Comparison of Automated Ribotyping, spa Typing, and MLST in 108 Clinical Isolates of Staphylococcus aureus from Orthopedic Infections
Abstract
:1. Introduction
2. Results
2.1. Results of the Automated Ribotyping Analysis
2.2. Results of the MLST Analysis
2.3. Relatedness between Ribogroups, STs, MLST CCs and eBURST Groups
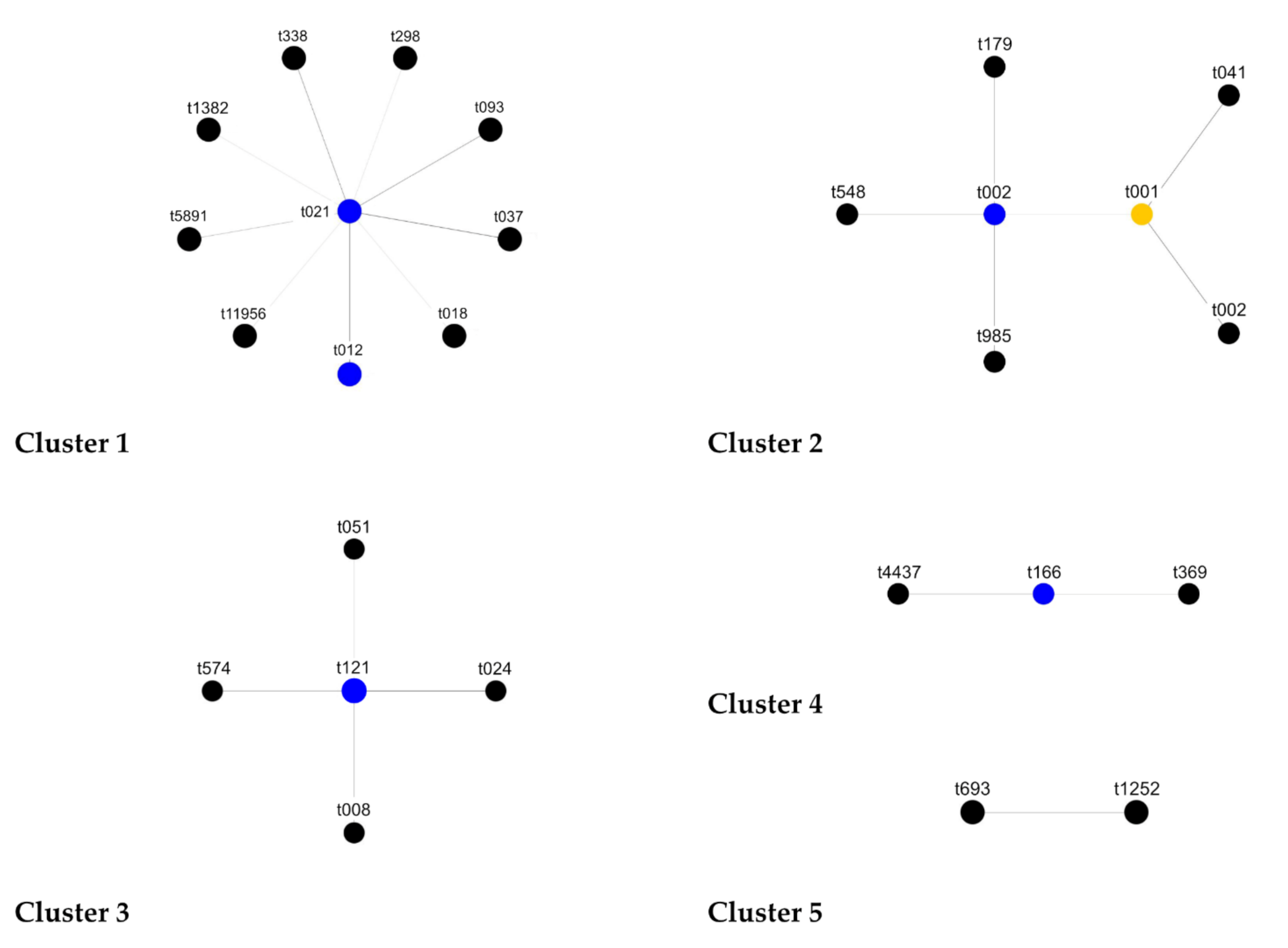
2.4. Relatedness between Ribogroups, spa Types, spa CCs and BURP Clusters
2.5. Ribogroups Association with agr Type, tst and mecA Genes, IS256 and Phenotypic Antibiotic Resistance
| Ribogroup | Prev. agr type | tst | IS256 | mecA | OXA | PEN | AMP | GEN | ERY | CLI | CHL | SXT | CIP | VAN | MAR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ribocluster | III 98% | 83% | 0% | 0% | 2% | 94% | 92% | 4% | 11% | 6% | 0% | 0% | 4% | 0% | 0.21 |
| cra-94-S7 | II 94% | 6% | 94% | 88% | 88% | 94% | 94% | 94% | 94% | 94% | 6% | 0% | 94% | 0% | 0.66 |
| cra-146-S8 | II 100% | 22% | 0% | 22% | 0% | 44% | 44% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0.11 |
| cra-137-S4 | I 100% | 0% | 50% | 79% | 86% | 100% | 100% | 57% | 57% | 62% | 21% | 7% | 86% | 0% | 0.57 |
| cra-157-S4 | I 85% | 0% | 0% | 0% | 0% | 77% | 77% | 8% | 8% | 8% | 8% | 0% | 0% | 0% | 0.18 |
| cra-147-S6 | III 100% | 25% | 50% | 25% | 25% | 75% | 75% | 13% | 25% | 25% | 0% | 0% | 0% | 0% | 0.24 |
2.6. Uncommon STs and spa Types Atypically Found within Ribogroups
2.7. Hunter-Gaston Discriminatory Power Analysis
3. Discussion
3.1. MLST vs. Automated Ribotyping
3.2. spa Typing vs. Automated Ribotyping
3.3. Clonal Complexes
4. Conclusions
5. Materials and Methods
5.1. Bacterial Isolates
5.2. Antibiotic Susceptibility
5.3. Automated Ribotyping
5.4. Bacterial DNA Isolation
5.5. Detection of mecA, tst, IS256, and agr Typing
5.6. spa Typing
5.7. MLST
5.8. Discriminatory Power
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arciola, C.R.; An, Y.H.; Campoccia, D.; Donati, M.E.; Montanaro, L. Etiology of implant orthopedic infections: A survey on 1027 clinical isolates. Int. J. Artif. Organs 2005, 28, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.; Cannella, B.A.; Crossett, L.S.; Yates, A.J.; McGough, R.L.; Hamilton, C.W. Preoperative screening/decolonization for Staphylococcus aureus to prevent orthopedic surgical site infection: Prospective cohort study with 2-year follow-up. J. Arthroplast. 2011, 26, 1501–1507. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, L.; Speziale, P.; Campoccia, D.; Ravaioli, S.; Cangini, I.; Pietrocola, G.; Giannini, S.; Arciola, C.R. Scenery of Staphylococcus implant infections in orthopedics. Future Microbiol. 2011, 6, 1329–1349. [Google Scholar] [CrossRef] [Green Version]
