Abstract
Lignocellulosic biomass from the secondary cell walls of plants has a veritable potential to provide some of the most appropriate raw materials for producing second-generation biofuels. Therefore, we must first understand how plants synthesize these complex secondary cell walls that consist of cellulose, hemicellulose, and lignin in order to deconstruct them later on into simple sugars to produce bioethanol via fermentation. Knotted-like homeobox (KNOX) genes encode homeodomain-containing transcription factors (TFs) that modulate various important developmental processes in plants. While Class I KNOX TF genes are mainly expressed in the shoot apical meristems of both monocot and eudicot plants and are involved in meristem maintenance and/or formation, Class II KNOX TF genes exhibit diverse expression patterns and their precise functions have mostly remained unknown, until recently. The expression patterns of Class II KNOX TF genes in Arabidopsis, namely KNAT3, KNAT4, KNAT5, and KNAT7, suggest that TFs encoded by at least some of these genes, such as KNAT7 and KNAT3, may play a significant role in secondary cell wall formation. Specifically, the expression of the KNAT7 gene is regulated by upstream TFs, such as SND1 and MYB46, while KNAT7 interacts with other cell wall proteins, such as KNAT3, MYB75, OFPs, and BLHs, to regulate secondary cell wall formation. Moreover, KNAT7 directly regulates the expression of some xylan synthesis genes. In this review, we summarize the current mechanistic understanding of the roles of Class II KNOX TFs in secondary cell wall formation. Recent success with the genetic manipulation of Class II KNOX TFs suggests that this may be one of the biotechnological strategies to improve plant feedstocks for bioethanol production.
1. Introduction
Increasing global demand for petroleum-based transportation fuels has created an imperative need for the search and development of alternative, sustainable, and renewable sources of bioenergy. First-generation biofuels are produced from starch and sugars that come from agricultural crops. Such applications are often associated with direct competition with food resources and are with some important ethical, ecological, and societal issues [1]. Second-generation biofuels are produced from lignocellulosic biomass derived from the secondary cell walls (SCWs) of plants that can be used to produce alternative transportation fuels, such as bioethanol. Despite its abundance, only a small portion of lignocellulosic biomass is presently used for bioethanol production, owing to its recalcitrance for the bioconversion to bioethanol. Additionally, converting woody biomass into fermentable sugars still requires high input technologies involving extensive pre-treatments and expensive enzymes [2,3]. Hence, a clear understanding of the plant metabolic processes influencing SCW properties will assist in improving plant feedstocks for bioethanol production [3].
Second-generation biofuels have become an important component of the global bioenergy agenda [2]. The use of plant biomass for the production of bioethanol has an enormous potential to revolutionize the global bioenergy outlook. Scientists have discovered novel ways of improving lignocellulosic biomass production in bioenergy crops and trees, such as switchgrass and poplars, that could be used for efficient biofuel production [4,5,6]. The SCW is mainly composed of cellulose, hemicelluloses, and lignin; one of the primary roles of SCW is to confer mechanical strength to plant tissues [6]. However, the main carbohydrate components of SCWs—cellulose and hemicellulose—can be deconstructed into simple sugars (saccharification), and these sugars can subsequently be fermented to produce bioethanol [3]. During the last two decades, several studies have been initiated to understand and modify the biosynthesis of cellulose, hemicellulose, and lignin through the manipulation of genes involved in these pathways [6]. In addition, several members of the NAC, MYB, and KNOX transcription factor (TF) families have been studied to elucidate their regulatory roles in SCW biosynthesis [7,8,9,10]. These TFs function by regulating the SCW biosynthetic genes that encode cellulose synthases (CesAs), xylan synthases, and lignin biosynthetic pathway enzymes. One of the Class II KNOTTED1-like homeodomain (KNOX) genes, KNAT7, has recently gained attention for its potential role in the transcriptional network regulating SCW biosynthesis [11,12,13,14,15,16,17]. This comprehensive review focuses on the recent developments in our understanding of the transcriptional networks involving Class II KNOX TFs in the regulation of SCW biosynthesis.
1.1. KNOX Genes and Encoded KNOX Proteins in Plants
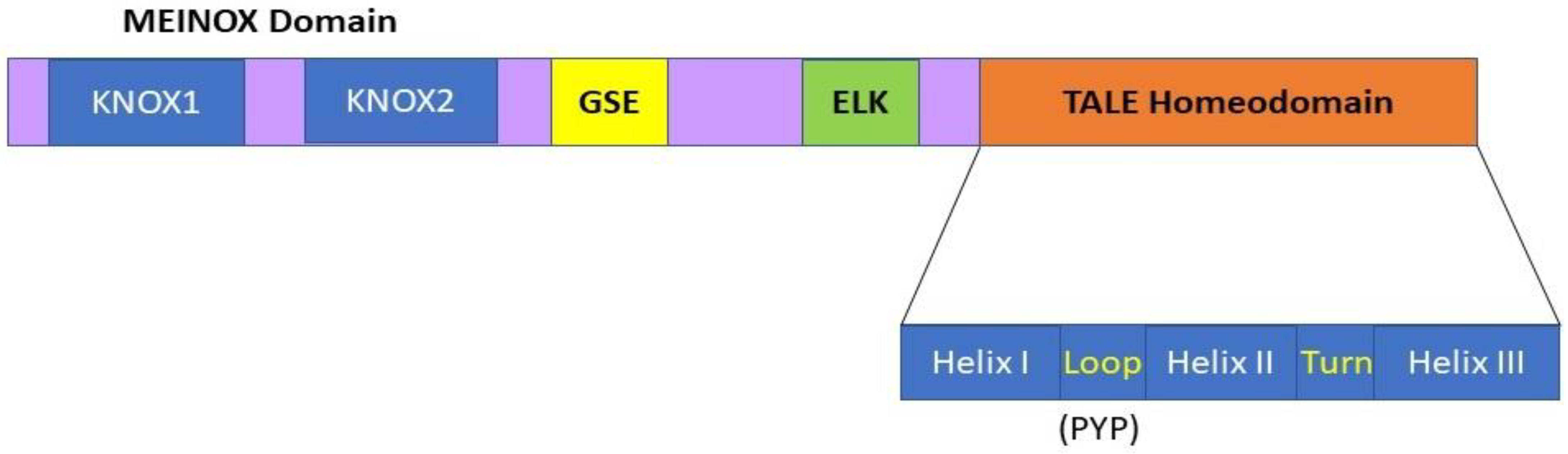
The KNOX genes are members of one of the ancestral gene families involved in the transition of plants from an aquatic to a terrestrial habitat during evolution [18]. KNOX genes encode homeodomain (HD)-containing TFs involved in various developmental processes. Typical HD proteins contain 60 amino acids, while the HD of KNOX proteins contains a highly conserved 63-amino acid stretch consisting of three ∝-helices that form a helix-turn-helix-type DNA binding motif [19] (Figure 1). Due to the presence of three extra amino acids between the first and second helices, all KNOX TF proteins are included in the TALE (three amino acid loop extension) superclass, the members of which are evolutionarily conserved from single-cell algae to higher plants [20]. The sequence immediately upstream of the HD, the ELK domain, has been suggested to function as a nuclear localization signal and be involved in protein–protein interactions [20]. In addition to the HD and ELK domains, a stretch of 100 amino acids located at the N terminus of almost all KNOX proteins, the MEINOX domain, also functions in protein–protein interactions [20]. This MEINOX domain in plants consists of two smaller domains, KNOX1 and KNOX2, separated by a poorly conserved linker sequence (Figure 1).
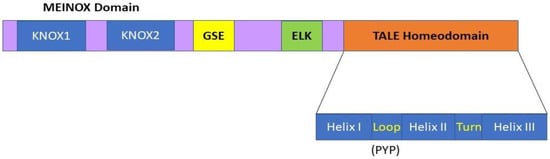
Figure 1.
