, and research directions. Annu. Rev. Entomol. 2015, 60, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Szczepaniec, A.; Varela, K.A.; Kiani, M.; Paetzold, L.; Rush, C.M. Incidence of resistance to neonicotinoid insecticides in Bactericera cockerelli across Southwest US. Crop Prot. 2019, 116, 188–195. [Google Scholar] [CrossRef]
- Butler, C.D.; Trumble, J.T. The potato psyllid, Bactericera cockerelli (Sulc) (Hemiptera: Triozidae): Life history, relationship to plant diseases, and management strategies. Terr. Arthropod Rev. 2012, 5, 87–111. [Google Scholar] [CrossRef]
- Munyaneza, J.; Crosslin, J.; Upton, J. Association of Bactericera cockerelli (Homoptera: Psyllidae) with “zebra chip”, a new potato disease in southwestern United States and Mexico. J. Econ. Entomol. 2007, 100, 656–663. [Google Scholar] [CrossRef]
- Brown, J.; Rehman, M.; Rogan, D.; Martin, R.; Idris, A. First report of “Candidatus Liberibacter psyllaurous” (synonym “Ca. L. solanacearum”) associated with ‘tomato vein-greening’and ‘tomato psyllid yellows’ diseases in commercial greenhouses in Arizona. Plant Dis. 2010, 94, 376. [Google Scholar] [CrossRef]
- Butler, C.D.; Trumble, J.T. Identification and impact of natural enemies of Bactericera cockerelli (Hemiptera: Triozidae) in Southern California. J. Econ. Entomol. 2012, 105, 1509–1519. [Google Scholar] [CrossRef] [Green Version]
- Wen, A.; Johnson, C.; Gudmestad, N.C. Development of a PCR assay for the rapid detection and differentiation of ‘Candidatus Liberibacter solanacearum’ haplotypes and their spatiotemporal distribution in the United States. Am. J. Potato Res. 2013, 90, 229–236. [Google Scholar] [CrossRef]
- Munyaneza, J.E. Zebra chip disease of potato: Biology, epidemiology, and management. Am. J. Potato Res. 2012, 89, 329–350. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Wang, R.; Ren, Y.; McKirdy, S. Potential distribution and the risks of Bactericera cockerelli and its associated plant pathogen Candidatus Liberibacter Solanacearum for global potato production. Insects 2020, 11, 298. [Google Scholar] [CrossRef]
- Swisher, K.D.; Munyaneza, J.E.; Crosslin, J.M. High resolution melting analysis of the cytochrome oxidase I gene identifies three haplotypes of the potato psyllid in the United States. Environ. Entomol. 2012, 41, 1019–1028. [Google Scholar] [CrossRef]
- Swisher, K.D.; Henne, D.C.; Crosslin, J.M. Identification of a fourth haplotype of Bactericera cockerelli (Hemiptera: Triozidae) in the United States. J. Insect Sci. 2014, 14, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustafa, T.; Horton, D.R.; Cooper, W.R.; Swisher, K.D.; Zack, R.S.; Pappu, H.R.; Munyaneza, J.E. Use of electrical penetration graph technology to examine transmission of ‘Candidatus Liberibacter solanacearum’ to potato by three haplotypes of potato psyllid (Bactericera cockerelli; Hemiptera: Triozidae). PLoS ONE 2015, 10, e0138946. [Google Scholar] [CrossRef] [PubMed]
- Greenway, G. Economic impact of zebra chip control costs on grower returns in seven US states. Am. J. Potato Res. 2014, 91, 714–719. [Google Scholar] [CrossRef]
- Greenway, G.; Rondon, S. Economic impacts of zebra chip in Idaho, Oregon, and Washington. Am. J. Potato Res. 2018, 95, 362–367. [Google Scholar] [CrossRef]
- Byrne, F.J.; Toscano, N.C. Uptake and persistence of imidacloprid in grapevines treated by chemigation. Crop Prot. 2006, 25, 831–834. [Google Scholar] [CrossRef]
- Prager, S.M.; Vindiola, B.; Kund, G.S.; Byrne, F.J.; Trumble, J.T. Considerations for the use of neonicotinoid pesticides in management of Bactericera cockerelli (Šulk) (Hemiptera: Triozidae). Crop Prot. 2013, 54, 84–91. [Google Scholar] [CrossRef]
- Tomizawa, M.; Casida, J.E. Neonicotinoid insecticide toxicology: Mechanisms of selective action. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 247–268. [Google Scholar] [CrossRef] [Green Version]