- Tande, A.J.; Patel, R. Prosthetic joint infection. Clin. Microbiol. Rev. 2014, 27, 302–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Chen, C.J.; Su, L.H.; Hu, S.; Yu, J.; Chiu, C.H. Evolution and pathogenesis of Staphylococcus aureus: Lessons learned from genotyping and comparative genomics. FEMS Microbiol. Rev. 2008, 32, 23–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francisco, A.P.; Bugalho, M.; Ramirez, M.; Carriço, J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 2009, 10, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mediavilla, J.R.; Chen, L.; Mathema, B.; Kreiswirth, B.N. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 2012, 15, 588–595. [Google Scholar] [CrossRef]
- Mellmann, A.; Weniger, T.; Berssenbrügge, C.; Keckevoet, U.; Friedrich, A.W.; Harmsen, D.; Grundmann, H. Characterization of clonal relatedness among the natural population of Staphylococcus aureus strains by using spa sequence typing and the BURP (based upon repeat patterns) algorithm. J. Clin. Microbiol. 2008, 46, 2805–2808. [Google Scholar] [CrossRef] [Green Version]
- Stefani, S.; Chung, D.R.; Lindsay, J.A.; Friedrich, A.W.; Kearns, A.M.; Westh, H.; Mackenzie, F.M. Meticillin-resistant Staphylococcus aureus (MRSA): Global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 2012, 39, 273–282. [Google Scholar] [CrossRef]
- Grimont, F.; Grimont, P.A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann. Inst. Pasteur Microbiol. 1986, 137B, 165–175. [Google Scholar] [CrossRef]
- van Belkum, A.; Tassios, P.T.; Dijkshoorn, L.; Haeggman, S.; Cookson, B.; Fry, N.K.; Fussing, V.; Green, J.; Feil, E.; Gerner-Smidt, P.; et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 2007, 13 (Suppl. 3), 1–46. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.; Schmidt, T.M. rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015, 43, D593–D598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klappenbach, J.A.; Saxman, P.R.; Cole, J.R.; Schmidt, T.M. rrndb: The Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001, 29, 181–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchet, V.; Huot, H.; Goldstein, R. Molecular genetic basis of ribotyping. Clin. Microbiol. Rev. 2008, 21, 262–273. [Google Scholar] [CrossRef] [Green Version]
- Andollina, A.; De Cesare, A.; Bertoni, G.; Modelli, L.; Manfreda, G. Identification and genetic characterisation of orthopaedic Staphylococcus isolates collected in Italy by automated EcoRI ribotyping. FEMS Microbiol. Lett. 2004, 234, 275–280. [Google Scholar] [CrossRef]
