Checkpoint-Dependent Sensitivities to Nucleoside Analogues Uncover Specific Patterns of Genomic Instability
Abstract
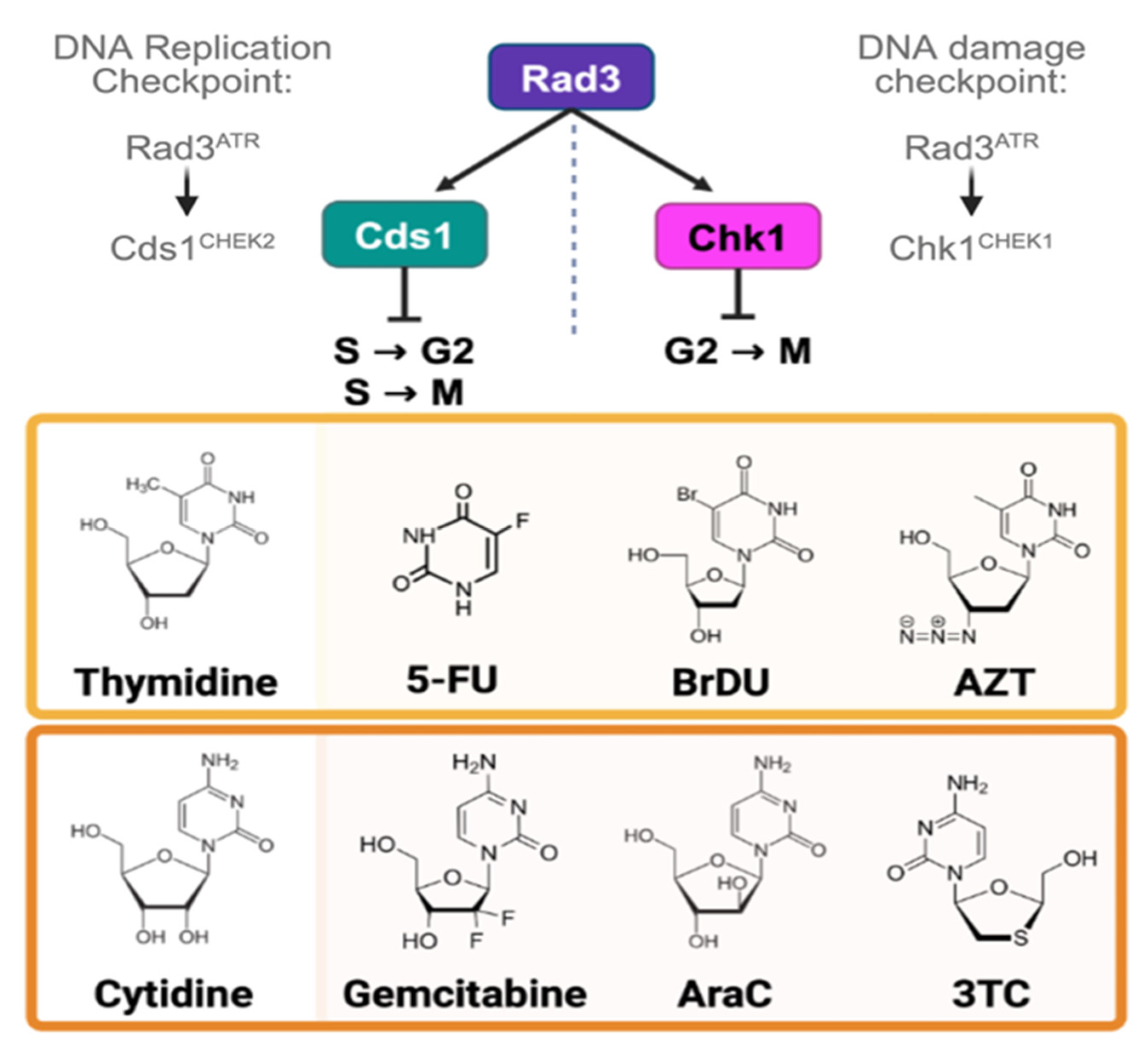
1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Drugs
2.3. Proliferation Assay and IC50
2.4. Microscopy
2.5. Statistical Analysis
3. Results and Discussion
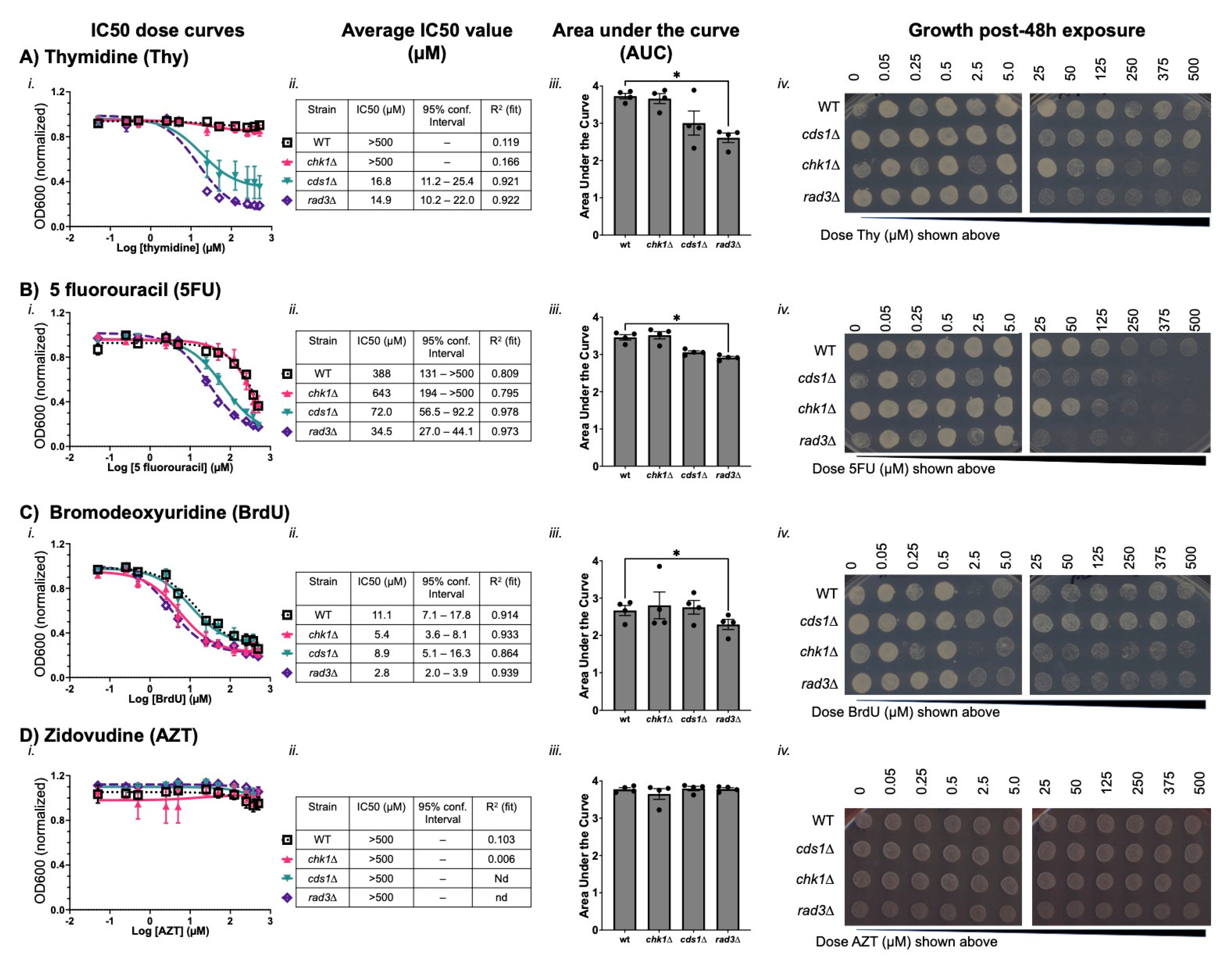
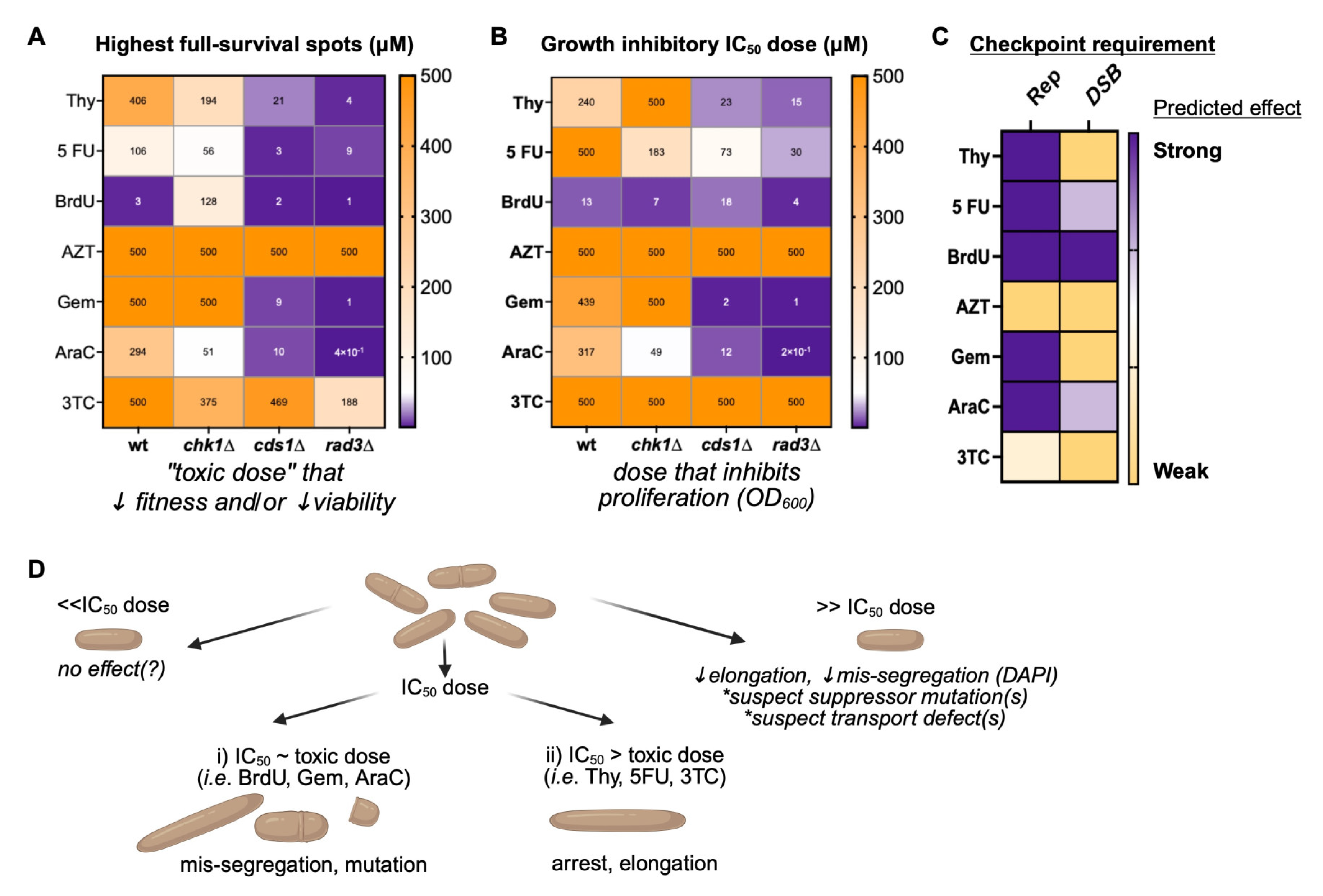
3.1. Thymidine Analogues Induce Both DNA Replication and DNA Damage Stress
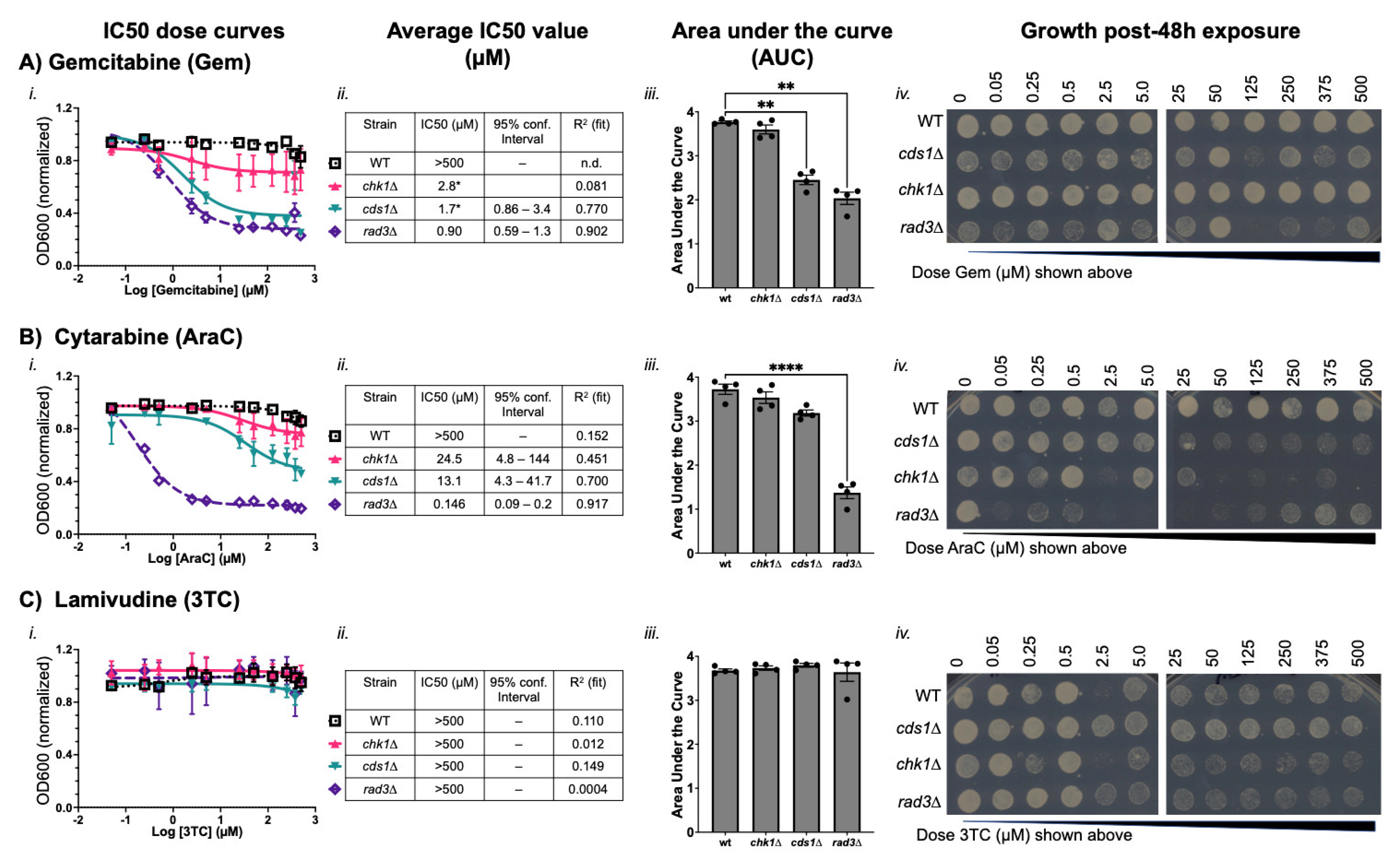
3.2. Cytidine Anticancer Analogues Generate Replication Instability and Not DNA Damage Arrest
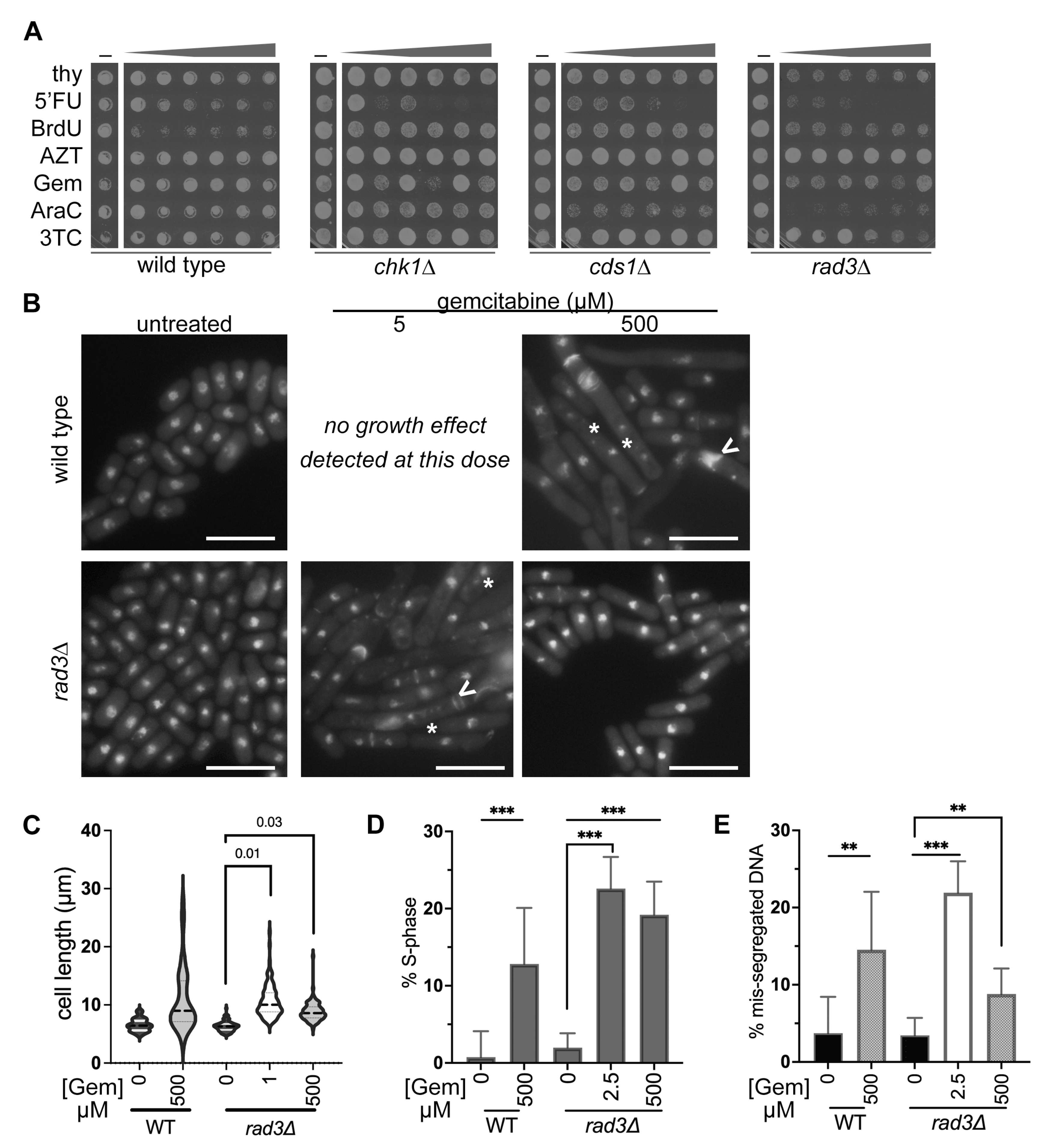
3.3. IC50 Doses of BrdU and Gemcitabine Induce DNA Mis-Segregation, Whilst Higher Doses Promote Drug-Resistant Morphologies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside analogues and nucleobases in cancer treatment. Lancet Oncol. 2002, 3, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 2008, 134, 124–146. [Google Scholar] [CrossRef]
- Tsesmetzis, N.; Paulin, C.B.J.; Rudd, S.G.; Herold, N. Nucleobase and Nucleoside Analogues: Resistance and Re-Sensitisation at the Level of Pharmacokinetics, Pharmacodynamics and Metabolism. Cancers 2018, 10, 240. [Google Scholar] [CrossRef]
- Peters, G.J. Novel Developments in the Use of Antimetabolites. Nucleosides Nucleotides Nucleic Acids 2014, 33, 358–374. [Google Scholar] [CrossRef]
- Dolnick, B.J.; Wu, X.P. Effects of 5-fluorouracil on mRNA. Adv. Exp. Med. Biol. 1993, 339, 57–63. [Google Scholar] [CrossRef]
- Silverstein, R.A.; de Valdivia, E.G.; Visa, N. The Incorporation of 5-Fluorouracil into RNA Affects the Ribonucleolytic Activity of the Exosome Subunit Rrp6. Mol. Cancer Res. 2011, 9, 332–340. [Google Scholar] [CrossRef]
- Chen, J.-K.; Merrick, K.A.; Kong, Y.W.; Izrael-Tomasevic, A.; Eng, G.; Handly, E.D.; Patterson, J.C.; Cannell, I.G.; Suarez-Lopez, L.; Hosios, A.M.; et al. An RNA damage response network mediates the lethality of 5-FU in colorectal cancer. Cell Rep. Med. 2024, 5, 101778. [Google Scholar] [CrossRef]
- Chalabi-Dchar, M.; Fenouil, T.; Machon, C.; Vincent, A.; Catez, F.; Marcel, V.; Mertani, H.C.; Saurin, J.-C.; Bouvet, P.; Guitton, J.; et al. A novel view on an old drug, 5-fluorouracil: An unexpected RNA modifier with intriguing impact on cancer cell fate. NAR Cancer 2021, 3, zcab032. [Google Scholar] [CrossRef]
