Discovery of Novel 3,4-Dihydro-2(1H)-Quinolinone Sulfonamide Derivatives as New Tubulin Polymerization Inhibitors with Anti-Cancer Activity
Abstract
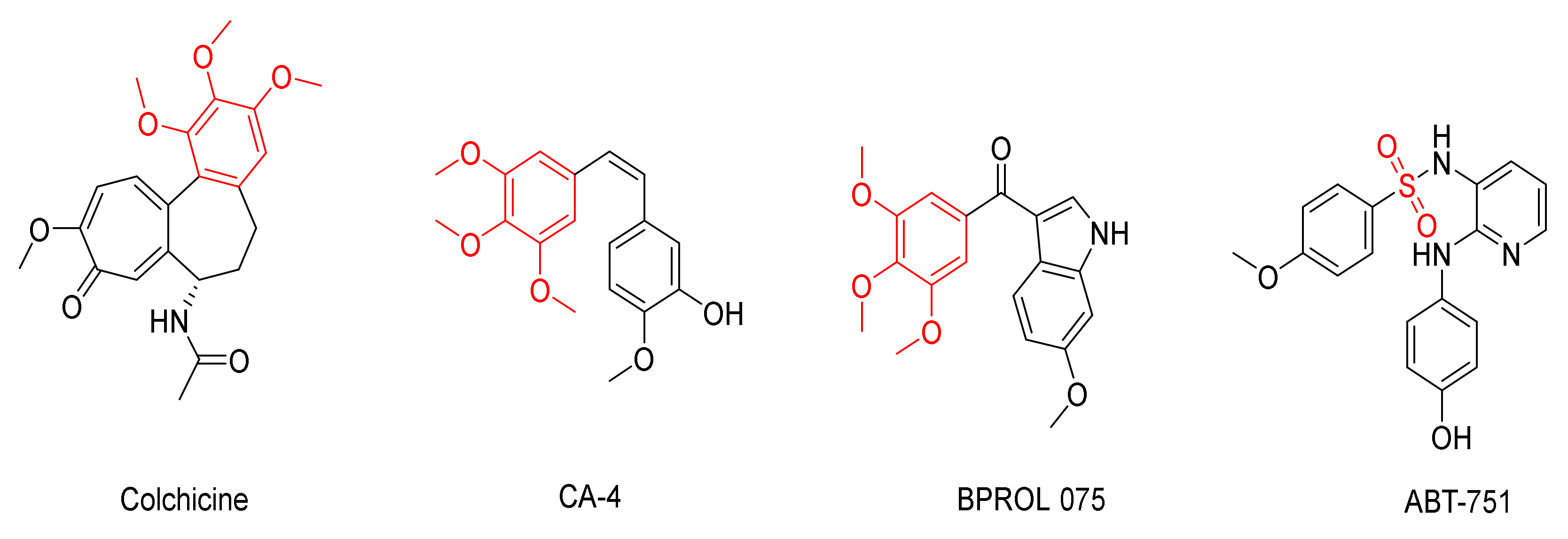
:1. Introduction
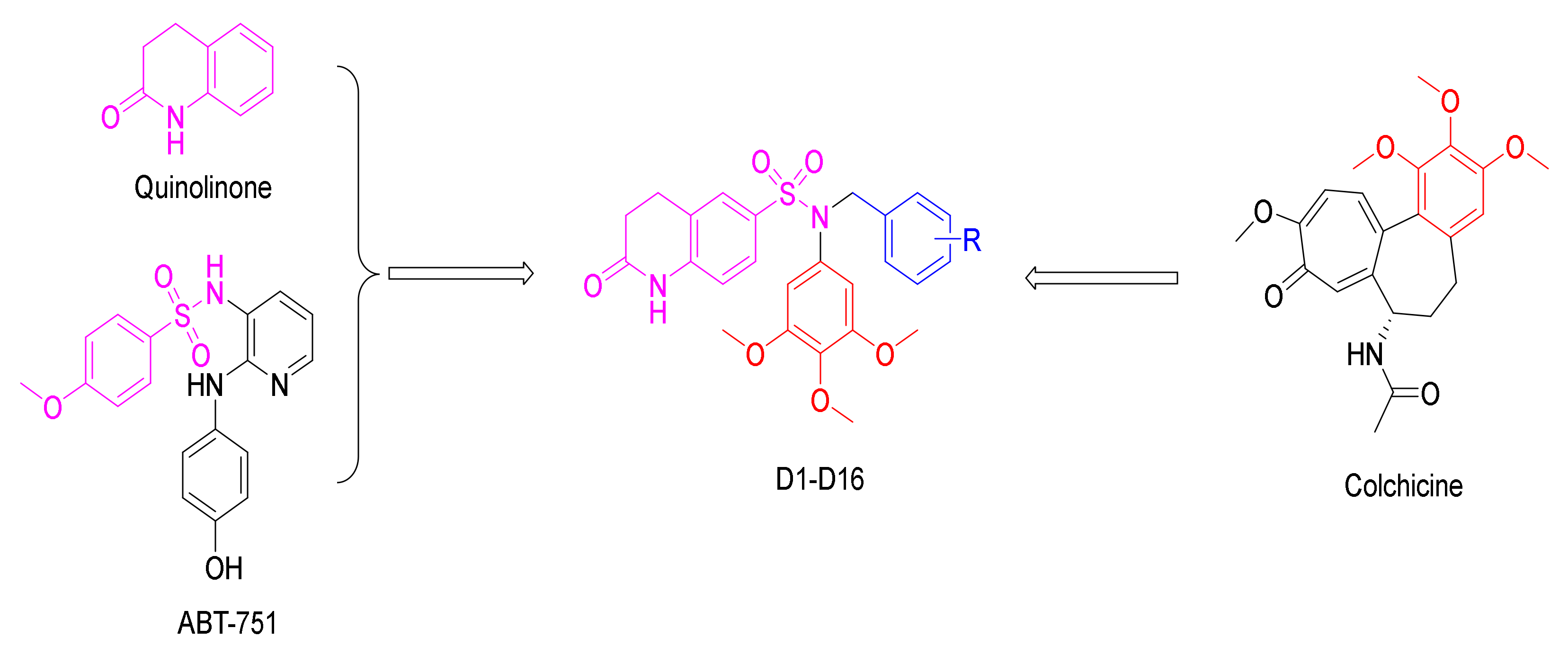
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. In Vitro Anticancer Activity
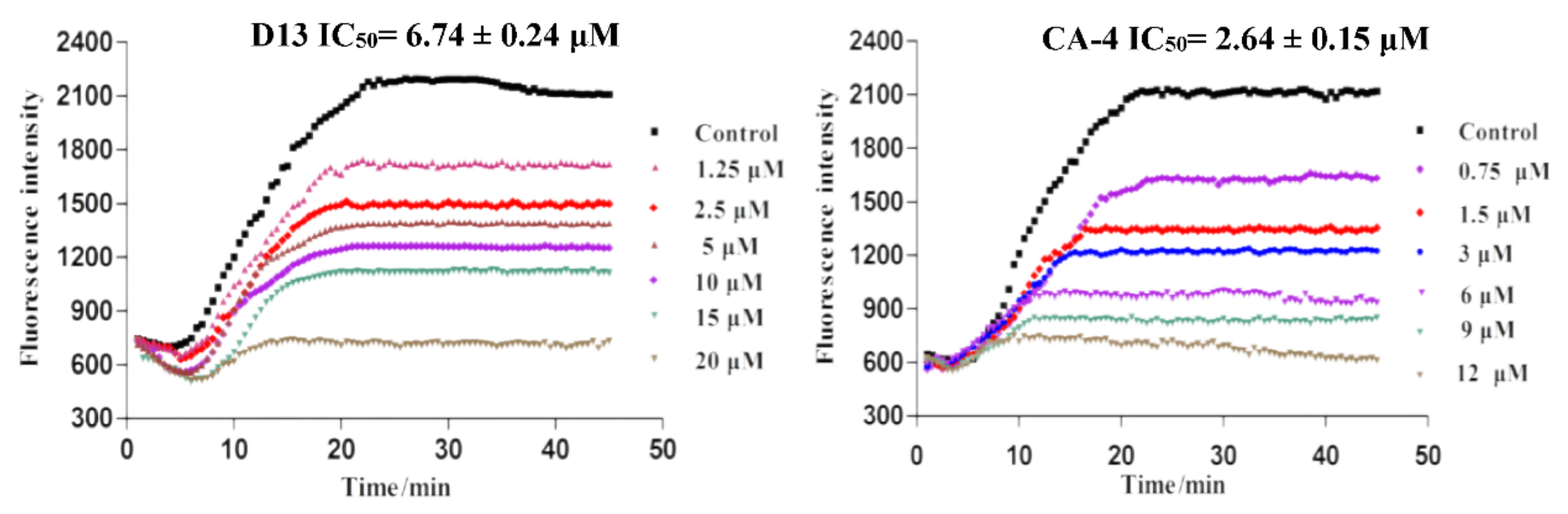
2.2.2. Inhibition of Tubulin Polymerization of D13
2.3. Docking Analysis
3. Conclusions
4. Experimental Section
4.1. Chemistry General Methods
4.2. Procedure for the Synthesis of Compound B
4.3. General Synthesis Process of Compounds C1–C16
4.4. General Synthesis Process of Compounds D1–D16
4.4.1. N-benzyl-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-7-sulfonamide (D1)
4.4.2. N-(4-bromobenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-7-sulfonamide (D2)
4.4.3. N-(4-fluorobenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D3)
4.4.4. N-(4-chlorobenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D4)
4.4.5. N-(4-methylbenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D5)
4.4.6. N-(4-(dimethylamino)benzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D6)
4.4.7. N-(3-methylbenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D7)
4.4.8. N-(2-fluorobenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D8)
4.4.9. N-(2-chlorobenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D9)
4.4.10. N-(3,4-dichlorobenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D10)
4.4.11. N-(5-chloro-2-fluorobenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-8-sulfonamide (D11)
4.4.12. N-(3-methoxybenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D12)
4.4.13. N-(4-methoxybenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D13)
4.4.14. N-(3,4-dimethoxybenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D14)
4.4.15. O-(2,5-dimethoxybenzyl)-2-oxo-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D15)
