Field-Deployable Detection of Chestnut Blight Pathogen Cryphonectria parasitica Using Enzyme-Mediated Duplex Exponential Amplification
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Sources
2.2. Genomic DNA Extraction from Strain Cultures and Infected Bark Tissues
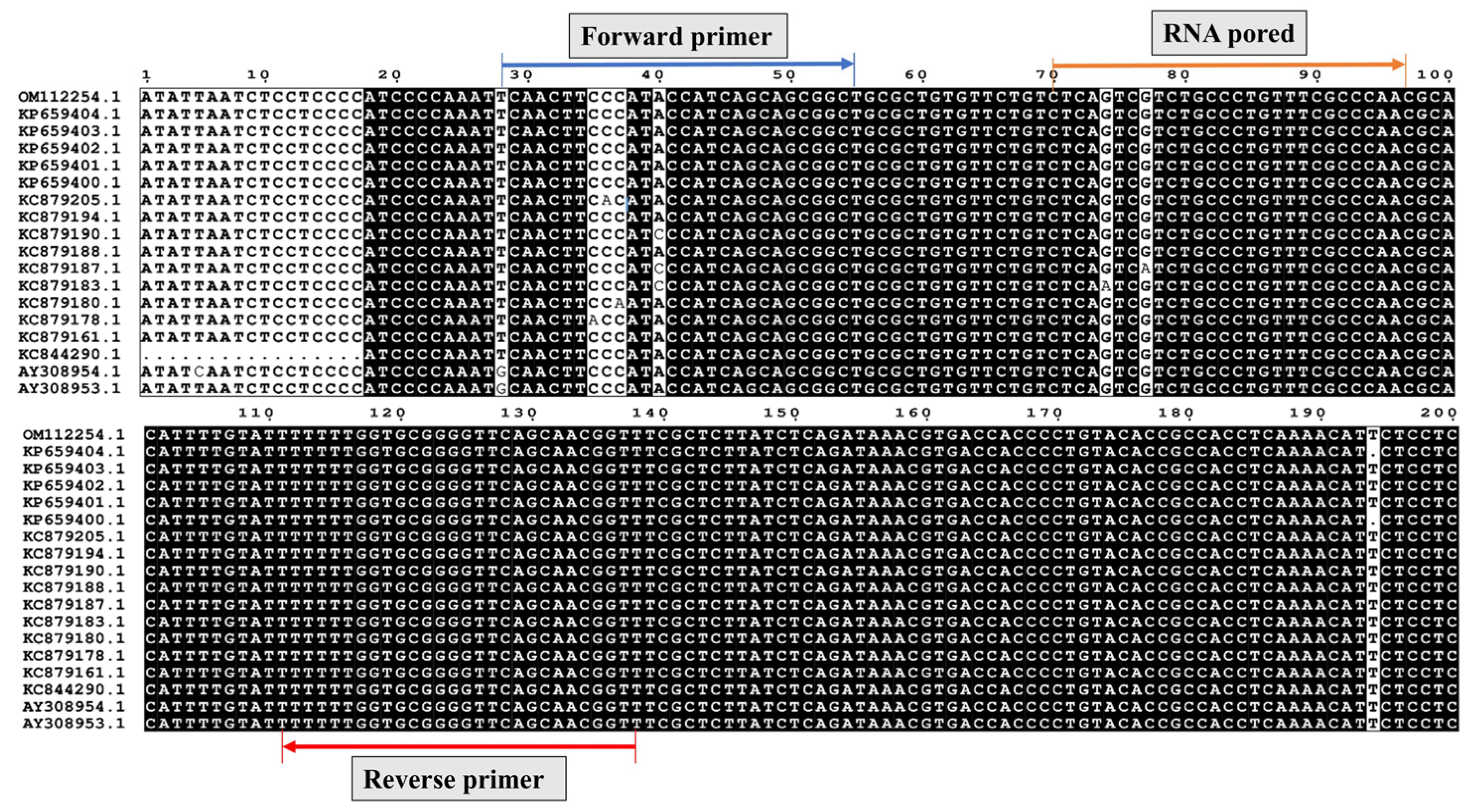
2.3. Primers and Probes
2.4. Isothermal Fluorescence Amplification Assay
2.5. RNA Probe Screening
2.6. Primer–Probe Screening and Optimization
2.7. Evaluation of EmDEA Assay Specificity
2.8. Evaluation of EmDEA Assay Sensitivity and Stability
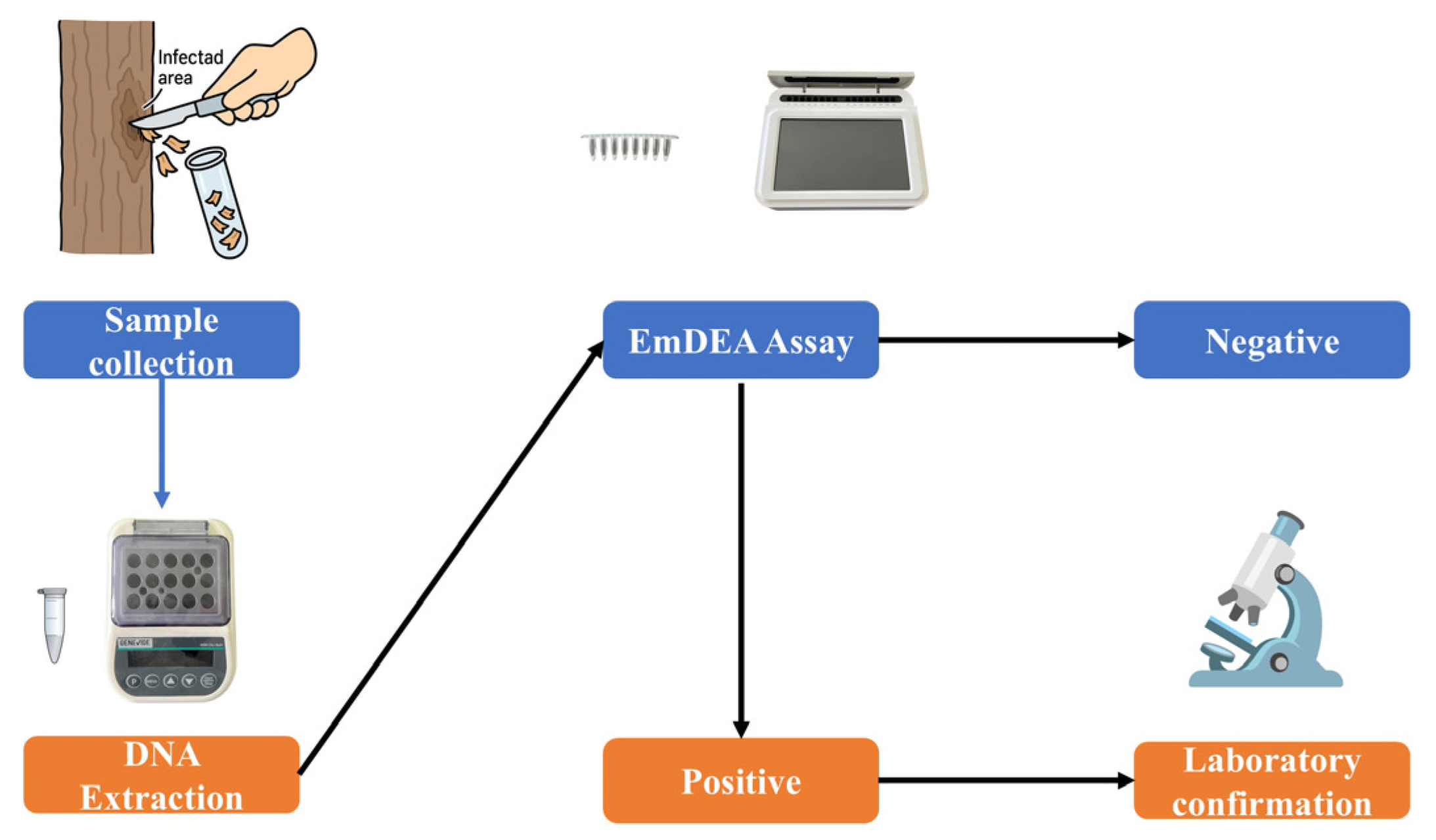
2.9. Field Evaluation of the EmDEA Assay
3. Results
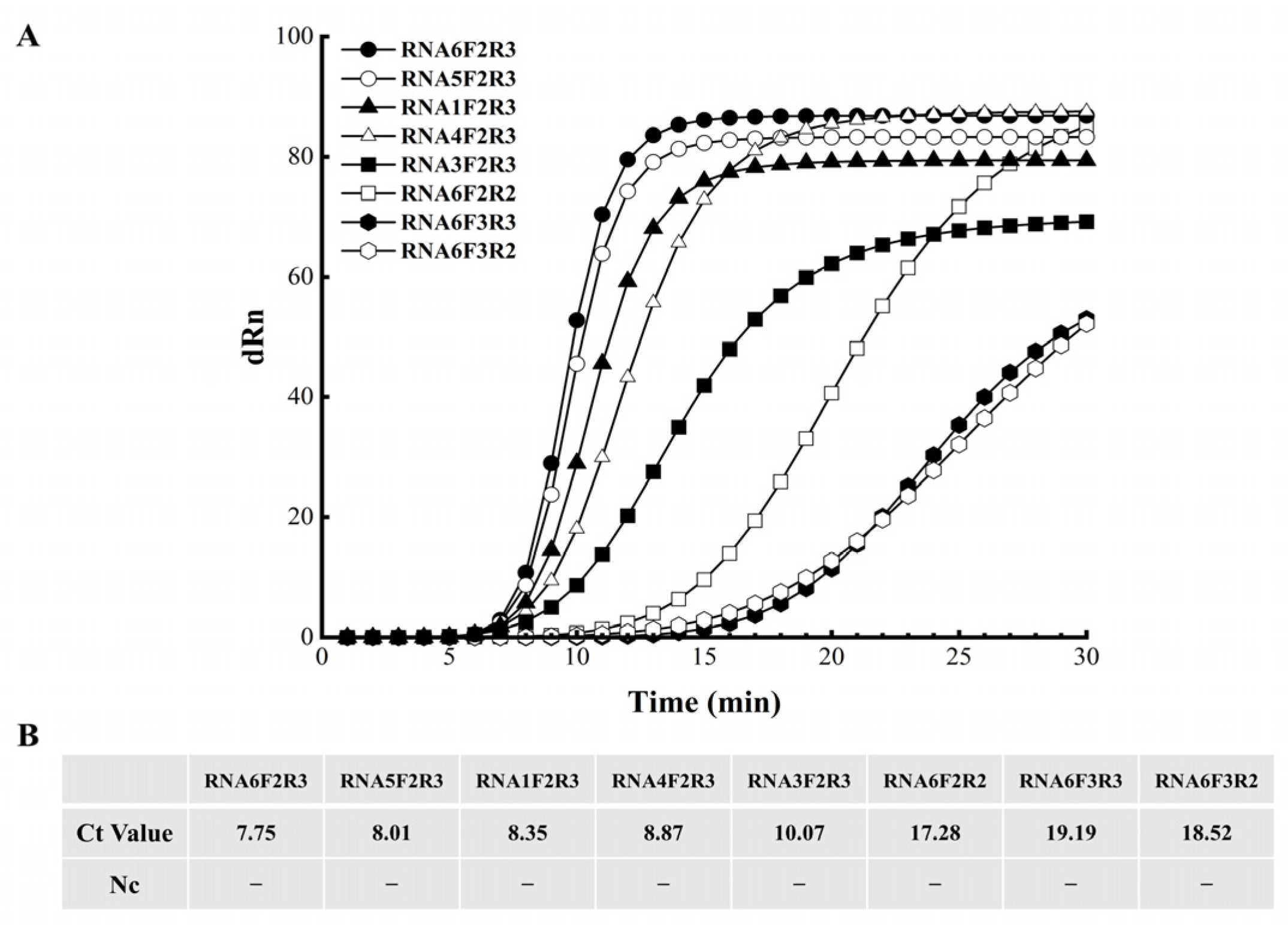
3.1. Identification of RNA Probes
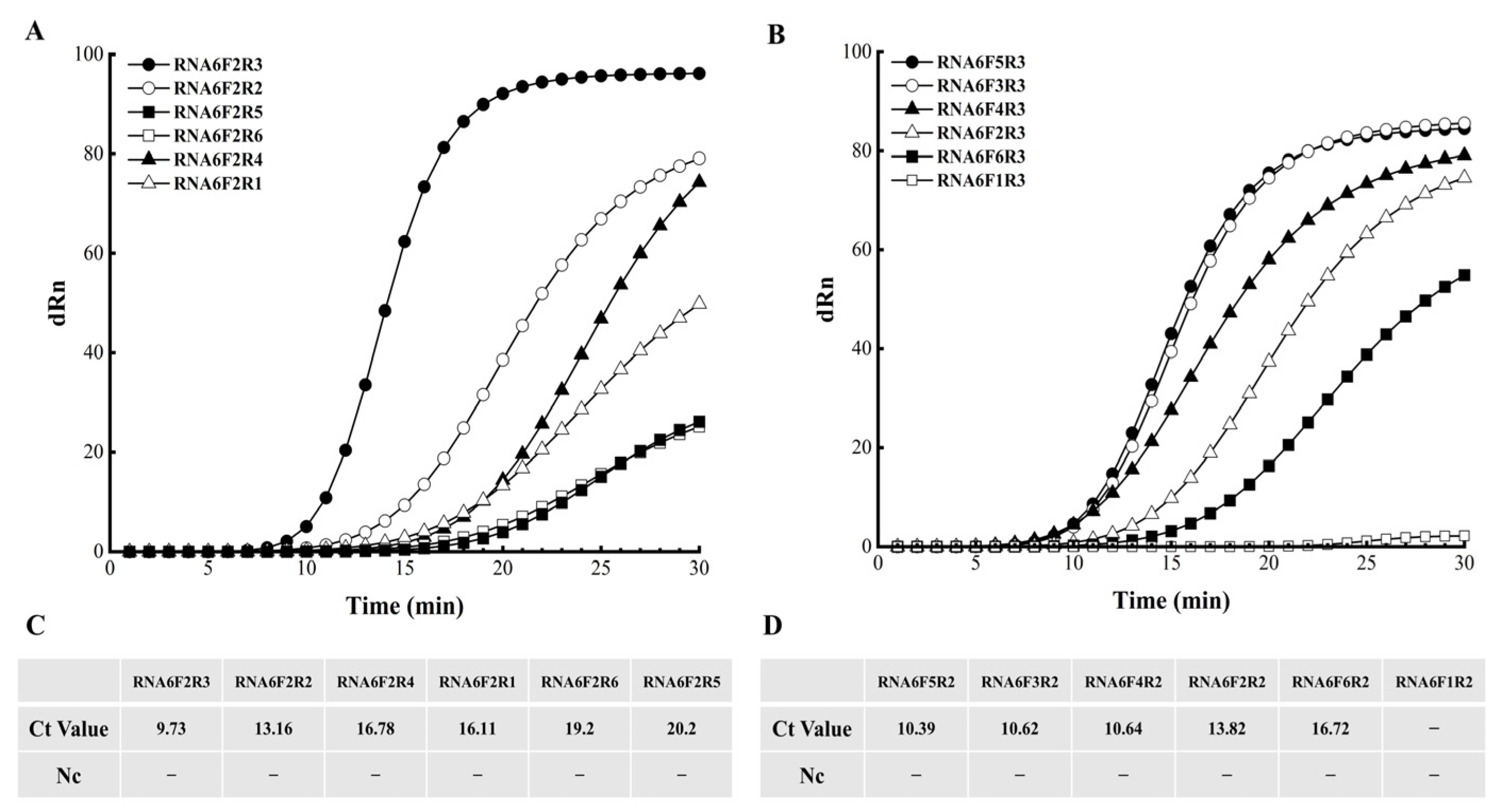
3.2. Optimization of Primer–Probe Combinations for EmDEA
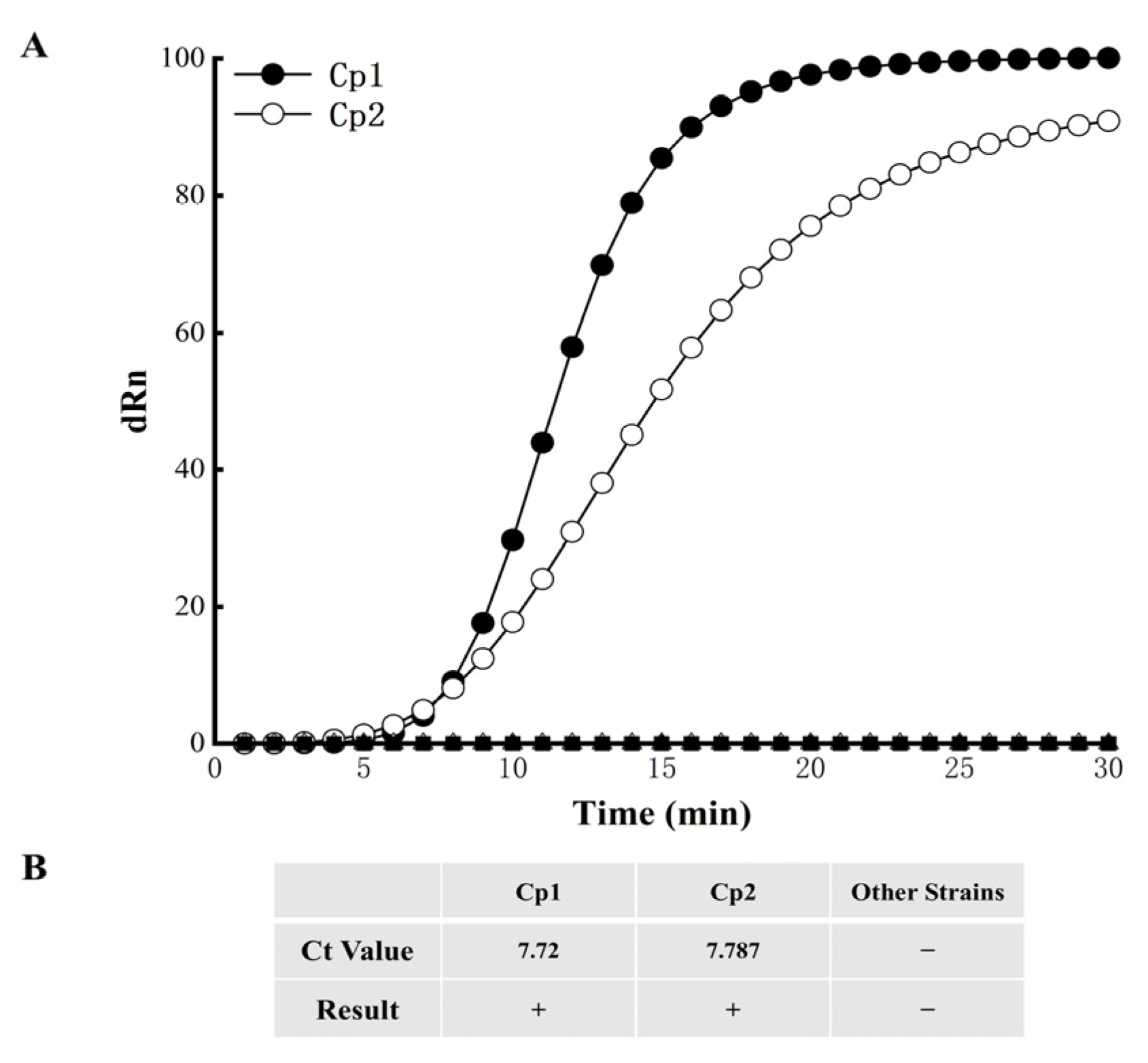
3.3. EmDEA Assay Specificity
3.4. EmDEA Assay Sensitivity
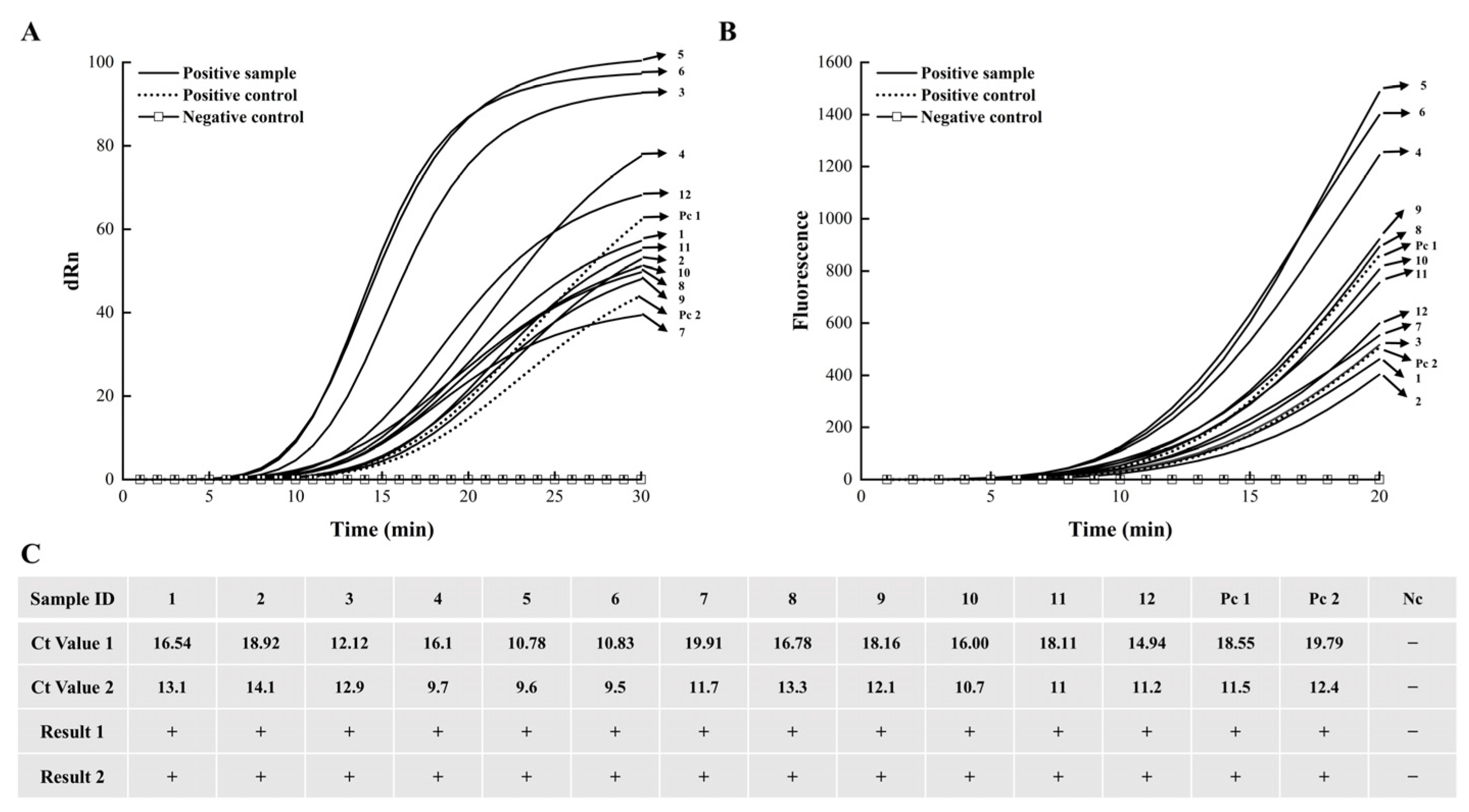
3.5. Field Evaluation Results of the EmDEA Assay on Chestnut Blight Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EmDEA | enzyme-mediated duplex exponential amplification |
References
- Anagnostakis, S.L. Chestnut blight: The classical problem of an introduced pathogen. Mycologia 1987, 79, 23–37. [Google Scholar] [CrossRef]