- Hollis, R.J.; Bruce, J.L.; Fritschel, S.J.; Pfaller, M.A. Comparative evaluation of an automated ribotyping instrument versus pulsed-field gel electrophoresis for epidemiological investigation of clinical isolates of bacteria. Diagn. Microbiol. Infect. Dis. 1999, 34, 263–268. [Google Scholar] [CrossRef]
- Campoccia, D.; Speziale, P.; Ravaioli, S.; Cangini, I.; Rindi, S.; Pirini, V.; Montanaro, L.; Arciola, C.R. The presence of both bone sialoprotein-binding protein gene and collagen adhesin gene as a typical virulence trait of the major epidemic cluster in isolates from orthopedic implant infections. Biomaterials 2009, 30, 6621–6628. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Kaspar, C.W.; Burgess, J.L.; Knight, I.T.; Colwell, R.R. Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can. J. Microbiol. 1990, 36, 891–894. [Google Scholar] [CrossRef]
- McAleese, F.; Murphy, E.; Babinchak, T.; Singh, G.; Said-Salim, B.; Kreiswirth, B.; Dunman, P.; O’Connell, J.; Projan, S.J.; Bradford, P.A. Use of ribotyping to retrospectively identify methicillin-resistant Staphylococcus aureus isolates from phase 3 clinical trials for tigecycline that are genotypically related to community-associated isolates. Antimicrob. Agents Chemother. 2005, 49, 4521–4529. [Google Scholar] [CrossRef] [Green Version]
- Klappenbach, J.A.; Dunbar, J.M.; Schmidt, T.M. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 2000, 66, 1328–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strommenger, B.; Braulke, C.; Heuck, D.; Schmidt, C.; Pasemann, B.; Nübel, U.; Witte, W. Spa Typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J. Clin. Microbiol. 2008, 46, 574–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benito, D.; Aspiroz, C.; Gilaberte, Y.; Sanmartín, R.; Hernández-Martin, Á.; Alonso, M.; Gómez, P.; Lozano, C.; Torres, C. Genetic lineages and antimicrobial resistance genotypes in Staphylococcus aureus from children with atopic dermatitis: Detection of clonal complexes CC1, CC97 and CC398. J. Chemother. 2016, 28, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.; Lin, C.Y.; Ho, M.W.; Lin, H.C.; Peng, C.T.; Lu, J.J. Concomitant genotyping revealed diverse spreading between methicillin-resistant Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus in central Taiwan. J. Microbiol. Immunol. Infect. 2014, 49, 363–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundmann, H.; Aanensen, D.M.; van den Wijngaard, C.C.; Spratt, B.G.; Harmsen, D.; Friedrich, A.W.; European Staphylococcal Reference Laboratory Working Group. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: A molecular-epidemiological analysis. PLoS Med. 2010, 7, e1000215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, M.; Hogan, P.G.; Satola, S.W.; Crispell, E.; Wylie, T.; Gao, H.; Sodergren, E.; Weinstock, G.M.; Burnham, C.D.; Fritz, S.A. Discriminatory Indices of Typing Methods for Epidemiologic Analysis of Contemporary Staphylococcus aureus Strains. Medicine 2015, 94, e1534. [Google Scholar] [CrossRef] [PubMed]