4.4.16. 2-oxo-N-(3,4,5-trimethoxybenzyl)-N-(3,4,5-trimethoxyphenyl)-1,2,3,4-tetrahydroquinoline-6-sulfonamide (D16)
4.5. In Vitro Anticancer Experiment
4.6. In Vitro Tubulin Polymerization Assay
4.7. Molecular Modeling
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lehotzky, A.; Tokesi, N.; Gonzalez-Alvarez, L.; Merino, V.; Bermejo, M.; Orosz, F.; Lau, P.; Kovacs, G.G.; Ovadi, J. Progress in the development of early diagnosis and a drug with unique pharmacology to improve cancer therapy. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2008, 366, 3599–3617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aly, H.A. Cancer therapy and vaccination. J. Immunol. Methods 2012, 382, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Nogales, E. Structural insights into microtubule function. Annu. Rev. Biochem. 2000, 69, 277–302. [Google Scholar] [CrossRef] [PubMed]
- Quasthoff, S.; Hartung, H.P. Chemotherapy-induced peripheral neuropathy. J. Neurol. 2002, 249, 9–17. [Google Scholar] [CrossRef]
- Ems-McClung, S.C.; Walczak, C.E. Kinesin-13s in mitosis: Key players in the spatial and temporal organization of spindle microtubules. Semin. Cell Dev. Biol. 2010, 21, 276–282. [Google Scholar] [CrossRef] [Green Version]
- Wittmann, T.; Hyman, A.; Desai, A. The spindle: A dynamic assembly of microtubules and motors. Nat. Cell. Biol. 2001, 3, E28–E34. [Google Scholar] [CrossRef]
- Horio, T.; Murata, T. The role of dynamic instability in microtubule organization. Front. Plant Sci. 2014, 5, 511. [Google Scholar] [CrossRef] [Green Version]
- Pellegrini, F.; Budman, D.R. Review: Tubulin function, action of antitubulin drugs, and new drug development. Cancer Investig. 2005, 23, 264–273. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [Green Version]