- Dallavalle, E.; Zambonelli, A. Epidemiological role of strains of Cryphonectria parasitica isolated from hosts other than chestnut. Eur. J. For. Pathol. 1999, 29, 97–102. [Google Scholar] [CrossRef]
- Stipes, R.J.; Appel, D.N.; Roane, M.K. Endothia species as pathogens of chestnut and oak. In Proceedings of the American Chestnut Symposium; MacDonald, W.L., Cech, F.C., Luchok, J., Smith, C., Eds.; West Virginia University: Morgantown, WV, USA, 1978; pp. 42–49. [Google Scholar]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef]
- Popov, A.; Tsvetkov, I.; Belov, A.; Konichev, A.; Ivanushkina, N.; Kochkina, G.; Ozerskaya, S. Molecular genetic identification of the phytopathogenic fungus Cryphonectria parasitica. Microbiology 2010, 79, 223–228. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Milgroom, M.G.; Lipari, S.E.; Powell, W. DNA fingerprinting and analysis of population structure in the chestnut blight fungus, Cryphonectria parasitica. Genetics 1992, 131, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Milgroom, M.G.; Lipari, S.E. Population differentiation in the chestnut blight fungus, Cryphonectria parasitica, in eastern North America. Phytopathology 1995, 85, 155–160. [Google Scholar] [CrossRef]