- Mead, T.J.; Lefebvre, V. Proliferation assays (BrdU and EdU) on skeletal tissue sections. Methods Mol Biol. 2014, 1130, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Sabatinos, S.A.; Mastro, T.L.; Green, M.D.; Forsburg, S.L. A Mammalian-Like DNA Damage Response of Fission Yeast to Nucleoside Analogs. Genetics 2013, 193, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, S. In vivo labeling of fission yeast DNA with thymidine and thymidine analogs. Methods 2004, 33, 213–219. [Google Scholar] [CrossRef]
- Anderson, P.L.; Rower, J.E. Zidovudine and Lamivudine for HIV Infection. Clin. Med. Rev. Ther. 2010, 2, a2004. [Google Scholar] [CrossRef]
- Weinert, T. DNA damage and checkpoint pathways: Molecular anatomy and interactions with repair. Cell 1998, 94, 555–558. [Google Scholar] [CrossRef]
- Alcasabas, A.A.; Osborn, A.J.; Bachant, J.; Hu, F.; Werler, P.J.H.; Bousset, K.; Furuya, K.; Diffley, J.F.X.; Carr, A.M.; Elledge, S.J. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 2001, 3, 958–965. [Google Scholar] [CrossRef]
- Enoch, T.; Carr, A.M.; Nurse, P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992, 6, 2035–2046. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, H.D.; Griffiths, D.J.; Edwards, R.J.; Christensen, P.U.; Murray, J.M.; Osman, F.; Walworth, N.; Carr, A.M. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998, 12, 382–395. [Google Scholar] [CrossRef]
- Boddy, M.N.; Furnari, B.; Mondesert, O.; Russell, P. Replication Checkpoint Enforced by Kinases Cds1 and Chk1. Science 1998, 280, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Brondello, J.-M.; Boddy, M.N.; Furnari, B.; Russell, P. Basis for the Checkpoint Signal Specificity That Regulates Chk1 and Cds1 Protein Kinases. Mol. Cell. Biol. 1999, 19, 4262–4269. [Google Scholar] [CrossRef]
- Zeng, Y.; Forbes, K.C.; Wu, Z.; Moreno, S.; Piwnica-Worms, H.; Enoch, T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature 1998, 395, 507–510. [Google Scholar] [CrossRef]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Alexandra, B.L.; Elena, V.I.; Pan, Y.F.; Luigi, N.; Alexei, P.; Dipanjan, C.; David, P. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef]
- Zhang, F.; Carvalho, C.M.; Lupski, J.R. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009, 25, 298–307. [Google Scholar] [CrossRef]