- Prota, A.E.; Bargsten, K.; Zurwerra, D.; Field, J.J.; Díaz, J.F.; Altmann, K.H.; Steinmetz, M.O. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science 2013, 339, 587–590. [Google Scholar] [CrossRef] [Green Version]
- Dumontet, C.; Jordan, M.A. Microtubule-binding agents: A dynamic field of cancer therapeutics. Nat. Rev. Drug Discov. 2010, 9, 790–803. [Google Scholar] [CrossRef] [Green Version]
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Nitika, V.; Kapil, K. Microtubule targeting agents: A benchmark in cancer therapy. Curr. Drug Ther. 2013, 8, 189–196. [Google Scholar] [CrossRef]
- Protaa, A.E.; Bargstena, K.; Diazb, J.F.; Marshc, M.; Cuevasd, C.; Linigere, M.; Neuhause, C.; Andreub, J.M.; Altmanne, K.-H.; Steinmetza, M.O. A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs. Proc. Natl. Acad. Sci. USA 2014, 111, 13817–13821. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Yu, Y.; Li, Y.; Yan, W.; Ye, H.; Niu, L.; Tang, M.; Wang, Z.; Yang, Z.; Pei, H.; et al. Cevipabulin-tubulin complex reveals a novel agent binding site on α-tubulin with tubulin degradation effect. Sci. Adv. 2021, 7, eabg4168. [Google Scholar] [CrossRef]
- Jordan, M. Mechanism of action of antitumor drugs that interact with mi-crotubules and tubulin Anti-Cancer Agents. Curr. Med. Chem. Anti-Cancer Agents 2002, 2, 1–17. [Google Scholar] [CrossRef]
- Vindya, N.G.; Sharma, N.; Yadav, M.; Ethiraj, K.R. Tubulins-the target for anticancer therapy. Curr. Top. Med. Chem. 2015, 15, 73–82. [Google Scholar] [CrossRef]
- Karki, R.; Mariani, M.; Andreoli, M.; He, S.; Scambia, G.; Shahabi, S.; Ferlini, C. βIII-Tubulin: Biomarker of taxane resistance or drug target? Expert Opin.Ther. Targets 2013, 17, 461–472. [Google Scholar] [CrossRef]
- Kaur, R.; Kaur, G.; Gill, R.K.; Soni, R.; Bariwal, J. Recent developments in tubulin polymerization inhibitors: An overview. Eur. J. Med. Chem. 2014, 87, 89–124. [Google Scholar] [CrossRef]