- Wronski, R.; Kudera, U.; Wlhelm, E. Characterization of Cryphonectria parasitica strains by random amplified polymorphic DNA (RAPD) technique and conventional methods. Eur. J. For. Pathol. 1997, 27, 95–103. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, J.; Zheng, L.; Liu, Y.; Zhu, T. A duplex PCR method for detection of Cryphonectria parasitica. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2015, 39, 7–13. [Google Scholar]
- Ma, W.; Zheng, L.; Zhang, J.; Liu, Y.; Zhu, T. Development of nested-PCR for detection of Cryphonectria parasitica based on the marker of sequence characterized amplified region. J. Zhejiang Univ. (Agric. Life Sci. Ed.) 2015, 41, 25–33. [Google Scholar]
- Rubio, S.; Barnes, A.; Webb, K.; Hodgetts, J. A real-time PCR assay for improved rapid, specific detection of Cryphonectria parasitica. Ann. Appl. Biol. 2017, 171, 52–61. [Google Scholar] [CrossRef]
- Chandelier, A.; Massot, M.; Fabreguettes, O.; Gischer, F.; Teng, F.; Robin, C. Early detection of Cryphonectria parasitica by real-time PCR. Eur. J. Plant Pathol. 2019, 153, 29–46. [Google Scholar] [CrossRef]