KNOX protein domain organization comprising MEINOX, ELK, and the TALE homeodomain (HD). The TALE homeodomain consists of three α-helices which comprise a helix-turn-helix type DNA binding motif, and contains three extra residues (PYP) in the loop between the first and second helices as compared to typical HDs. The MEINOX domain is present at the N terminus of the KNOX proteins, and it functions during protein–protein interactions. This MEINOX domain in plants is made of two smaller domains, KNOX1 and KNOX2, separated by a linker sequence. The ELK domain has been suggested to function as a nuclear localization signal and be involved in protein–protein interactions. The relatively small and less well-conserved amino acid motif located between the MEINOX and ELK domains is called the GSE domain; its function is not well understood.
Plant KNOX genes are divided into three subclasses based on their sequence similarity within the HD encoding regions, intron positions, expression patterns, and phylogenetic analysis [21,22,23,24]. Class I KNOX genes are similar to the knotted1 gene of maize [25] and are mainly expressed in the shoot apical meristems (SAMs) of both monocot and eudicot plants. The Class I KNOX genes STM, KNAT1/BP, KNAT2, and KNAT6 in Arabidopsis play an important role in the transcriptional regulation of meristem development, leaf shape control, and hormone homeostasis [26]. Loss-of-function mutations in these genes affect meristem maintenance and/or formation [27]. The only member of Class III KNOX gene, KNATM, is involved in the regulation of leaf polarity, leaf shape, and compound leaf development [28]. Four Class II KNOX genes (KNAT3, KNAT4, KNAT5, and KNAT7) in Arabidopsis form a separate monophyletic group and have several orthologues in higher plant genomes with few known functions [23,24,29]. Interestingly, Class II KNOX genes have been suggested to regulate the haploid-to-diploid morphological transition in land plants [18]. The first plant homeobox gene was discovered over 25 years ago; however, we only recently began to decipher the roles of Class II KNOX genes in higher plant growth and development. This review focuses on the functions of Class II KNOX genes and their encoded proteins in higher plants.
1.2. The Expression Patterns of Class II KNOX Genes in Plants Provide Some Clues about Their Functionality in SCW Formation
The only Class II KNOX gene that has recently been well characterized and extensively studied is KNAT7 [12,13,14,15,16,30,31]. The role of KNAT7 TF as a regulator in SCW biosynthesis was first reported in Arabidopsis through the observation of the irx (irregular xylem) phenotype that occurred in a loss-of-function knat7 mutant, irx11 [30]. At the same time, the tight co-expression of the KNAT7 TF gene along with SCW-specific CesA genes was reported using microarrays of Arabidopsis inflorescence stems undergoing SCW formation [11,32]. Promoter-GUS expression studies of AtKNAT7 in Arabidopsis showed that it is highly expressed in developing xylem, phloem fibers, and cambium cells of inflorescence stems [13]. Wang et al. [15] recently examined whether several Class II KNOX genes from Arabidopsis, KNAT3, KNAT4, KNAT5, and KNAT7, were expressed during SCW deposition. All these Class II KNOX gene promoters regulated GUS expression in the vascular bundles in younger stems and intrafascicular fibers and vessel cells in older stems. These observations suggest that these Class II KNOX genes have similar expression patterns during the deposition of the SCWs. Qin et al. [16] also showed that while KNAT7 expression was much higher in stem tissues, KNAT3 expression remained similar in all tissues examined. Promoter-GUS fusions confirmed that KNAT3 and KNAT7 genes are co-expressed in developing xylem and interfascicular fibers in the Arabidopsis stem.
In poplar (Populus balsamifera), the expression of PtKNAT7 gradually increases from the primary cell wall expansion stage to the mature xylem tissue formation stage, and from the youngest to the older internodes of stem [13]. Cotton GhKNL1 was reported to be preferentially expressed in developing cotton fibers during SCW biosynthesis [33]. Switchgrass KNAT7 also appears to be a functional ortholog of Arabidopsis KNAT7, based on its expression patterns [34]. In our laboratory, we studied the expression patterns of two Class II KNOX genes, KNAT3 and KNAT7, in tobacco (Nicotiana benthamiana) [14]. Higher expression of NbKNAT7 was seen in older stems of tobacco showing secondary growth followed by young stems and old leaves, while NbKNAT3 displayed higher expression in older leaves followed by roots and young leaves. These two Class II KNOX genes were also found to be highly expressed during tension wood formation in aspen. The expression of NbKNAT3 and NbKNAT7 in young and old stems indicates that they play a role in wood formation. Thus, Class II KNOX genes are associated with SCW formation during xylem and fiber development.
1.3. Genetic Mutations in Class II KNOX Genes Further Clarify Their Role in SCW Formation
Until 2005, KNAT7 was not often discussed in mutation studies of the Class II KNOX genes; however, a number of Class II KNOX mutations have recently been studied in detail (Table 1). A T-DNA insertion in the intron of the KNAT7 gene resulted in a loss-of-function mutant, irx11, that showed only a moderately weak growth phenotype. The irx11 mutant also exhibited the typical irx phenotype in xylem vessels that were collapsed due to weak SCW formation. The irx11 mutant did not have significantly altered cellulose or xylan content compared to controls. No lignin content of these mutants was reported at that time. While discovering a set of novel TFs involved in SCW biosynthesis, Zhong et al. [12] associated KNAT7 expression with SCW formation, and the dominant repression of KNAT7 (DR-KNAT7 mutants) affected SCW formation in both xylem and fiber cells (Table 1). Curiously, they did not observe the typical irx phenomenon in these DR-KNAT7 mutants, a tell-tale sign of weak SCW formation; however, the cell wall thicknesses of both xylem vessels and fibers were reduced compared to controls (28% down in interfascicular fibers (IF), 26% down in vessels (V), and 80% down in xylary fibers (XF)). Several monosaccharides from the cell walls of DR-KNAT7 mutants were reduced by 20–30%, except for arabinose, which was increased by 18%. The overexpression of KNAT7 did not increase the SCW thickness of fibers and vessels. These results indicated that KNAT7 could be a positive regulator of SCW formation in Arabidopsis. However, Li et al. [13] reported a contrasting observation that loss-of-function mutants in the AtKNAT7 gene resulted in differential thicknesses of interfascicular and xylary fibers compared to vessels (58% up in IF, 35% down in V, and 31% up in XF; Table 1). The vessels walls were thinner, resulting in collapsed xylem vessels that showed the irx phenotype (similar to [30]); however, the interfascicular fibers were significantly thicker than in the wild type control, suggesting that KNAT7 is a transcriptional repressor of fiber SCW formation (but a transcriptional activator of vessel SCW formation). KNAT7 overexpression lines exhibited thinner fiber walls (57% down in IF) with normal vessel and xylary fiber cell walls. Interestingly, even though many SCW-specific cellulose and xylan synthesis genes were upregulated in these mutants, no quantitative changes in cellulose or xylan were reported. All ten lignin synthesis genes tested were upregulated along with an 11% increase in lignin content of cell walls from the stem. Li et al. [30] speculated that KNAT7 interacts with different partner proteins in different cell types to form functionally distinct complexes. Recently, the regulatory roles of other members of the Class II KNOX gene family, KNAT3, KNAT4, and KNAT5, in SCW formation were explored in Arabidopsis inflorescence stems [15,16] (Table 1). Loss-of-function mutants of knat3, knat4, and knat5 did not produce any irx phenotype, as observed in the case of loss-of-function mutants of knat7 [15]. This could be due to the functional redundancy of KNOX II genes. However, knat3/knat7 double mutants displayed an enhanced irx phenotype compared to single knat7 mutants. These double mutants had thinner interfascicular fiber cell walls compared to the single mutants and wild-type plants (40% down in IF) indicating a potentially positive regulatory role of KNAT3 in combination with KNAT7 in xylem SCW development. Even though many SCW genes were highly expressed in the knat3/knat7 double mutants, the cellulose and xylan contents of their cell walls were reduced by 19% and 43%, respectively, and the changes in lignin content were not significant. The Syringyl to Guaicyl (S/G) lignin ratio was down by 83%; however, it was not possible to correlate all these cell wall content changes with the changes in gene expression patterns. In addition, the severe irx phenotype in these double mutants indicated the overlapping roles and partial functional redundancy of KNAT3 and KNAT7 in xylem vessel development during SCW formation. Furthermore, KNAT3 overexpression in Arabidopsis resulted in thickened interfascicular fibers in the SCW of inflorescence stems [15]. This study described KNAT3 as a potential transcriptional activator, working together with KNAT7 to promote SCW biosynthesis in xylem vessels. A synergistic interaction of KNAT3 and KNAT7 to regulate monolignol biosynthesis in Arabidopsis was also reported in another study [16]. Most importantly, they attempted to link S-lignin formation with KNAT3 and KNAT7 expression; however, they could not show the direct transcriptional regulation of a key gene, ferulate 5-hydroxylase (F5H), involved in S-lignin formation by KNAT3 or KNAT7. Similar to the earlier observation by Wang et al. [15], the overexpression of KNAT3 also caused thickening in the interfascicular fiber walls, indicating the positive regulation of interfascicular fiber wall development by KNAT3. These studies by Wang et al. and Qin et al. [15,16] reconciled the paradoxical observations about KNAT7 mutants in Arabidopsis and indicated that KNAT3 and KNAT7 might be working synergistically in fibers, but antagonistically in vessels, during the regulation of SCW biosynthesis (Table 1).