- Holland, A.J.; Cleveland, D.W. Chromoanagenesis and cancer: Mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 2012, 18, 1630–1638. [Google Scholar] [CrossRef]
- Shoshani, O.; Bakker, B.; de Haan, L.; Tijhuis, A.E.; Wang, Y.; Kim, D.H.; Maldonado, M.; Demarest, M.A.; Artates, J.; Zhengyu, O.; et al. Transient genomic instability drives tumorigenesis through accelerated clonal evolution. Genes Dev. 2021, 35, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Closas, M.; Hall, P.; Nevanlinna, H.; Pooley, K.; Morrison, J.; Richesson, D.A.; Bojesen, S.E.; Nordestgaard, B.G.; Axelsson, C.K.; Arias, J.I.; et al. Heterogeneity of Breast Cancer Associations with Five Susceptibility Loci by Clinical and Pathological Characteristics. PLoS Genet. 2008, 4, e1000054. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Rondón, N.; Villegas, V.E.; Rondón-Lagos, M. The Role of Chromosomal Instability in Cancer and Therapeutic Responses. Cancers 2017, 10, 4. [Google Scholar] [CrossRef]
- Lepage, C.C.; Morden, C.R.; Palmer, M.C.L.; Nachtigal, M.W.; McManus, K.J. Detecting Chromosome Instability in Cancer: Approaches to Resolve Cell-to-Cell Heterogeneity. Cancers 2019, 11, 226. [Google Scholar] [CrossRef]
- Sahin, I.H.; Lowery, M.A.; Stadler, Z.K.; Salo-Mullen, E.; Iacobuzio-Donahue, C.A.; Kelsen, D.P.; O’reilly, E.M. Genomic instability in pancreatic adenocarcinoma: A new step towards precision medicine and novel therapeutic approaches. Expert. Rev. Gastroenterol. Hepatol. 2016, 10, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Wu, L.; Tannock, I.F. Repopulation in murine breast tumors during and after sequential treatments with cyclophosphamide and 5-fluorouracil. Cancer Res. 2003, 63, 2134–2138. [Google Scholar]
- Davis, A.J.; Tannock, I.F. Tumor physiology and resistance to chemotherapy: Repopulation and drug penetration. Cancer Treat. Res. 2002, 112, 1–26. [Google Scholar]
- Vukovic, B.; Beheshti, B.; Park, P.; Lim, G.; Bayani, J.; Zielenska, M.; Squire, J. Correlating breakage-fusion-bridge events with the overall chromosomal instability and in vitro karyotype evolution in prostate cancer. Cytogenet. Genome Res. 2007, 116, 1–11. [Google Scholar] [CrossRef]
- Bianchi, V.; Pontis, E.; Reichard, P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J. Biol. Chem. 1986, 261, 16037–16042. [Google Scholar] [CrossRef]