- Liu, Y.M.; Chen, H.L.; Lee, H.Y.; Liou, J.P. Tubulin inhibitors: A patent review. Expert Opin. Ther. Pat. 2014, 24, 69–88. [Google Scholar] [CrossRef]
- Meng, T.; Wang, W.; Zhang, Z.; Ma, L.; Zhang, Y.; Miao, Z.; Shen, J. Synthesis and biological evaluation of 6H-pyrido[2′,1′:2,3]imidazo[4,5-c]isoquinolin5(6H)-ones as antimitotic agents and inhibitors of tubulin polymerization. Bioorg. Med. Chem. 2014, 22, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Tokali, F.S.; Taslimi, P.; Usanmaz, H.; Karaman, M.; Sendil, K. Synthesis, characterization, biological activity and molecular docking studies of novel schiff bases derived from thiosemicarbazide: Biochemical and computational approach. J. Mol. Struct. 2021, 1231, 129666. [Google Scholar] [CrossRef]
- Hijji, Y.; Benjamin, E.; Butcher, R. Crystal structure, spectral, thermal and experimental/computational investigation of Anthracen-benzo[d]thiazol-2-amine new Schiff base derivative. J. Mol. Struct. 2020, 1229, 129824. [Google Scholar] [CrossRef]
- Tashima, T.; Murata, H.; Kodama, H. Design and synthesis of novel and highly-active pan-histone deacetylase (pan-HDAC) inhibitors. Bioorg. Med. Chem. 2014, 22, 3720–3731. [Google Scholar] [CrossRef]



| Compd. | R | (μM) IC50 b | ||||
|---|---|---|---|---|---|---|
| HeLa | A549 | HCT116 | HepG-2 | L02 | ||
| D1 | -H | 13.46 ± 2.15 | 14.15 ±1.26 | 25.90 ± 0.98 | 9.87 ± 2.49 | 20.73 ± 3.12 |
| D2 | 4-Br | 3.98 ± 0.25 | 4.19 ± 1.27 | 1.95 ± 0.77 | 7.53 ± 0.27 | 10.56 ± 1.26 |
| D3 | 4-F | 7.54 ± 1.48 | 18.24 ± 4.11 | 3.87 ± 0.12 | 6.31 ± 1.21 | 5.31 ± 1.08 |
| D4 | 4-Cl | 9.67 ± 2.1 | 8.49 ± 1.77 | 4.21 ± 0.5 | 4.12 ± 0.55 | 6.35 ± 2.03 |
| D5 | 4-CH3 | 1.34 ± 0.3 | 2.96 ± 0.13 | 1.13 ± 0.01 | 2.36 ± 0.11 | 3.05 ± 0.19 |
| D6 | 4-N(CH3)2 | 3.63 ± 0.98 | 2.42 ± 0.46 | 2.05 ± 0.11 | 4.74 ± 1.56 | 5.31 ± 1.02 |
| D7 | 3-CH3 | 36.49 ± 3.59 | 34.99 ± 6.74 | 28.17 ± 4.26 | 44.31 ± 4.31 | 30.21 ± 2.56 |