Table 1.
Gene Mutations in Class II KNOX genes and their effect on SCW formation.
Similar observations regarding the function of KNAT7 in SCW formation were reported recently in rice (Table 1). Wang et al. [35] characterized the KNAT7 transcription factor gene that controls SCW wall thickening in the stem. Interestingly, KNAT7 also regulates cell expansion in rice grains. An Osknat7 CRISPR/CAS9 mutant had a thicker wall in fiber cells than that in the wild type, similar to the Arabidopsis knat7 mutant, and transgenic rice plants overexpressing KNAT7 had opposite effects. Interestingly, the Osknat7 mutant also exhibited a larger grain size due to cell expansion in spikelet bracts. The authors proposed that KNAT7 plays a negative regulatory role in SCW formation, similar to Arabidopsis [13]. The negative regulation of SCW formation by KNAT7 TF was also supported by Gong et al. [33] in cotton fibers (Table 1). The dominant repression of cotton KNAT7 (GhKNL1) resulted in abnormal fibers of shorter length in the cotton mutant compared to the controls, suggesting that cell elongation and SCW formation are also related to the function of KNAT7 in cotton. The overexpression of cotton KNAT7 in Arabidopsis produced thinner interfascicular fibers without any changes in the vessel or xylary fiber thickness. Thus, these Class II KNOX genes are involved in SCW formation in various plant species.
1.4. Targeted Genetic Manipulations in Class II KNOX Genes Confirm Their Role in SCW Formation
Apart from the detailed study of Class II KNOX gene mutants, targeted genetic manipulations of Class II KNOX genes, especially, KNAT7 genes have offered some additional clues regarding the functions of these genes (Table 2). While the overexpression of KNAT7 in Arabidopsis did not produce any specific SCW phenotype [12], subsequently, Li et al. [13] reported that such experiments produced thin interfascicular fibers without any changes in wall thickness of vessels suggesting that KNAT7 TF is indeed a regulator of SCW formation.

Table 2.
Genetic manipulation of Class II KNOX genes in different plant species.
The successful complementation of Arabidopsis knat7 mutants with the overexpression of the cotton GhKNL1 gene [33] and poplar PtKNAT7 [13] rescued the defective irx phenotype of the knat7 mutants, suggesting the functional conservation of KNAT7 genes among Arabidopsis, cotton, and poplar. The overexpression of cotton GhKNL1 in Arabidopsis resulted in thinner interfascicular fibers and slightly thinner vessels walls without any change in the xylary fibers compared to control plants [33]. The overexpression of cotton GhKNAT7 significantly reduced the deposition of lignocellulose in the interfascicular fibers of Arabidopsis [24]. However, the SCWs of the xylem fibers and vessels in the transgenic plants did not show any difference from the control plants. The dominant repression of the same cotton KNAT7 orthologue in Arabidopsis produced thinner interfascicular fibers, but thicker vessels and xylary fiber walls, suggesting that KNAT7 can act as a negative or positive regulator of SCW formation in different cell types.
In our laboratory, we generated RNAi lines of tobacco (N. benthamiana) that exhibited reduced expression of KNAT7 [14]. NbKNAT7 downregulated through a transient virus-induced gene silencing (VIGS) system resulted in increased xylem proliferation with thin-walled fiber cells. The glycome analyses of the cell walls showed increased polysaccharide extractability in 1 M KOH extracts of the VIGS-NbKNAT7 lines, suggestive of SCW loosening. In addition, there were increased saccharification rates (40% higher than control) in stems of VIGS-NbKNAT7 lines, which indicated the alteration of cell wall composition in VIGS lines downregulated for the NbKNAT7 gene. Similar to the VIGS results, the stems of stable RNAi lines also showed increased xylem area in their stems as compared to control stems [14]. The cell walls of xylem fibers were thinner (over 50%) in the RNAi lines as compared to vector control lines. The stems of KNAT7 repression lines in tobacco showed reduced expression of SCW genes that resulted in thinner fiber cell walls with altered cell wall composition [14]. All these results suggested that KNAT7 TF might act as a positive regulator of SCW formation in tobacco.
In a recent study performed in our laboratory by Ahlawat et al. [17], transgenic poplar plants overexpressing PtKNAT7 and AtKNAT7 genes showed enhanced expression of the SCW genes CesA8, IRX9, PAL, and CCR, and reduced expression of the same genes in the poplar PtKNAT7 antisense plants. These results further suggested a positive regulatory role of KNAT7 in SCW formation in poplars. In addition, the genetic suppression of KNAT7 in transgenic poplar stems reduced lignin content by about 6% and altered the lignin composition (S/G ratio) of poplar wood with increased saccharification ability (44–53% higher saccharification efficiency over control plants). Yoo et al. [36] also reported a negative correlation between lignin content and the saccharification efficiency of woody tissues and a positive correlation between the S/G ratio and the saccharification efficiency of SCW biomass. Therefore, a change in the S/G ratio and reduction in lignin content might be important for improving the saccharification efficiency of SCW biomass. All the studies reported so far in Arabidopsis and other higher plants suggest that KNAT7 acts differentially as a negative and positive regulator of SCW biosynthesis in different cell types of the same plant or in different plant species.
1.5. Transcriptional Network of the Class II KNOX Genes Involved in SCW Formation
A complex network of transcription factors regulates SCW biosynthesis in plants [8,9,37,38,39,40]. Among these, some Class II KNOX TFs also regulate SCW biogenesis. The major constituents of the SCW are cellulose, lignin, and hemicelluloses [6]. Cellulose is a polymer of glucose synthesized at the plasma membrane by the cellulose synthase (CesA) complex [41], while lignin is composed of guaiacyl (G), syringyl (S), and p-hydroxyphenyl (H) units that are synthesized through the phenylpropanoid pathway [42]. Xylan is the major hemicellulose component in the SCW and consists of a linear backbone of β-(1–4)-linked D-xylosyl (Xyl) residues and α-linked (OMe(methyl)) glucuronic acid (GlcA) side branches [43]. Many specific genes involved in cellulose, hemicellulose, and lignin biosynthesis pathways have previously been identified in plants (e.g., [43,44,45]) and it was anticipated that Class II KNOX TF proteins might directly regulate the expression of some of these genes. The first direct evidence of KNAT7-mediated regulation of xylan biosynthesis in the SCW was reported only recently by He et al. [31], who demonstrated that KNAT7 physically binds to the promoters of the xylan biosynthetic genes, IRREGULAR XYLEM 9 (IRX9), IRX10, IRX14L, and FRAGILE FIBER 8 (FRA8; Figure 2). Wang et al. [46] also reported the involvement of KNAT7 in xylan synthesis during mucilage production. While various cellulose and lignin biosynthesis genes have been shown to be differentially expressed in various knat7 mutants and during the ectopic expression of the KNAT7 gene in transgenic plants, the direct regulation of any of these SCW genes by KNAT7 TF has not yet been reported. In addition, no information is currently available on transcriptional regulation by the TFs encoded by the other three Class II KNOX genes, namely KNAT3, KNAT4, and KNAT5, or their orthologs in any other plant species.