- Yadav, S.; Ramasamy, S.; Sundararajan, G.; Ramaraj, R.; Ramasamy, S. Advancements and applications of loop-mediated isothermal amplification technology: A comprehensive overview. Front. Microbiol. 2024, 15, 1406632. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from plant tissues. In Plant Molecular Biology Manual; Gelvin, S.B., Schilperoort, R.A., Eds.; Kluwer Academic: Dordrecht, The Netherlands, 1989; pp. 73–83. [Google Scholar]
- Sayers, E.W.; Cavanaugh, M.; Frisse, L.; Pruitt, K.D.; Schneider, V.A.; Underwood, B.A.; Yankie, L.; Karsch-Mizrachi, I. GenBank 2025 update. Nucleic Acids Res. 2025, 53, D56–D61. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B.; List, F.B.C.A.; Bolchacova, E. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Tacke, B.K.; Casper, H.H. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 2000, 97, 7905–7910. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.; Boonham, N.; Hughes, K.; Griffin, R.; Barker, I. On-site DNA extraction and real-time PCR for detection of Phytophthora ramorum in the field. Appl. Environ. Microbiol. 2005, 71, 6702–6710. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.J.; Tomlinson, J.A.; Griffin, R.L.; Boonham, N.; Inman, A.J.; Lane, C.R. Development of a one-step real-time polymerase chain reaction assay for diagnosis of Phytophthora ramorum. Phytopathology 2006, 96, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Kox, L.; Brouwershaven, I.v.; Vossenberg, B.v.d.; Beld, H.v.d.; Bonants, P.; Gruyter, J.d. Diagnostic values and utility of immunological, morphological, and molecular methods for in planta detection of Phytophthora ramorum. Phytopathology 2007, 97, 1119–1129. [Google Scholar] [CrossRef]
- Schaad, N.W.; Frederick, R.D. Real-time PCR and its application for rapid plant disease diagnostics. Can. J. Plant Pathol. 2002, 24, 250–258. [Google Scholar] [CrossRef]
- Atkins, S.D.; Clark, I.M. Fungal molecular diagnostics: A mini review. J. Appl. Genet. 2004, 45, 3–15. [Google Scholar] [PubMed]


| No. | Scientific Name | Source | Host | Detection Result |
|---|---|---|---|---|
| Cp1 | Cryphonectria parasitica | Shanxi, China | Castanea mollissima | + |
| Cp2 | Cryphonectria parasitica | Zhejiang, China | Castanea mollissima | + |
| Gd | Gnomoniopsis daii | Shandong, China | Castanea mollissima | − |
| Nc | Neopestalotiopsis clavispora | Zhejiang, China | Castanea mollissima | − |
| Nc-1 | Neopestalotiopsis clavispora | Shanxi, China | Castanea mollissima | − |
| Nc-2 | Neopestalotiopsis clavispora | Shandong, China | Castanea mollissima | − |
| Lt | Lasiodiplodia theobromae | Shandong, China | Castanea mollissima | − |