- Alvino, G.M.; Collingwood, D.; Murphy, J.M.; Delrow, J.; Brewer, B.J.; Raghuraman, M.K. Replication in hydroxyurea: It’s a matter of time. Mol. Cell. Biol. 2007, 27, 6396–6406. [Google Scholar] [CrossRef]
- Errico, A.; Costanzo, V.; Hunt, T. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA 2007, 104, 14929–14934. [Google Scholar] [CrossRef]
- Kurose, A.; Tanaka, T.; Huang, X.; Traganos, F.; Dai, W.; Darzynkiewicz, Z. Effects of hydroxyurea and aphidicolin on phosphorylation of ataxia telangiectasia mutated on Ser. 1981 and histone H2AX on Ser. 139 in relation to cell cycle phase and induction of apoptosis. Cytom. Part A 2006, 69, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Seifer, M.; Hamatake, R.K.; Colonno, R.J.; Standring, D.N. In Vitro Inhibition of Hepadnavirus Polymerases by the Triphosphates of BMS-200475 and Lobucavir. Antimicrob. Agents Chemother. 1998, 42, 3200–3208. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, C.; Pizzul, P.; Longhese, M.P.; Bonetti, D. Sensing R-Loop-Associated DNA Damage to Safeguard Genome Stability. Front. Cell Dev. Biol. 2021, 8, 618157. [Google Scholar] [CrossRef] [PubMed]
- Promonet, A.; Padioleau, I.; Liu, Y.; Sanz, L.; Biernacka, A.; Schmitz, A.-L.; Skrzypczak, M.; Sarrazin, A.; Mettling, C.; Rowicka, M.; et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Zimmer, J.; Tacconi, E.M.C.; Folio, C.; Badie, S.; Porru, M.; Klare, K.; Tumiati, M.; Markkanen, E.; Halder, S.; Ryan, A.; et al. Targeting BRCA1 and BRCA2 Deficiencies with G-Quadruplex-Interacting Compounds. Mol. Cell 2016, 61, 449–460. [Google Scholar] [CrossRef]
- Chang, E.Y.-C.; Tsai, S.; Aristizabal, M.J.; Wells, J.P.; Coulombe, Y.; Busatto, F.F.; Chan, Y.A.; Kumar, A.; Zhu, Y.D.; Wang, A.Y.-H.; et al. MRE11-RAD50-NBS1 promotes Fanconi Anemia R-loop suppression at transcription–replication conflicts. Nat. Commun. 2019, 10, 4265. [Google Scholar] [CrossRef]
- Lisby, M.; Barlow, J.H.; Burgess, R.C.; Rothstein, R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 2004, 118, 699–713. [Google Scholar] [CrossRef]
- Du, L.L.; Nakamura, T.M.; Russell, P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006, 20, 1583–1596. [Google Scholar] [CrossRef]
- Jazayeri, A.; Falck, J.; Lukas, C.; Bartek, J.; Smith, G.C.M.; Lukas, J.; Jackson, S.P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006, 8, 37–45. [Google Scholar] [CrossRef]
- Hicks, W.M.; Yamaguchi, M.; Haber, J.E. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc. Natl. Acad. Sci. USA 2011, 108, 3108–3115. [Google Scholar] [CrossRef]