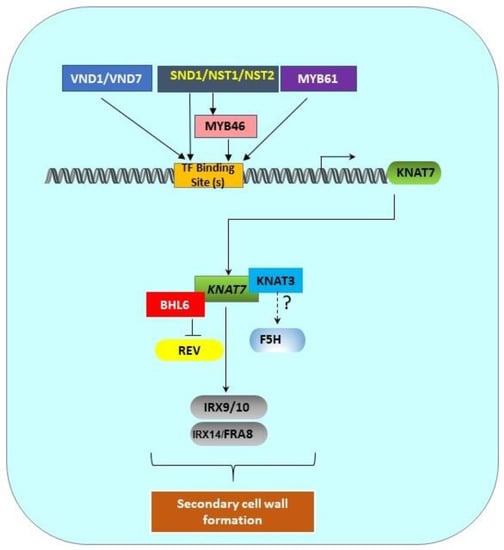
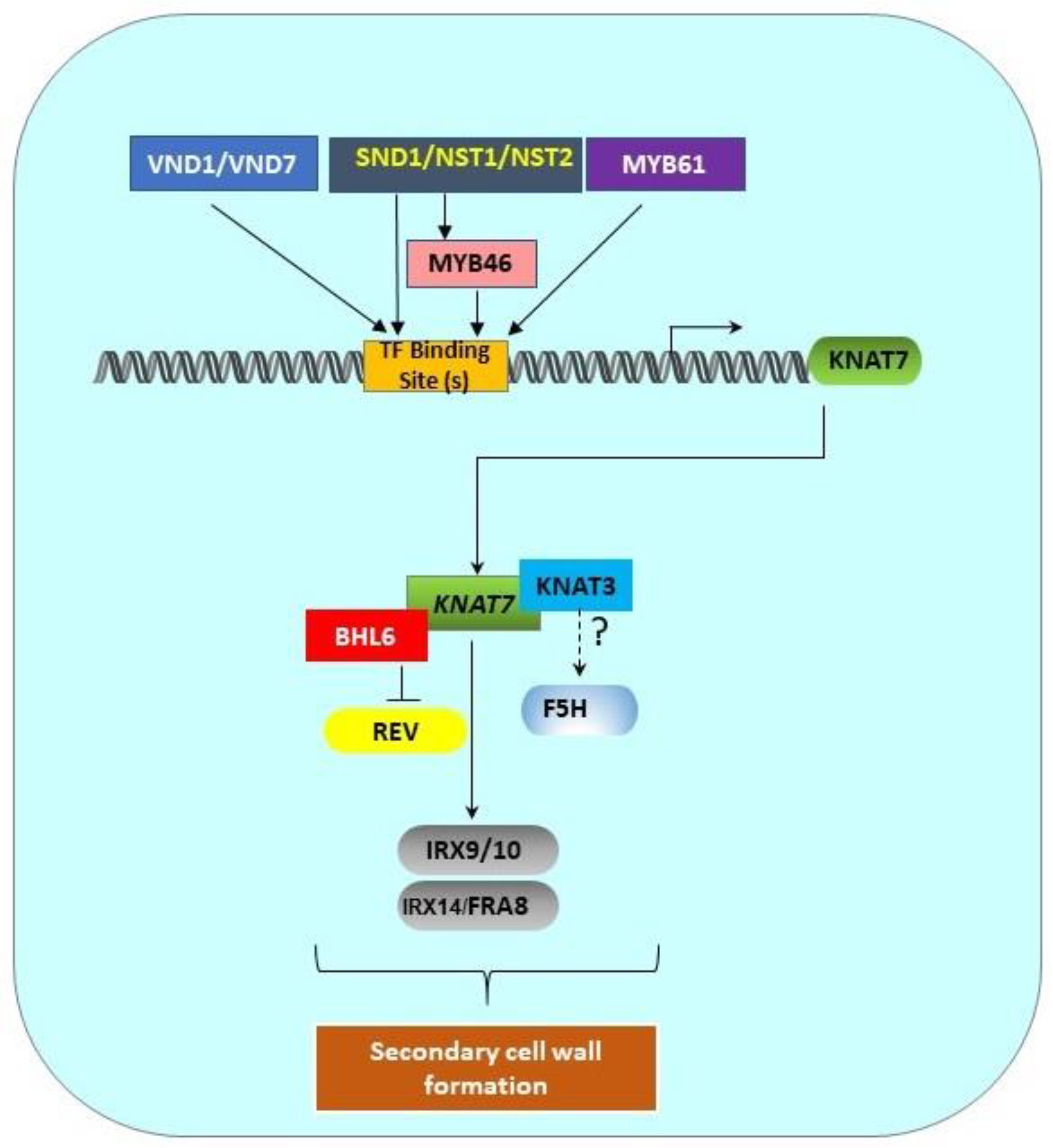
Figure 2.
Transcriptional regulation pathway of KNAT7 gene. SCW-associated upstream transcription factors (MYB61, SND1/NST1/NST2, VND1/VND7) and MYB46 directly bind the binding sites in the KNAT7 gene promoter to regulate the expression of the KNAT7 gene. KNAT7 positively regulates the expression of various xylan synthesis genes (IRX9/10 and IRX14L/FRA8). Interactions between KNAT7 and KNAT3 TFs might regulate F5H expression, and the interactions between KNAT7 and BLH6 negatively regulate the expression of the homeodomain-ZIP (HD-ZIP) TF gene Revoluta. All these interactions ultimately regulate SCW formation in higher plants. All genes are shown as rounded rectangles and proteins are indicated by rectangles.
1.6. Upstream Top- and Mid-Level Master Switches Control the Expression of KNAT7
The expression of KNAT7, a lower-level TF, is directly regulated by top and middle-level upstream TFs, such as NAC and MYB proteins [9,38]. SCW-associated NACs, such as SND1, NST1, NST2, VND6, and VND7, are top-level master switches that directly control KNAT7 expression [12] (Figure 2). Zhong et al. [47] reported that SND1, a master switch of SCW formation in Arabidopsis fibers, directly controls KNAT7 expression. The downregulation of SND1 and its homolog NST1 also caused the downregulation of KNAT7 expression [48]. Zhong et al. [12] identified the direct targets of SND1, and they discovered that the expressions of KNAT7, MYB46, MYB103, and SND3 are directly under the control of SND1. In addition, the SND1 homologs NST1, NST2, VND6, and VND7 were also found to directly activate KNAT7, among many other TFs. Zhong et al. [49] also identified 19-bp- secondary wall NAC binding elements (SNBEs); the KNAT7 promoter has three such SNBEs at -616, -507, and -331 positions relative to the start codon. Interestingly, Zhong et al. [50] further showed that SND1 also directly regulates the expression of another mid-level master switch TF, MYB46 (Figure 2). Furthermore, a recombinant SND1 protein was able to bind to the MYB46 promoter fragment and caused a mobility shift. Chromatin immunoprecipitation assays (ChIP) also enriched MYB46 promoter fragments 3–5-fold, suggesting that SND1 directly binds to the MYB46 promoter. Finally, the overexpression of MYB46 also upregulated KNAT7, and MYB46 appears to be a common target of secondary wall-associated SND1 homologs, including NST1, NST2, VND6, and VND7. Thus, in addition to SND1 and its homologs, MYB46 is also able to directly induce the expression of KNAT7 (Figure 2).