- Tavares, A.; Faria, N.A.; de Lencastre, H.; Miragaia, M. Population structure of methicillin-susceptible Staphylococcus aureus (MSSA) in Portugal over a 19-year period (1992–2011). Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Miko, B.A.; Hafer, C.A.; Lee, C.J.; Sullivan, S.B.; Hackel, M.A.; Johnson, B.M.; Whittier, S.; Della-Latta, P.; Uhlemann, A.C.; Lowy, F.D. Molecular characterization of methicillin-susceptible Staphylococcus aureus clinical isolates in the United States, 2004 to 2010. J. Clin. Microbiol. 2013, 51, 874–879. [Google Scholar] [CrossRef] [Green Version]
- Deurenberg, R.H.; Stobberingh, E.E. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 2008, 8, 747–763. [Google Scholar] [CrossRef] [PubMed]
- Shakeri, F.; Ghaemi, E.A. New Spa Types among MRSA and MSSA Isolates in North of Iran. Adv. Microbiol. 2014, 4, 899–905. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Rhman, S.H.; Rizk, D.E. Comparative Assessment of Different PCR-Based Typing Methods of Pseudomonas aeruginosa Isolates. Infect. Drug Resist. 2021, 14, 1019–1035. [Google Scholar] [CrossRef] [PubMed]
- Pavlic, M.; Griffiths, M.W. Principles, applications, and limitations of automated ribotyping as a rapid method in food safety. Foodborne Pathog. Dis. 2009, 6, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, L.; Speziale, P.; Campoccia, D.; Pirini, V.; Ravaioli, S.; Cangini, I.; Visai, L.; Arciola, C.R. Polymorphisms of agr locus correspond to distinct genetic patterns of virulence in Staphylococcus aureus clinical isolates from orthopedic implant infections. J. Biomed. Mater. Res. A 2010, 94, 825–832. [Google Scholar]
- Blasi, F.; Lovito, C.; Albini, E.; Bano, L.; Dalmonte, G.; Drigo, I.; Maresca, C.; Massacci, F.R.; Orsini, S.; Primavilla, S.; et al. Clostridioides difficile in Calves in Central Italy: Prevalence, Molecular Typing, Antimicrobial Susceptibility and Association with Antibiotic Administration. Animals 2021, 11, 515. [Google Scholar] [CrossRef] [PubMed]
- NCCLS. Performance Standards for Antimicrobial Susceptibility Testing: Twelfth Informational Supplement NCCLS Document M100-S12 NCCLS; NCCLS: Wayne, PA, USA, 2002. [Google Scholar]
- Campoccia, D.; Baldassarri, L.; Pirini, V.; Ravaioli, S.; Montanaro, L.; Arciola, C.R. Molecular epidemiology of Staphylococcus aureus from implant orthopaedic infections: Ribotypes, agr polymorphism, leukocidal toxins and antibiotic resistance. Biomaterials 2008, 29, 4108–4116. [Google Scholar] [CrossRef]