- Mehta, A.; Haber, J.E. Sources of DNA Double-Strand Breaks and Models of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef]
- Hsiang, Y.H.; Lihou, M.G.; Liu, L.F. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989, 49, 5077–5082. [Google Scholar] [PubMed]
- Wan, S.; Capasso, H.; Walworth, N.C. The topoisomerase I poison camptothecin generates a Chk1-dependent DNA damage checkpoint signal in fission yeast. Yeast 1999, 15, 821–828. [Google Scholar] [CrossRef]
- Tsao, Y.P.; D’Arpa, P.; Liu, L.F. The involvement of active DNA synthesis in camptothecin-induced G2 arrest: Altered regulation of p34cdc2/cyclin B. Cancer Res. 1992, 52, 1823–1829. [Google Scholar] [PubMed]
- Acilan, C.; Potter, D.M.; Saunders, W.S. DNA repair pathways involved in anaphase bridge formation. Genes Chromosom. Cancer 2007, 46, 522–531. [Google Scholar] [CrossRef]
- Liu, Y.; Nielsen, C.F.; Yao, Q.; Hickson, I.D. The origins and processing of ultra fine anaphase DNA bridges. Curr. Opin. Genet. Dev. 2014, 26, 1–5. [Google Scholar] [CrossRef]
- Karpenshif, Y.; Bernstein, K.A. From yeast to mammals: Recent advances in genetic control of homologous recombination. DNA Repair. 2012, 11, 781–788. [Google Scholar] [CrossRef]
- Alabert, C.; Bianco, J.N.; Pasero, P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 2009, 28, 1131–1141. [Google Scholar] [CrossRef]
- Aylon, Y.; Kupiec, M. New insights into the mechanism of homologous recombination in yeast. Mutat. Res. Mol. Mech. Mutagen. 2004, 566, 231–248. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair. 2007, 6, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Sogo, J.M.; Lopes, M.; Foiani, M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 2002, 297, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Sun, L.; Shen, F.; Chen, Y.; Hua, Y.; Liu, Y.; Zhang, M.; Hu, Y.; Wang, Q.; Xu, W.; et al. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 2012, 149, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, C.S.; Wood, V.; Fantes, P.A. An Ancient Yeast for Young Geneticists: A Primer on the Schizosaccharomyces pombe Model System. Genetics 2015, 201, 403–423, Erratum in Genetics 2016, 202, 1241. [Google Scholar] [CrossRef]
- Sipiczki, M. Where does fission yeast sit on the tree of life? Genome Biol. 2000, 1, reviews1011.1. [Google Scholar] [CrossRef] [PubMed]