Zhong and Ye [50] showed that a 7-bp long consensus secondary wall MYB-responsive element (SMRE), ACC(A/T)A(A/C)(T/C) in KNAT7 is directly involved in the MYB mediated activation of KNAT7, and the KNAT7 promoter has three such SMREs located at the -802, -763, and -656 positions upstream of the translation initiation codon. Kim et al. [51] identified an eight bp motif that has one additional (T/C) before the SMRE and named it M46RE; KNAT7 promoters have two such motifs. Ko et al. [52] comprehensively reviewed the literature regarding the functions of MYB46 and its close homolog, MYB83, and concluded that the expressions of many important genes involved in cellulose, hemicellulose, and lignin synthesis are directly regulated by MYB46/MYB83. Therefore, it appears that the defects in SCWs of the knat7 mutant might also be due to complex regulation by SND1 homologs and MYB46/83-regulated KNAT7 activities.
KNAT7 functions as a common hub in several transcriptional networks that are involved in xylem differentiation and mucilage production, including the network that involves AtMYB61 [53]. The loss of AtMYB61 function in a mutant resulted in defective xylem production, and it was shown that the MYB61 protein binds directly to an AC-rich element (ACC(A/T)A(A/C/T) in the promoter of AtKNAT7; there are three such AC-rich elements in the KNAT7 promoter at the -704, -665, and -558 positions upstream of the transcription start site. Interestingly, there are high similarities between these AC-rich elements, SMREs, and M46REs. Thus, MYB61 appears to be another upstream regulator of KNAT7 in Arabidopsis (Figure 2).
1.7. Physical Interactions of Class II KNOX TF Proteins with Other Proteins
As homeodomain proteins, Class II KNOX proteins possess a DNA binding capacity, and many accessory proteins are known to physically interact with them (Table 3). Studies have reported that in at least four different plant species, namely Arabidopsis, rice, cotton, and poplars, the general scheme of interactions remains similar, while a number of species-specific variations have also been reported (Table 3).

Table 3.
Protein–protein interactions among Class II KNOX proteins and other TFs.
In Arabidopsis, BEL1 encodes a TALE homeodomain-containing TF that heterodimerizes with KNOX proteins via interactions between the N-terminal region and the homeodomain and MEINOX domain of KNOX proteins [54]. Interestingly, no interactions were evident between BEL1 and KNAT3, KNAT4, and KNAT7 proteins; however, positive interactions were observed between BEL1 and KNAT5 and a few Class I KNOX proteins [54]. Furthermore, the C-terminal domain of BEL1, including the homeodomain, appears to be important for such specific interactions. However, Hackbush et al. [55] subsequently discovered that BEL1 and KNAT5 do not interact, but BLH (BELL-LIKE HOMEODOMAIN) proteins, such as BLH9/KNAT3 and BLH9/KNAT7, interact. In fact, out of four Class II KNOX proteins, eight BLH-proteins interacted with KNAT3, only BLH6 interacted with KNAT4, nine BLH proteins interacted with KNAT5, and two BLH proteins (BLH5 and BLH7) interacted with KNAT7. Another group of proteins that are known to interact with KNOX proteins are the ovate family proteins (OFPs) that are repressors of transcription and are involved in plant growth and development [55,56,57,58,59]. According to Hackbush et al. [55], five OFPs interact with KNAT3 and four different OFPs interact with KNAT4, KNAT5, and KNAT7. However, Li et al. [57] reported that only OFP4 showed strong Y2H interactions with KNAT7, while OFP1 showed only weak interactions. However, bimolecular fluorescence complementation (BiFC) and mutation data confirmed that KNAT7, OFP1, and OFP4 interact and play an important transcriptional repressor role during SCW formation (Table 3). Thus, interactions among BLH, OFPs, and KNOX proteins appear to play some major roles in plant development, including in SCW formation. Contrary to some of these findings, Liu et al. [60] showed that BLH6 specifically interacts with KNAT7, which represses commitment to SCW formation, and this interaction of TFs modulates the expression of the homeodomain-ZIP (HD-ZIP) TF gene, Revoluta (Figure 2). KNAT7 is a putative transcriptional repressor in Arabidopsis leaf protoplasts, and its repression is enhanced by physical interaction with OFP1 and OFP4 [57]. This was confirmed by the presence of irx vessels and altered fiber cell wall phenotypes displayed by ofp4 single and ofp4/ knat7 double mutants, similar to knat7 single mutants. OFP1 and OFP4 are also components of the BLH6–KNAT7 multi-protein complex and may modulate the activity of the BLH6–KNAT7 complex [58]. KNAT7 also physically interacts and forms functional complexes with MYB75 and BLH6, which are involved in SCW formation [61,62] (Table 3). blh6 knockout mutants displayed thicker cell walls in their interfascicular fibers, similar to knat7 mutants [62], suggesting its role as a transcriptional repressor controlling SCW formation in interfascicular fibers through its interactions with KNAT7.
Arabidopsis Class II KNOX TFs are also known to interact with each other. The Y2H data from Hackbush et al. [55] showed that KNAT3 physically interacts with KNAT4, KNAT4 interacts with KNAT7, and KNAT5 interacts with KNAT7. Recently, KNAT7 was reported to form a functional heteromeric complex with KNAT3 and regulate SCW formation, possibly via F5H, a syringyl lignin gene in Arabidopsis (Figure 2 and Table 3). Qin et al. [16] showed that a significant downregulation of the F5H gene, a key gene known to play a significant role in S-lignin formation, occurred in a KNAT3/KNAT7 double mutant and as a result, the S/G lignin ratio was reduced by 83–84% compared to wild type stems. However, yeast-one-hybrid (Y1H) experiments did not show direct binding of either the KNAT7 or KNAT3 protein with the F5H gene promoter. They suggested that a larger complex (including KNAT3 and/or KNAT7) might regulate F5H gene expression in Arabidopsis. They also reported that KNAT3, but not KNAT7, can physically interact with the known top-level master regulators NST1 and NST2, but not SND1 (Table 3). However, neither NST1 nor NST2 alone or in combination with KNAT3 could activate the F5H promoter, suggesting that some other factors required for the formation of this complex were still missing in their experiments. In the poplar genome, there are two closely related F5H (or CAld5H) genes [63,64]. Wang et al. [64] discovered that 12 TFs are co-expressed with CAld5H genes. Only BLH6a and BLH6b are specifically bound to the CAld5H2 promoter, and BLH2 is bound to both the promoters. BLH6 is also a transcriptional repressor. No mention of any KNOX II TFs in the BLH complex was made in this work. Bhargava et al. [65] showed that MYB75 and MYB5 both interact with KNAT7 (Table 3).
Ma et al. [24] reported interactions among GhBEL1-like and GhKNOX II proteins from cotton (Table 3). GhBEL1, GhBLH1, and GhBLH6 interact with GhKNAT7. Moreover, GhKNAT7 interacts with GhMYB75, GhOFP1/5/4, and GhBLH1/5/6, forming heteromers. KNAT7 can form heterodimeric interactions (KNAT7–BLH and KNAT7–MYB) and at the same time can form trimeric interactions (KNAT7-BLH-OFP) to regulate SCW biosynthesis, and the functional conservation of these interactions in different plant species will help us to understand the complex regulatory network of SCW formation. Wang et al. [66] recently showed that the microtubule-associated GhIQD14 protein also interacts with the GhKNL1 protein (GhKNAT7) to regulate SCW formation; Arabidopsis and rice have similar genes encoding similar IQD14 proteins (Table 3).