- Arciola, C.R.; Baldassarri, L.; Von Eiff, C.; Campoccia, D.; Ravaioli, S.; Pirini, V.; Becker, K.; Montanaro, L. Prevalence of genes encoding for staphylococcal leukocidal toxins among clinical isolates of Staphylococcus aureus from implant orthopedic infections. Int. J. Artif. Organs 2007, 30, 792–797. [Google Scholar] [CrossRef] [Green Version]
- Montanaro, L.; Ravaioli, S.; Ruppitsch, W.; Campoccia, D.; Pietrocola, G.; Visai, L.; Speziale, P.; Allerberger, F.; Arciola, C.R. Molecular Characterization of a Prevalent Ribocluster of Methicillin-Sensitive Staphylococcus aureus from Orthopedic Implant Infections. Correspondence with MLST CC30. Front. Cell. Infect. Microbiol. 2016, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- von Eiff, C.; Friedrich, A.W.; Peters, G.; Becker, K. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 2004, 49, 157–162. [Google Scholar] [CrossRef]
- Schmid, D.; Simons, E.; Ruppitsch, W.; Hrivniaková, L.; Stoeger, A.; Wechsler-Fördös, A.; Peter, L.; Geppert, F.; Allerberger, F. Limited value of routine spa typing: A cross-sectional study of methicillin-resistant Staphylococcus aureus-positive patients in an Austrian hospital. Am. J. Infect. Control 2013, 41, 617–624. [Google Scholar] [CrossRef]
- Harmsen, D.; Claus, H.; Witte, W.; Rothganger, J.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [Green Version]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Ribogroup | ST (%) | MLST CC Founder (Breaker) | eBURST Groups ** |
|---|---|---|---|
| Ribocluster (48) | ST30 (64.6%) ST34 (20.8%) ST2954 * (6.2%) ST2957 * (2.1%) ST2960 * (2.1%) ST untypeable (2.1%) ST243 (2.1%) | CC30 (SLV) | Group 1 |
| cra-94-S-7 (16) | ST228 (87.5%) ST2956 * (6.25%) ST46 (6.25%) | CC5 (DLV) CC5 (DLV) CC45 † (SLV) | Group 2 Group 2 ST46 singleton |
| cra-146-S-8 (9) | ST5 (88.9%) ST2626 (11.1%) | CC5 CC5 (SLV) | Group 2 |
| cra-137-S-4 (14) | ST8 (64.3%) ST247 (28.6%) ST241 (7.1%) | CC8 CC8 (DLV) CC8 (DLV) | Group 3 |
| cra-157-S-4 (13) | ST7 (69.2%) ST2955 * (7.7%) ST109 (15.4%) ST101 (7.7%) | CC7 CC7 (DLV) CC1 † (TLV) CC not delivered† | Group 4 Group 4 ST109 singleton ST101 singleton |
| cra-147-S-6 (8) | ST1 (100%) | CC1 | ST1 singleton |
| Ribogroup | MLST CC | MLST | spa Type | spa CC | Cluster |
|---|---|---|---|---|---|
| Ribocluster (48) | CC30 (48) | ST30 (31) | t012 (12) t021 (8) t018 (3) t093 (1) t338 (3) t1382 (2) t5891 (1) t11956 (1) | CC021/012 (37) | Cluster 1 |
| ST2954 ** (3) | t298 (2) t012 (1) | ||||
| ST2957 ** (1) ST2960 ** (1) ST untyp. (1) | t021 (1) t021 (1) t338 (1) | ||||
| ST34 (8) | t166 (4) t4437 (1) t369 (3) | CC166 (8) | Cluster 4 | ||