- Vyas, A.; Freitas, A.V.; Ralston, Z.A.; Tang, Z. Fission Yeast Schizosaccharomyces pombe: A Unicellular “Micromammal” Model Organism. Curr. Protoc. 2021, 1, e151, Erratum in Curr. Protoc. 2021, 1, e225. [Google Scholar] [CrossRef]
- Chhipa, M.A.; Sanayhie, S.A.; Sabatinos, S.A. Assessing Drug Sensitivity in Fission Yeast Using Half-Maximal Inhibitory Concentration (IC50) Assays. Methods Mol. Biol. 2025, 2862, 241–253. [Google Scholar] [CrossRef]
- Boeckemeier, L.; Kraehenbuehl, R.; Keszthelyi, A.; Gasasira, M.U.; Vernon, E.G.; Beardmore, R.; Vågbø, C.B.; Chaplin, D.; Gollins, S.; Krokan, H.E.; et al. Mre11 exonuclease activity removes the chain-terminating nucleoside analog gemcitabine from the nascent strand during DNA replication. Sci. Adv. 2020, 6, eaaz4126. [Google Scholar] [CrossRef]
- Alyahya, M.Y.; Khan, S.; Bhadra, S.; Samuel, R.E.; Xu, Y.-J. Replication stress induced by the ribonucleotide reductase inhibitor guanazole, triapine and gemcitabine in fission yeast. FEMS Yeast Res. 2022, 22, foac014. [Google Scholar] [CrossRef]
- Fleck, O.; Fahnøe, U.; Løvschal, K.V.; Gasasira, M.-F.U.; Marinova, I.N.; Kragelund, B.B.; Carr, A.M.; Hartsuiker, E.; Holmberg, C.; Nielsen, O. Deoxynucleoside Salvage in Fission Yeast Allows Rescue of Ribonucleotide Reductase Deficiency but Not Spd1-Mediated Inhibition of Replication. Genes 2017, 8, 128. [Google Scholar] [CrossRef]
- Hodson, J.A.; Bailis, J.M.; Forsburg, S.L. Efficient labeling of fission yeast Schizosaccharomyces pombe with thymidine and BUdR. Nucleic Acids Res. 2003, 31, 134e. [Google Scholar] [CrossRef]
- Sabatinos, S.A.; Forsburg, S.L. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 2010, 470, 759–795. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Russell, P. Growth and the Environment of Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2016, pdb.top079764, Erratum in Cold Spring Harb. Protoc. 2016, pdb.err093682. Erratum in Cold Spring Harb. Protoc. 2016. [Google Scholar] [CrossRef] [PubMed]
- Karam, E.; Sabatinos, S.A. Investigating Fission Yeast Mutagenesis Using Canavanine Sensitivity Assays. Methods Mol. Biol. 2025, 2862, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Rhind, N.; Russell, P. Chk1 and Cds1: Linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 2000, 113, 3889–3896. [Google Scholar] [CrossRef]