| Dh | Diaporthe hongkongensis | Zhejiang, China | Castanea mollissima | − |
| Cj | Cryphonectria japonica | Beijing, China | Quercus sp. | − |
| Cj-1 | Cryphonectria japonica | Shandong, China | Quercus sp. | − |
| Cq1 | Cryphonectria quercicola | Shanxi, China | Quercus sp. | − |
| Cq2 | Cryphonectria quercus | Shandong, China | Quercus sp. | − |
| Cf | Colletotrichum fructicola | Zhejiang, China | Citrus reticulata | − |
| Cs | Colletotrichum siamense | Zhejiang, China | Ilex chinensis | − |
| Lp | Lasiodiplodia pseudotheobromae | Zhejiang, China | Torreya grandis | − |
| Bc | Botrytis cinerea | Zhejiang, China | Vitis vinifera | − |
| Aa | Alternaria alternata | Zhejiang, China | Juniperus chinensis | − |
| Pc | Phytophthora cinnamomi | Zhejiang, China | Carya cathayensis | − |
| Pv | Pythium vexans | Zhejiang, China | Carya cathayensis | − |
| Bd | Botryosphaeria dothidea | Zhejiang, China | Carya cathayensis | − |
| Bd-1 | Botryosphaeria dothidea | Zhejiang, China | Castanea mollissima | − |
| Fo | Fusarium oxysporum | Zhejiang, China | Carya cathayensis | − |
| Fo-1 | Fusarium oxysporum | Zhejiang, China | Camellia oleifera | − |
| Fo-2 | Fusarium oxysporum | Shanxi, China | Castanea mollissima | − |