| t3906 (1) | Singleton (1) | - | |||
| ST34 (1) ST30 (1) | t13129 * (1) t13132 * (1) | spatypes with missing alignments (2) | - | ||
| cra-94-S-7 (16) | CC5 (15) | ST228 (7) ST228 (6) ST2956 ** (1) | t001 (7) t041 (6) t109 (1) | CC002 (14) | Cluster 2 |
| ST228 (1) | t1382 (1) | CC021/012 (1) | Cluster 1 | ||
| CC45 (1) | ST46 (1) | t1646 (1) | Singleton (1) | - | |
| cra-146-S-8 (9) | CC5 (9) | ST5 (8) | t002 (2) t179 (1) t548 (2) t985 (1) | CC002 (6) | Cluster 2 |
| t045 (1) | Singleton (2) | - | |||
| t5349 (1) | |||||
| ST2616 (1) | t13130 * (1) | spatype with missing alignments (1) | - | ||
| cra-137-S-4 (14) | CC8 (14) | ST8 (9) | t008 (5) t024 (2) t121 (1) t574 (1) | CC121 (13) | Cluster 3 |
| ST247 (4) | t051 (4) | ||||
| ST241 (1) | t037 †(1) | CC021/012 (1) | Cluster 1 | ||
| cra-157-S-4 (13) | CC7 (10) | ST7 (9) ST2955 ** (1) | t091 (9) t091 (1) | Singleton (13) | - |
| CC1 (2) | ST109 (2) | t209 (2) | |||
| CC n.d. | ST101 (1) | t7956 (1) | |||
| cra-147-S-6 (8) | CC1 | ST1 (8) | t127 (5) t2478 (1) | Singleton (6) | - |
| t693 (1) t1252 (1) | Excluded CC no founder (2) | Cluster 5 |
| Ribogroup | STs | MLST Allele Succession (arc-aroe-glpf-gmk-pta-tpi-yiql) | agr Type | spa Types-spa CC | spa Repeats Succession |
|---|---|---|---|---|---|
| cra-94-S-7 (16) | ST228 (14) ST46 (1) | 01-04-01-04-12-24-29 10–14-08–06-14-03-02 | II I | t001-CC002 (7) t041-CC002 (6) t1646–singleton (1) | 26-30-17-34-17-20-17-12-17-16 26-30-17-34-17-20-17-34-17-20-17-12-17-16 09-20-16-34-13-17-34-16-34 |
| cra-157-S-4 (13) | ST7 (9) ST109 (2) ST101 (1) | 05-04–01-04-04-06-03 03-27-01-01–01-01-1003-01-14-15-11-19-03 | I II I | t091-singleton (9) t209-singleton (2) t7956-singleton (1) | 07-23-21-17-34-12-23-02-12-23 07-16-12-23-3404-13-21-12-17-20-17-12-17-17-17 |
| cra-137-S-4 (14) | ST8 (9) ST241 (1) | 03-03-01-01-04–04–03 02-03-01-01-04–04–03 | I I | t008 (5 CC121) t037 (1 CC021/012) | 11-19-12-21-17-34-24-34-22-25 15-12-16-02-25-17-24 |
| Spa Typing | MLST | Ribotyping | spa CC | MLST CC | |
|---|---|---|---|---|---|
| MSSA (79) | 0.936 | 0.802 * | 0.596 | 0.746 † | 0.590 ** |
| MRSA (29) | 0.879 | 0.751 | 0.636 | 0.594 | 0.567 |
| TOTAL (108) | 0.955 | 0.869 * | 0.744 | 0.817 † | 0.721 ** |
| Ribogroup 1 | MLST CC 1 | MLST ST | spa Type | spa CC | IS256 |
|---|---|---|---|---|---|
| Ribocluster (48/48) | CC30 (48/48) | ST30 (31), ST34 (10), ST2954 (3), ST2957 (1), ST2960 (1), ST243 (1), ST non-typeable (1) | t012 (13), t021 (10), t018 (3), t093 (1), t166 (4), t298 (2), t338 (4), t369 (3), t1382 (2), t3906(1), t4437 (1), t5891 (1), t11956 (1), t13129 * (1), t13132 * (1) | CC021/012 (37) CC166 (8) singleton and missing alignments (3) | neg (44) |