- Bentley, N.J.; Holtzman, D.A.; Flaggs, G.; Keegan, K.S.; DeMaggio, A.; Ford, J.C.; Hoekstra, M.; Carr, A.M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996, 15, 6641–6651. [Google Scholar] [CrossRef]
- Sabatinos, S.A.; Green, M.D.; Forsburg, S.L. Continued DNA Synthesis in Replication Checkpoint Mutants Leads to Fork Collapse. Mol. Cell. Biol. 2012, 32, 4986–4997. [Google Scholar] [CrossRef]
- Yanagida, M. Fission yeast cut mutations revisited: Control of anaphase. Trends Cell Biol. 1998, 8, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Deng, X. Cell Synchronization by Double Thymidine Block. Bio-Protocol 2018, 8, e2994. [Google Scholar] [CrossRef]
- Hung, C.-W.; Martínez-Márquez, J.Y.; Javed, F.T.; Duncan, M.C. A simple and inexpensive quantitative technique for determining chemical sensitivity in Saccharomyces cerevisiae. Sci. Rep. 2018, 8, 11919. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Alvin, W.T.-N.; Yang, W.; Arnoud, B.; Kyle, R.C.; Dmitry, A.G.; Erik, N.B.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101, Erratum in Nature. 2023, 614, E41. [Google Scholar] [CrossRef]
- Bohnert, K.A.; Gould, K.L. Cytokinesis-based constraints on polarized cell growth in fission yeast. PLoS Genet. 2012, 8, e1003004, Erratum in PLoS Genet. 2013, 9. [Google Scholar] [CrossRef]
- Kippert, F.; Lloyd, D. The aniline blue fluorochrome specifically stains the septum of both live and fixed Schizosaccharomyces pombe cells. FEMS Microbiol. Lett. 1995, 132, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, V. Gemcitabine: Progress in the Treatment of Pancreatic Cancer. Oncology 2000, 60, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Xie, J. Promising molecular mechanisms responsible for gemcitabine resistance in cancer. Genes Dis. 2015, 2, 299–306. [Google Scholar] [CrossRef]
- Höfer, S.; Frasch, L.; Brajkovic, S.; Putzker, K.; Lewis, J.; Schürmann, H.; Leone, V.; Sakhteman, A.; The, M.; Bayer, F.P.; et al. Gemcitabine and ATR inhibitors synergize to kill PDAC cells by blocking DNA damage response. Mol. Syst. Biol. 2025, 21, 231–253. [Google Scholar] [CrossRef]
- Masterson, J.C.; O’DEa, S. 5-Bromo-2-deoxyuridine activates DNA damage signalling responses and induces a senescence-like phenotype in p16-null lung cancer cells. Anti-Cancer Drugs 2007, 18, 1053–1068. [Google Scholar] [CrossRef]