MYB61 is one of the TFs that directly regulates the expression of KNAT7 in Arabidopsis [53], (Figure 2). In rice, a gibberellin-mediated DELLA-NAC signaling pathway regulates cellulose synthesis [67], and KNAT7, BELL, and OFP2 are known to interact during vasculature development [68]. NAC29/31directly regulates the expression of MYB61, which in turn activates CesA expression (Table 3). Wang et al. [35] recently showed that interactions between KNAT7 and NAC31 suppress the activation of MYB61 expression, suggesting that the order of signal transduction in SCW formation may have changed during the evolution of dicots and monocots [40]. Similarly, biochemical and gene expression studies in rice revealed that KNAT7 negatively regulates cellulose biosynthesis and cell expansion by interacting with NAC31 and a cell growth master regulator, growth regulating factor 4 (GRF4), which is known to control the expression of the expansin genes that regulate grain size (Table 3).
In a recent report, it was shown that KNAT7 and MYB6 heterodimers repressed SCW development in poplar and Arabidopsis while promoting anthocyanin synthesis [69]. The overexpression of MYB6 in transgenic poplar resulted in reduced SCW deposition, accompanied by the repressed expression of SCW biosynthetic genes. MYB6 has a DNA binding domain and interacts with the bHLH protein. KNAT7 also interacts with MYB6, MYB75, and MYB115 based on Y2H and BiFC data (Table 3). Therefore, it appears that the complex interactions of KNAT7 proteins with other cellular proteins play a major role in SCW formation in higher plants.
2. Concluding Remarks and Future Perspectives
Recent studies have indicated that Class II KNOX genes are expressed during SCW formation in Arabidopsis and other higher plants. The expression of these genes in tissues undergoing SCW thickening and the effects of mutations in KNAT3 and KNAT7 genes on SCW synthesis clearly suggests their role in the transcriptional regulation of the genes involved in SCW formation. A clear understanding of the role of KNAT4 and KNAT5 in this process still awaits; if those genes are redundant in function, then their functions need to be ascertained using mutant complementation analysis with other Class II KNOX genes. Although this review focused on SCW formation, a few other metabolic processes in the life cycle of a plant, such as mucilage production, have been associated with Class II KNOX genes. As Romano et al. [53] suggested, KNAT7 appears to be a major hub where several pathways converge to coordinate multiple aspects of resource allocation in plants.
Some ambiguity still exists around whether KNAT7 acts as a transcriptional activator or suppressor in SCW development. The suppression of KNAT7 function increased SCW formation in interfascicular fibers but resulted in reduced cell wall synthesis in xylary fibers with collapsed vessels, suggesting that it is a transcriptional suppressor [13,15]. Quite contrasting results were observed by other authors, who suggested that KNAT7 is a transcriptional activator [12,14,15,17,31,33]. Recent reports by Wang et al. and Qin et al. [15,16] reconciled these observations, suggesting that KNAT7 acts as a suppressor in interfascicular fibers but as an activator in vessels and xylary fibers. How the same TF plays these two contrasting roles is still unknown. These studies showed a differential regulatory role for KNAT7 depending on the tissue and cell type and its interacting partners. It is also possible that there are species-specific variations in KNAT7 function in SCW biosynthesis. More studies are required to answer the questions addressing the functional ambiguity of KNAT7 and how it interacts with cell wall TFs and other KNOX II proteins to regulate SCW formation. Furthermore, a detailed investigation into the regulatory network and downstream targets of Class II KNOX TF proteins is required to understand the transcriptional regulation of SCW formation. These studies will help us to modify cell wall formation in transgenic plants and enhance saccharification, as we recently showed [14,17]. Our understanding of the molecular controls of the deposition of each call wall component will help us to design cell walls for improved biomass production and reduced recalcitrance to bioconversion to ethanol. The modification of identified TFs through genetic engineering could help in overcoming some of the current bottlenecks leading to the realization of renewable bioenergy.
Author Contributions
Conceptualization, data curation, writing and editing, and visualization: A.N.; S.K.P.; Y.K.A. and C.P.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding and the APC was waived.
Acknowledgments
Y.K.A. thanks the Government of India for the National Overseas Scholarship and the Department of Biological Sciences, Michigan Technological University for supporting his doctoral research. We dedicate this review to the loving memory of Carl Douglas, University of British Columbia, Canada for his pioneering work with KNAT7. He was not only a brilliant scientist and dedicated researcher but also a wonderful and caring human being.
Conflicts of Interest
A.N. is currently employed by Kaveri Seeds, India. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
| BLH | BEL-like homeodomain |
| BiFc | Bimolecular fluorescence complementation |
| CesA | Cellulose synthase |
| ChIP | Chromatin immunoprecipitation assays |
| F5H | Ferulate 5-hydroxylase |
| HD | Homeodomain |
| irx | Irregular xylem |
| KNOX | Knotted-like homeobox |
| OFPs | Ovate family proteins |
| SCW | Secondary cell wall |
| S/G | Syringyl to Guaicyl lignin ratio |
| TALE | Three amino acid loop extension |
| TFs | Transcription factors |
| VIGS | Virus-induced gene silencing |
| Y1H | Yeast-one hybrid |
| Y2H | Yeast-two hybrid |
References
- Mohr, A.; Raman, S. Lessons from First Generation Biofuels and Implications for the Sustainability Appraisal of Second Generation Biofuels. Energy Policy 2013, 63, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Vega-Sánchez, M.E.; Ronald, P.C. Genetic and Biotechnological Approaches for Biofuel Crop Improvement. Curr. Opin. Biotechnol. 2010, 21, 218–224. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mansfield, S.D. Solutions for Dissolution—Engineering Cell Walls for Deconstruction. Curr. Opin. Biotechnol. 2009, 20, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.; Hatcher, C.; Mazarei, M.; Haynes, E.; Baxter, H.; Kim, K.; Hamilton, C.; Sykes, R.; Turner, G.; Davis, M.; et al. Development and Field Assessment of Transgenic Hybrid Switchgrass for Improved Biofuel Traits. Euphytica 2020, 216, 25. [Google Scholar] [CrossRef]
- Bryant, N.D.; Pu, Y.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Kalluri, U.C.; Yoo, C.G.; Ragauskas, A.J. Transgenic Poplar Designed for Biofuels. Trends Plant Sci. 2020, 25, 881–896. [Google Scholar] [CrossRef]
- de Vries, L.; Guevara-Rozo, S.; Cho, M.; Liu, L.-Y.; Renneckar, S.; Mansfield, S.D. Tailoring Renewable Materials via Plant Biotechnology. Biotechnol. Biofuels 2021, 14, 167. [Google Scholar] [CrossRef]