- Crossley, M.S.; Rondon, S.I.; Schoville, S.D. A comparison of resistance to imidacloprid in Colorado potato beetle (Leptinotarsa decemlineata Say) populations collected in the Northwest and Midwest US. Am. J. Potato Res. 2018, 95, 495–503. [Google Scholar] [CrossRef]
- Liu, D.; Trumble, J.T. Comparative fitness of invasive and native populations of the potato psyllid (Bactericera cockerelli). Entomol. Exp. Appl. 2007, 123, 35–42. [Google Scholar] [CrossRef]
- Prager, S.M.; Trumble, J.T. Psyllids: Biology, ecology, and management. In Sustainable Management of Arthropod Pests of Tomato, 1st ed.; Wakil, W., Brust, G.E., Perring, T.D., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 163–176. [Google Scholar]
- Rinkevich, F.D.; Du, Y.; Dong, K. Diversity and convergence of sodium channel mutations involved in resistance to pyrethroids. Pestic. Biochem. Physiol. 2013, 106, 93–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Yang, C.; Yang, Y.; Li, J.; Lin, K.; Li, M.; Qiu, X. Distribution and frequency of G119S mutation in ace-1 gene within Anopheles sinensis populations from Guangxi, China. Malar. J. 2015, 14, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claudianos, C.; Ranson, H.; Johnson, R.; Biswas, S.; Schuler, M.; Berenbaum, M.; Feyereisen, R.; Oakeshott, J.G. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006, 15, 615–636. [Google Scholar] [CrossRef] [Green Version]
- Ingham, V.A.; Jones, C.M.; Pignatelli, P.; Balabanidou, V.; Vontas, J.; Wagstaff, S.C.; Moore, J.D.; Ranson, H. Dissecting the organ specificity of insecticide resistance candidate genes in Anopheles gambiae: Known and novel candidate genes. BMC Genom. 2014, 15, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Liang, P.; Fang, K.; Chu, D. Silence of inositol 1,4,5-trisphosphate receptor expression decreases cyantraniliprole susceptibility in Bemisia tabaci. Pestic. Biochem. Physiol. 2017, 142, 162–169. [Google Scholar] [CrossRef]
- Fournier-Level, A.; Good, R.T.; Wilcox, S.A.; Rane, R.V.; Schiffer, M.; Chen, W.; Battlay, P.; Perry, T.; Batterham, P.; Hoffmann, A.A.; et al. The spread of resistance to imidacloprid is restricted by thermotolerance in natural populations of Drosophila melanogaster. Nat. Ecol. Evol. 2019, 3, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Glunt, K.D.; Coetzee, M.; Huijben, S.; Koffi, A.A.; Lynch, P.A.; N’Guessan, R.; Oumbouke, W.A.; Sternberg, E.D.; Thomas, M.B. Empirical and theoretical investigation into the potential impacts of insecticide resistance on the effectiveness of insecticide-treated bed nets. Evol. Appl. 2018, 11, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [Green Version]
- Foll, M.; Gaggiotti, O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, s13742-015. [Google Scholar] [CrossRef] [PubMed]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
- Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Liu, H.; Miller, M.S.; Swank, D.M.; Kronert, W.A.; Maughan, D.W.; Bernstein, S.I. Paramyosin phosphorylation site disruption affects indirect flight muscle stiffness and power generation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2005, 102, 10522–10527. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.S.; Jackson, S.P. Ubiquitylation, neddylation and the DNA damage response. Open Biol. 2015, 5, 150018. [Google Scholar] [CrossRef] [Green Version]
- George, S.; Palli, S.R. Histone Deacetylase 11 Knockdown blocks larval development and metamorphosis in the red flour beetle, Tribolium castaneum. Front. Genet. 2020, 11, 683–695. [Google Scholar] [CrossRef]
- Tauszig, S.; Jouanguy, E.; Hoffmann, J.A.; Imler, J.-L. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 97, 10520–10525. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, A.; Habineza, P.; Wang, X.; Xiao, R.; Ji, T.; Hou, Y.; Shi, Z. Spätzle homolog-mediated Toll-like pathway regulates innate immune responses to maintain the homeostasis of gut microbiota in the red palm weevil, Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Front. Microbiol. 2020, 11, 846–857. [Google Scholar] [CrossRef]