| Fg | Fusarium graminearum | Zhejiang, China | Carya cathayensis | − |
| Fs | Fusarium solani | Zhejiang, China | Carya cathayensis | − |
| Fs-1 | Fusarium solani | Zhejiang, China | Torreya grandis | − |
| Fs-2 | Fusarium solani | Shanxi, China | Castanea mollissima | − |
| Primer and Probe | Name | Sequence (5′→3′) |
|---|---|---|
| Forward primer | F1 | AAGCTAATACGACTCACTATAGGGTTAATCTCCTCCCCATCCCCAAATTCAA |
| F2 | AAGCTAATACGACTCACTATAGGGTCCTCCCCATCCCCAAATTCAACTTCCC | |
| F3 | AAGCTAATACGACTCACTATAGGGCCATCCCCAAATTCAACTTCCCATACCA | |
| F4 | AAGCTAATACGACTCACTATAGGGCCAAATTCAACTTCCCATACCATCAGCA | |
| F5 | AAGCTAATACGACTCACTATAGGGTCAACTTCCCATACCATCAGCAGCGGCT | |
| F6 | AAGCTAATACGACTCACTATAGGGTCCCATACCATCAGCAGCGGCTGCGCTG | |
| Reverse primer | R1 | TGCTGAACCCCGCACCAAAAAAATACAA |
| R2 | GAGCGAAACCGTTGCTGAACCCCGCACC | |
| R3 | AACCGTTGCTGAACCCCGCACCAAAAAA | |
| R4 | AGATAAGAGCGAAACCGTTGCTGAACCC | |
| R5 | TATCTGAGATAAGAGCGAAACCGTTGCT | |
| R6 | AAACCGTTGCTGAACCCCGCACCA | |
| RNA probe | RNA1 | FAM-GCAGCCGCUGCUGAUGGUAUGGGAAGUU-BHQ1 |
| RNA2 | FAM-AACACAGCGCAGCCGCUGCUGAUGGUAU-BHQ1 | |
| RNA3 | FAM-UGAGACAGAACACAGCGCAGCCGCUGCU-BHQ1 | |
| RNA4 | FAM-CAGACGACUGAGACAGAACACAGCGCAG-BHQ1 | |
| RNA5 | FAM-AAACAGGGCAGACGACUGAGACAGAACA-BHQ1 | |