- Demura, T.; Ye, Z.-H. Regulation of Plant Biomass Production. Curr. Opin. Plant Biol. 2010, 13, 298–303. [Google Scholar] [CrossRef]
- Ko, J.-H.; Kim, W.-C.; Han, K.-H. Ectopic Expression of MYB46 Identifies Transcriptional Regulatory Genes Involved in Secondary Wall Biosynthesis in Arabidopsis. Plant J. 2009, 60, 649–665. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. Complexity of the Transcriptional Network Controlling Secondary Wall Biosynthesis. Plant Sci. 2014, 229, 193–207. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. Secondary Cell Walls: Biosynthesis, Patterned Deposition and Transcriptional Regulation. Plant Cell Physiol. 2015, 56, 195–214. [Google Scholar] [CrossRef]
- Ehlting, J.; Mattheus, N.; Aeschliman, D.S.; Li, E.; Hamberger, B.; Cullis, I.F.; Zhuang, J.; Kaneda, M.; Mansfield, S.D.; Samuels, L.; et al. Global Transcript Profiling of Primary Stems from Arabidopsis Thaliana Identifies Candidate Genes for Missing Links in Lignin Biosynthesis and Transcriptional Regulators of Fiber Differentiation: Global Transcript Profiling of Stems. Plant J. 2005, 42, 618–640. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX Gene KNAT7 Negatively Regulates Secondary Wall Formation in Arabidopsis and Is Functionally Conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.K.; Nookaraju, A.; Fujino, T.; Pattathil, S.; Joshi, C.P. Virus-Induced Gene Silencing (VIGS)-Mediated Functional Characterization of Two Genes Involved in Lignocellulosic Secondary Cell Wall Formation. Plant Cell Rep. 2016, 35, 2353–2367. [Google Scholar] [CrossRef]
- Wang, S.; Yamaguchi, M.; Grienenberger, E.; Martone, P.T.; Samuels, A.L.; Mansfield, S.D. The Class II KNOX Genes KNAT3 and KNAT7 Work Cooperatively to Influence Deposition of Secondary Cell Walls That Provide Mechanical Support to Arabidopsis Stems. Plant J. 2020, 101, 293–309. [Google Scholar] [CrossRef]
- Qin, W.; Yin, Q.; Chen, J.; Zhao, X.; Yue, F.; He, J.; Yang, L.; Liu, L.; Zeng, Q.; Lu, F.; et al. The Class II KNOX Transcription Factors KNAT3 and KNAT7 Synergistically Regulate Monolignol Biosynthesis in Arabidopsis. J. Exp. Bot. 2020, 71, 5469–5483. [Google Scholar] [CrossRef]
- Ahlawat, Y.K.; Nookaraju, A.; Harman-Ware, A.E.; Doeppke, C.; Biswal, A.K.; Joshi, C.P. Genetic Modification of KNAT7 Transcription Factor Expression Enhances Saccharification and Reduces Recalcitrance of Woody Biomass in Poplars. Front. Plant Sci. 2021, 12, 762067. [Google Scholar] [CrossRef]
- Sakakibara, K.; Ando, S.; Yip, H.K.; Tamada, Y.; Hiwatashi, Y.; Murata, T.; Deguchi, H.; Hasebe, M.; Bowman, J.L. KNOX2 Genes Regulate the Haploid-to-Diploid Morphological Transition in Land Plants. Science 2013, 339, 1067–1070. [Google Scholar] [CrossRef]
- Desplan, C.; Theis, J.; O’Farrell, P.H. The Sequence Specificity of Homeodomain-DNA Interaction. Cell 1988, 54, 1081–1090. [Google Scholar] [CrossRef]
- Burglin, T.R. Analysis of TALE Superclass Homeobox Genes (MEIS, PBC, KNOX, Iroquois, TGIF) Reveals a Novel Domain Conserved between Plants and Animals. Nucleic Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef]
- Kerstetter, R.; Vollbrecht, E.; Lowe, B.; Veit, B.; Yamaguchi, J.; Hake, S. Sequence Analysis and Expression Patterns Divide the Maize Knotted1-like Homeobox Genes into Two Classes. Plant Cell 1994, 6, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, G.; Janssen, B.J.; Kellogg, E.A.; Sinha, N. Phylogenetic Relationships and Evolution of the KNOTTED Class of Plant Homeodomain Proteins. Mol. Biol. Evol. 1999, 16, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Shi, A.; Wu, D.; Weng, Y.; Qin, J.; Ravelombola, W.S.; Shu, X.; Zhou, W. Genome-Wide Identification, Classification and Evolutionary Expansion of KNOX Gene Family in Rice (Oryza Sativa) and Populus (Populustrichocarpa). Am. J. Plant Sci. 2018, 9, 1071–1092. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, N.; Hao, P.; Sun, H.; Wang, C.; Ma, L.; Wang, H.; Zhang, X.; Wei, H.; Yu, S. Genome-Wide Identification and Characterization of TALE Superfamily Genes in Cotton Reveals Their Functions in Regulating Secondary Cell Wall Biosynthesis. BMC Plant Biol. 2019, 19, 432. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The Developmental Gene Knotted-1 Is a Member of a Maize Homeobox Gene Family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Hake, S.; Smith, H.M.S.; Holtan, H.; Magnani, E.; Mele, G.; Ramirez, J. The Role of Knox Genes in Plant Development. Annu. Rev. Cell Dev. Biol. 2004, 20, 125–151. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Sato, Y.; Matsuoka, M. Function of KNOX Homeodomain Proteins in Plant Development. Plant Biotechnol. 2001, 18, 85–92. [Google Scholar] [CrossRef]
- Kimura, S.; Koenig, D.; Kang, J.; Yoong, F.Y.; Sinha, N. Natural Variation in Leaf Morphology Results from Mutation of a Novel KNOX Gene. Curr. Biol. 2008, 18, 672–677. [Google Scholar] [CrossRef]
- Di Giacomo, E.; Sestili, F.; Iannelli, M.A.; Testone, G.; Mariotti, D.; Frugis, G. Characterization of KNOX Genes in Medicago Truncatula. Plant Mol. Biol. 2008, 67, 135–150. [Google Scholar] [CrossRef]
- Brown, D.M.; Zeef, L.A.H.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of Novel Genes in Arabidopsis Involved in Secondary Cell Wall Formation Using Expression Profiling and Reverse Genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef]
- He, J.-B.; Zhao, X.-H.; Du, P.-Z.; Zeng, W.; Beahan, C.T.; Wang, Y.-Q.; Li, H.-L.; Bacic, A.; Wu, A.-M. KNAT7 Positively Regulates Xylan Biosynthesis by Directly Activating IRX9 Expression in Arabidopsis: KNAT7 Positively Regulates Xylan Biosynthesis. J. Integr. Plant Biol. 2018, 60, 514–528. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Wei, H.; Milne, J.; Page, G.P.; Somerville, C.R. Identification of Genes Required for Cellulose Synthesis by Regression Analysis of Public Microarray Data Sets. Proc. Natl. Acad. Sci. USA 2005, 102, 8633–8638. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.-Y.; Huang, G.-Q.; Sun, X.; Qin, L.-X.; Li, Y.; Zhou, L.; Li, X.-B. Cotton KNL1, Encoding a Class II KNOX Transcription Factor, Is Involved in Regulation of Fibre Development. J. Exp. Bot. 2014, 65, 4133–4147. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Yuan, Y.; Spiekerman, J.J.; Guley, J.T.; Egbosiuba, J.C.; Ye, Z.-H. Functional Characterization of NAC and MYB Transcription Factors Involved in Regulation of Biomass Production in Switchgrass (Panicum Virgatum). PLoS ONE 2015, 10, e0134611. [Google Scholar] [CrossRef]
- Wang, S.; Yang, H.; Mei, J.; Liu, X.; Wen, Z.; Zhang, L.; Xu, Z.; Zhang, B.; Zhou, Y. Rice Homeobox Protein KNAT7 Integrates the Pathways Regulating Cell Expansion and Wall Stiffness. Plant Physiol. 2019, 181, 669–682. [Google Scholar] [CrossRef]
- Yoo, C.G.; Dumitrache, A.; Muchero, W.; Natzke, J.; Akinosho, H.; Li, M.; Sykes, R.W.; Brown, S.D.; Davison, B.; Tuskan, G.A.; et al. Significance of Lignin S/G Ratio in Biomass Recalcitrance of Populus Trichocarpa Variants for Bioethanol Production. ACS Sustain. Chem. Eng. 2018, 6, 2162–2168. [Google Scholar] [CrossRef]