- Jiang, D.; Du, X.; Wang, C.; Zhang, S.; Wang, G. CD109 antigen-like gene is induced by ecdysone signaling and involved in the cellular immunity of Helicoverpa armigera. Biosci. Biotechnol. Biochem. 2020, 84, 1183–1190. [Google Scholar] [CrossRef]
- Vézilier, J.; Nicot, A.; De Lorgeril, J.; Gandon, S.; Rivero, A. The impact of insecticide resistance on Culex pipiens immunity. Evol. Appl. 2013, 6, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, R.D.; Mitchell, R.D., III; Deguenon, J.M.; Ponnusamy, L.; Reisig, D.; Pozo-Valdivia, A.D.; Kurtz, R.W.; Roe, R.M. Multiple known mechanisms and a possible role of an enhanced immune system in Bt-resistance in a field population of the bollworm, Helicoverpa zea: Differences in gene expression with RNAseq. Int. J. Mol. Sci. 2020, 21, 6528. [Google Scholar] [CrossRef] [PubMed]
- Martelli, F.; Zhongyuan, Z.; Wang, J.; Wong, C.O.; Karagas, N.E.; Roessner, U.; Rupasinghe, T.; Venkatachalam, K.; Perry, T.; Bellen, H.J.; et al. Low doses of the neonicotinoid insecticide imidacloprid induce ROS triggering neurological and metabolic impairments in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 117, 25840–25850. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Feyereisen, R. Evolution of insect P450. Biochem. Soc. Trans. 2006, 34, 1252–1255. [Google Scholar] [CrossRef] [Green Version]
- Feyereisen, R. Insect CYP genes and P450 enzymes. In Insect Molecular Biology and Biochemistry, 1st ed.; Gilbert, L.I., Ed.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 236–316. [Google Scholar]
- Feyereisen, R.; Dermauw, W.; Van Leeuwen, T. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic. Biochem. Physiol. 2015, 121, 61–77. [Google Scholar] [CrossRef]
- Corbel, V.; N’guessan, R.; Brengues, C.; Chandre, F.; Djogbenou, L.; Martin, T.; Akogbeto, M.; Hougard, J.M.; Rowland, M. Multiple insecticide resistance mechanisms in Anopheles gambiae and Culex quinquefasciatus from Benin, West Africa. Acta Trop. 2007, 101, 207–216. [Google Scholar] [CrossRef]
- de Little, S.C.; Edwards, O.; van Rooyen, A.R.; Weeks, A.; Umina, P.A. Discovery of metabolic resistance to neonicotinoids in green peach aphids (Myzus persicae) in Australia. Pest Manag. Sci. 2017, 73, 1611–1617. [Google Scholar] [CrossRef]
- Weetman, D.; Wilding, C.S.; Neafsey, D.E.; Müller, P.; Ochomo, E.; Isaacs, A.T.; Steen, K.; Rippon, E.J.; Morgan, J.C.; Mawejje, H.D.; et al. Candidate-gene based GWAS identifies reproducible DNA markers for metabolic pyrethroid resistance from standing genetic variation in East African Anopheles gambiae. Sci. Rep. 2018, 8, 2920–2931. [Google Scholar] [CrossRef]
- Traverso, L.; Lavore, A.; Sierra, I.; Palacio, V.; Martinez-Barnetche, J.; Latorre-Estivalis, J.M.; Mougabure-Cueto, G.; Francini, F.; Lorenzo, M.G.; Rodríguez, M.H.; et al. Comparative and functional triatomine genomics reveals reductions and expansions in insecticide resistance-related gene families. PLoS Negl. Trop. Dis. 2017, 11, e0005313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Umina, P.A.; Rašić, G.; Bell, N.; Fang, J.; Lord, A.; Hoffmann, A.A. Origin of resistance to pyrethroids in the redlegged earth mite (Halotydeus destructor) in Australia: Repeated local evolution and migration. Pest Manag. Sci. 2020, 76, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Epstein, B.; Kelley, J.L.; Zheng, Q.; Bergland, A.O.; Castillo Carrillo, C.I.; Jensen, A.S.; Dahan, J.; Karasev, A.V.; Snyder, W.E. Using NextRAD sequencing to infer movement of herbivores among host plants. PLoS ONE 2017, 12, e0177742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.; Meier, A.R.; Epstein, B.; Bergland, A.O.; Castillo Carrillo, C.I.; Cooper, W.R.; Cruzado, R.K.; Horton, D.R.; Jensen, A.S.; Kelley, J.L.; et al. Host plants and Wolbachia shape the population genetics of sympatric herbivore populations. Evol. Appl. 2020, 13, 2740–2753. [Google Scholar] [CrossRef]
- Toews, D.P.; Brelsford, A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012, 21, 3907–3930. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).