| D8 | 2-F | 21.62 ± 3.56 | >100 | 17.54 ± 1.33 | 50.91 ± 6.77 | 60.21 ± 4.21 |
| D9 | 2-Cl | 16.94 ± 4.11 | 31.68 ± 4.55 | 12.16 ± 2.89 | 27.46 ± 3.28 | 36.25 ± 4.21 |
| D10 | 3,4-Cl | 75.29 ± 10.32 | >100 | 51.24 ± 13.49 | - | 48.26 ± 6.64 |
| D11 | 2-Cl-5-F | 38.41 ± 4.57 | 19.27 ± 2.38 | 25.48 ± 5.23 | 58.37 ± 6.93 | 65.31 ± 4.25 |
| D12 | 3-OCH3 | 54.64 ± 8.15 | - | 48.23 ± 7.46 | 64.12 ± 7.56 | 56.26 ± 2.15 |
| D13 | 4-OCH3 | 1.46 ± 0.11 | 1.57 ± 0.25 | 0.94 ± 0.31 | 1.82 ± 0.97 | 3.61 ± 1.02 |
| D14 | 3,4-OCH3 | >100 | 49.28 ± 12.53 | 87.81 ± 15.23 | - | 66.52 ± 7.81 |
| D15 | 2,5-OCH3 | 86.36 ± 14.23 | - | >100 | 75.36 ± 15.44 | >100 |
| D16 | 3,4,5-OCH3 | 4.91 ± 0.76 | 7.34 ± 1.44 | 2.08 ± 0.24 | 5.48 ± 1.23 | 4.25 ± 0.87 |
| 5-Fu | - | 29.57 ± 3.21 | 8.71 ± 0.23 | 11.67 ± 2.37 | 18.31 ± 0.76 | 20.04 ± 1.25 |
| CA-4 | - | 0.02 ± 0.001 | 0.12 ± 0.023 | 0.04 ± 0.016 | 0.05 ± 0.021 | 0.15 ± 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Gong, G.-H. Discovery of Novel 3,4-Dihydro-2(1H)-Quinolinone Sulfonamide Derivatives as New Tubulin Polymerization Inhibitors with Anti-Cancer Activity. Molecules 2022, 27, 1537. https://doi.org/10.3390/molecules27051537
Ma J, Gong G-H. Discovery of Novel 3,4-Dihydro-2(1H)-Quinolinone Sulfonamide Derivatives as New Tubulin Polymerization Inhibitors with Anti-Cancer Activity. Molecules. 2022; 27(5):1537. https://doi.org/10.3390/molecules27051537
Chicago/Turabian StyleMa, Juan, and Guo-Hua Gong. 2022. "Discovery of Novel 3,4-Dihydro-2(1H)-Quinolinone Sulfonamide Derivatives as New Tubulin Polymerization Inhibitors with Anti-Cancer Activity" Molecules 27, no. 5: 1537. https://doi.org/10.3390/molecules27051537
APA StyleMa, J., & Gong, G.-H. (2022). Discovery of Novel 3,4-Dihydro-2(1H)-Quinolinone Sulfonamide Derivatives as New Tubulin Polymerization Inhibitors with Anti-Cancer Activity. Molecules, 27(5), 1537. https://doi.org/10.3390/molecules27051537