| cra-94-S7 (2/16) cra-146-S8 (7/9) | CC5 (8/25) CC45 (1/25) | ST5 (6), ST228 (1), ST2626 (1) ST46 (1) | t1382 (1), t1646 (1), t002 (1), t045 (1), t179(1), t548 (2), t5349 (1), t13130 * (1) | CC002 (5) CC021/012 (1) singletons (3) | neg (8) pos (1) |
| cra-137-S4 (3/14) | CC8 (3/14) | ST8 (3) | t008 (1), t024 (1), t574 (1) | CC121 (3) | neg (3) |
| cra-157-S4 (13/13) | CC7 (10/13) CC1 (2/13) CC not delivered (1/13) | ST7 (9)–ST2955 (1) ST109 (2) ST101 (1) | t091 (10), t209 (2), t7956 (1) | singletons (13) | neg (13) |
| cra-147-S6 (6/8) | CC1 (6/8) | ST1 (6) | t127 (3), t693 (1), t1252 (1), t2478 (1) | excluded (2) singletons (4) | neg (4) pos (2) |
| Ribogroup 1 | MLST CC 1 | MLST ST | spa Type | spa CC | IS256 |
|---|---|---|---|---|---|
| cra-94-S7 (14/16) cra-146-S8 (2/9) | CC5 (16/25) | ST228 (13) ST5 (2) ST2956 (1) | t001 (7), t041 (6), t109 (1), t002 (1), t985 (1) | CC002 (16) | pos (14) neg (2) |
| cra-137-S4 (11/14) | CC8 (11/14) | ST8 (6) ST247 (4) ST241 (1) | t008 (4), t051 (4), t024 (1), t037 (1), t121 (1) | CC121 (10) singleton (1) | pos (7) neg (4) |
| cra-147-S6 (2/8) | CC1 (2/8) | ST1 (2) | t127 (2) | singletons (2) | pos (2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravaioli, S.; Campoccia, D.; Ruppitsch, W.; Allerberger, F.; Poggi, A.; Chisari, E.; Montanaro, L.; Arciola, C.R. Comparison of Automated Ribotyping, spa Typing, and MLST in 108 Clinical Isolates of Staphylococcus aureus from Orthopedic Infections. Int. J. Mol. Sci. 2022, 23, 1660. https://doi.org/10.3390/ijms23031660
Ravaioli S, Campoccia D, Ruppitsch W, Allerberger F, Poggi A, Chisari E, Montanaro L, Arciola CR. Comparison of Automated Ribotyping, spa Typing, and MLST in 108 Clinical Isolates of Staphylococcus aureus from Orthopedic Infections. International Journal of Molecular Sciences. 2022; 23(3):1660. https://doi.org/10.3390/ijms23031660
Chicago/Turabian StyleRavaioli, Stefano, Davide Campoccia, Werner Ruppitsch, Franz Allerberger, Alessandro Poggi, Emanuele Chisari, Lucio Montanaro, and Carla Renata Arciola. 2022. "Comparison of Automated Ribotyping, spa Typing, and MLST in 108 Clinical Isolates of Staphylococcus aureus from Orthopedic Infections" International Journal of Molecular Sciences 23, no. 3: 1660. https://doi.org/10.3390/ijms23031660
APA StyleRavaioli, S., Campoccia, D., Ruppitsch, W., Allerberger, F., Poggi, A., Chisari, E., Montanaro, L., & Arciola, C. R. (2022). Comparison of Automated Ribotyping, spa Typing, and MLST in 108 Clinical Isolates of Staphylococcus aureus from Orthopedic Infections. International Journal of Molecular Sciences, 23(3), 1660. https://doi.org/10.3390/ijms23031660