- Pimrote, K. Transcriptional Regulatory Network Controlling Secondary Cell Wall Biosynthesis and Biomass Production in Vascular Plants. Afr. J. Biotechnol. 2012, 11, 13928–13937. [Google Scholar] [CrossRef]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-Based Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Land Plants. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Rao, X.; Dixon, R.A. Current Models for Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Grasses. Front. Plant Sci. 2018, 9, 399. [Google Scholar] [CrossRef]
- Yu, Y. OsKNAT7 Bridges Secondary Cell Wall Formation and Cell Growth Regulation. Plant Physiol. 2019, 181, 385–386. [Google Scholar] [CrossRef]
- Joshi, C.P.; Bhandari, S.; Ranjan, P.; Kalluri, U.C.; Liang, X.; Fujino, T.; Samuga, A. Genomics of Cellulose Biosynthesis in Poplars. New Phytol. 2004, 164, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin Biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Teng, Q.; Zhong, R.; Ye, Z.-H. Molecular Dissection of Xylan Biosynthesis during Wood Formation in Poplar. Mol. Plant 2011, 4, 730–747. [Google Scholar] [CrossRef] [PubMed]
- Polko, J.K.; Kieber, J.J. The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Guerriero, G.; Grima-Pettenati, J.; Baucher, M. A Molecular Blueprint of Lignin Repression. Trends Plant Sci. 2019, 24, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Y.; Pei, S.; Lu, M.; Kong, Y.; Zhou, G.; Hu, R. KNAT7 Regulates Xylan Biosynthesis in Arabidopsis Seed-Coat Mucilage. J. Exp. Bot. 2020, 71, 4125–4139. [Google Scholar] [CrossRef]
- Zhong, R.; Demura, T.; Ye, Z.-H. SND1, a NAC Domain Transcription Factor, Is a Key Regulator of Secondary Wall Synthesis in Fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 Transcription Factor Is a Direct Target of SND1 and Regulates Secondary Wall Biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.-H. Global Analysis of Direct Targets of Secondary Wall NAC Master Switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. MYB46 and MYB83 Bind to the SMRE Sites and Directly Activate a Suite of Transcription Factors and Secondary Wall Biosynthetic Genes. Plant Cell Physiol. 2012, 53, 368–380. [Google Scholar] [CrossRef]
- Kim, W.-C.; Kim, J.-Y.; Ko, J.-H.; Kang, H.; Han, K.-H. Identification of Direct Targets of Transcription Factor MYB46 Provides Insights into the Transcriptional Regulation of Secondary Wall Biosynthesis. Plant Mol. Biol. 2014, 85, 589–599. [Google Scholar] [CrossRef]
- Ko, J.-H.; Jeon, H.-W.; Kim, W.-C.; Kim, J.-Y.; Han, K.-H. The MYB46/MYB83-Mediated Transcriptional Regulatory Programme Is a Gatekeeper of Secondary Wall Biosynthesis. Ann. Bot. 2014, 114, 1099–1107. [Google Scholar] [CrossRef]
- Romano, J.M.; Dubos, C.; Prouse, M.B.; Wilkins, O.; Hong, H.; Poole, M.; Kang, K.; Li, E.; Douglas, C.J.; Western, T.L.; et al. At MYB61, an R2R3-MYB Transcription Factor, Functions as a Pleiotropic Regulator via a Small Gene Network. New Phytol. 2012, 195, 774–786. [Google Scholar] [CrossRef]
- Bellaoui, M.; Pidkowich, M.S.; Samach, A.; Kushalappa, K.; Kohalmi, S.E.; Modrusan, Z.; Crosby, W.L.; Haughn, G.W. The Arabidopsis BELL1 and KNOX TALE Homeodomain Proteins Interact through a Domain Conserved between Plants and Animals. Plant Cell 2001, 13, 2455–2470. [Google Scholar] [CrossRef]
- Hackbusch, J.; Richter, K.; Muller, J.; Salamini, F.; Uhrig, J.F. A Central Role of Arabidopsis Thaliana Ovate Family Proteins in Networking and Subcellular Localization of 3-Aa Loop Extension Homeodomain Proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Guo, J.; Zeng, Q.; Ellis, B.E.; Chen, J.-G. Arabidopsis Ovate Family Proteins, a Novel Transcriptional Repressor Family, Control Multiple Aspects of Plant Growth and Development. PLoS ONE 2011, 6, e23896. [Google Scholar] [CrossRef]
- Li, E.; Wang, S.; Liu, Y.; Chen, J.-G.; Douglas, C.J. OVATE FAMILY PROTEIN4 (OFP4) Interaction with KNAT7 Regulates Secondary Cell Wall Formation in Arabidopsis Thaliana: KNAT7-OFP Complex Regulates Secondary Wall Formation. Plant J. 2011, 67, 328–341. [Google Scholar] [CrossRef]
- Liu, Y.; Douglas, C.J. A Role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 Multi-Protein Complex Regulating Secondary Cell Wall Formation in Arabidopsis Thaliana. Plant Signal. Behav. 2015, 10, e1033126. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Ellis, B. Overview of OVATE FAMILY PROTEINS, A Novel Class of Plant-Specific Growth Regulators. Front. Plant Sci. 2016, 7, 417. [Google Scholar] [CrossRef]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Li, W.L.; Schuetz, M.; Brady, S.M.; Douglas, C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 Interact and Regulate Secondary Cell Wall Formation via Repression of REVOLUTA. Plant Cell 2015, 26, 4843–4861. [Google Scholar] [CrossRef]
- Bhargava, A.; Mansfield, S.D.; Hall, H.C.; Douglas, C.J.; Ellis, B.E. MYB75 Functions in Regulation of Secondary Cell Wall Formation in the Arabidopsis Inflorescence Stem. Plant Physiol. 2010, 154, 1428–1438. [Google Scholar] [CrossRef]
- Liu, Y. Investigation of a KNAT7-BLH-OFP Transcription Factor Complex Involved in Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis Thaliana. Master’s Thesis, University of British Columbia, Vancouver, BC, Canada, 2010. [Google Scholar] [CrossRef]
- Osakabe, K.; Tsao, C.C.; Li, L.; Popko, J.L.; Umezawa, T.; Carraway, D.T.; Smeltzer, R.H.; Joshi, C.P.; Chiang, V.L. Coniferyl Aldehyde 5-Hydroxylation and Methylation Direct Syringyl Lignin Biosynthesis in Angiosperms. Proc. Natl. Acad. Sci. USA 1999, 96, 8955–8960. [Google Scholar] [CrossRef]
- Wang, Q.; Dai, X.; Pang, H.; Cheng, Y.; Huang, X.; Li, H.; Yan, X.; Lu, F.; Wei, H.; Sederoff, R.R.; et al. BEL1-like Homeodomain Protein BLH6a Is a Negative Regulator of CAld5H2 in Sinapyl Alcohol Monolignol Biosynthesis in Poplar. Front. Plant Sci. 2021, 12, 695223. [Google Scholar] [CrossRef]
- Bhargava, A.; Ahad, A.; Wang, S.; Mansfield, S.D.; Haughn, G.W.; Douglas, C.J.; Ellis, B.E. The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta 2013, 237, 1199–1211. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Liu, C.; Ge, C.; Xu, F.; Luo, M. Ectopic expression of GhIQD14 (cotton IQ67 domain-containing protein 14) causes twisted organ and modulates secondary wall formation in Arabidopsis. Plant Physiol. Biochem. 2021, 163, 276–284. [Google Scholar] [CrossRef]
- Huang, D.; Wang, S.; Zhang, B.; Shang-Guan, K.; Shi, Y.; Zhang, D.; Liu, X.; Wu, K.; Xu, Z.; Fu, X.; et al. A Gibberellin-Mediated DELLA-NAC Signaling Cascade Regulates Cellulose Synthesis in Rice. Plant Cell 2015, 27, 1681–1696. [Google Scholar] [CrossRef]
- Schmitz, A.J.; Begcy, K.; Sarath, G.; Walia, H. Rice Ovate Family Protein 2 (OFP2) Alters Hormonal Homeostasis and Vasculature Development. Plant Sci. 2015, 241, 177–188. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3- MYB Transcription Factor MYB 6 Promotes Anthocyanin and Proanthocyanidin Biosynthesis but Inhibits Secondary Cell Wall Formation in Populus Tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).