| RNA6 | FAM-GUUGGGCGAAACAGGGCAGACGACUGAG-BHQ1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Feng, Z.; Liu, Y.; Tang, C.; Guo, K.; Hu, J. Field-Deployable Detection of Chestnut Blight Pathogen Cryphonectria parasitica Using Enzyme-Mediated Duplex Exponential Amplification. Curr. Issues Mol. Biol. 2025, 47, 762. https://doi.org/10.3390/cimb47090762
Wang S, Feng Z, Liu Y, Tang C, Guo K, Hu J. Field-Deployable Detection of Chestnut Blight Pathogen Cryphonectria parasitica Using Enzyme-Mediated Duplex Exponential Amplification. Current Issues in Molecular Biology. 2025; 47(9):762. https://doi.org/10.3390/cimb47090762
Chicago/Turabian StyleWang, Shuai, Zhongwei Feng, Yiming Liu, Changyun Tang, Kai Guo, and Jiafu Hu. 2025. "Field-Deployable Detection of Chestnut Blight Pathogen Cryphonectria parasitica Using Enzyme-Mediated Duplex Exponential Amplification" Current Issues in Molecular Biology 47, no. 9: 762. https://doi.org/10.3390/cimb47090762
APA StyleWang, S., Feng, Z., Liu, Y., Tang, C., Guo, K., & Hu, J. (2025). Field-Deployable Detection of Chestnut Blight Pathogen Cryphonectria parasitica Using Enzyme-Mediated Duplex Exponential Amplification. Current Issues in Molecular Biology, 47(9), 762. https://doi.org/10.3390/cimb47090762