- Duong, H.; Bin Hong, Y.; Kim, J.S.; Lee, H.; Yi, Y.W.; Kim, Y.J.; Wang, A.; Zhao, W.; Cho, C.H.; Seong, Y.; et al. Inhibition of checkpoint kinase 2 (CHK2) enhances sensitivity of pancreatic adenocarcinoma cells to gemcitabine. J. Cell. Mol. Med. 2013, 17, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Kianfard, Z.; Chhipa, M.A.; Kianfard, Z.; Karam, E.; Magalage, S.P.; Sabatinos, S.A. Genetic profiles of gemcitabine sensitivity in Schizosaccharomyces pombe. bioRxiv 2025. [CrossRef]
- Dixon, S.J.; Andrews, B.; Boone, C. Exploring the conservation of synthetic lethal genetic interaction networks. Commun. Integr. Biol. 2009, 2, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S.; O’CArrigan, B.; Jackson, S.P.; Yap, T.A. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov. 2017, 7, 20–37. [Google Scholar] [CrossRef]
- Turk, A.A.; Wisinski, K.B. PARP inhibitors in breast cancer: Bringing synthetic lethality to the bedside. Cancer 2018, 124, 2498–2506. [Google Scholar] [CrossRef] [PubMed]
- Min, A.; Im, S.-A.; Yoon, Y.-K.; Song, S.-H.; Nam, H.-J.; Hur, H.-S.; Kim, H.-P.; Lee, K.-H.; Han, S.-W.; Oh, D.-Y.; et al. RAD51C-Deficient Cancer Cells Are Highly Sensitive to the PARP Inhibitor Olaparib. Mol. Cancer Ther. 2013, 12, 865–877. [Google Scholar] [CrossRef]
- Zhao, L.; So, C.W.E. PARP-inhibitor-induced synthetic lethality for acute myeloid leukemia treatment. Exp. Hematol. 2016, 44, 902–907. [Google Scholar] [CrossRef]
- Fordham, S.E.; Blair, H.J.; Elstob, C.J.; Plummer, R.; Drew, Y.; Curtin, N.J.; Heidenreich, O.; Pal, D.; Jamieson, D.; Park, C.; et al. Inhibition of ATR acutely sensitizes acute myeloid leukemia cells to nucleoside analogs that target ribonucleotide reductase. Blood Adv. 2018, 2, 1157–1169. [Google Scholar] [CrossRef]
- Bradbury, A.; Hall, S.; Curtin, N.; Drew, Y. Targeting ATR as Cancer Therapy: A new era for synthetic lethality and synergistic combinations? Pharmacol. Ther. 2020, 207, 107450. [Google Scholar] [CrossRef] [PubMed]
- Lynx, M.D.; Bentley, A.T.; McKee, E.E. 3′-Azido-3′-deoxythymidine (AZT) inhibits thymidine phosphorylation in isolated rat liver mitochondria: A possible mechanism of AZT hepatotoxicity. Biochem. Pharmacol. 2006, 71, 1342–1348. [Google Scholar] [CrossRef][Green Version]
- Francisco, B.; García-Benayas, T.; Juan, J.d.l.C.; Juan, G.-L.; Vincent, S. First-Line Therapy and Mitochondrial Damage: Different Nucleosides, Different Findings. HIV Clin. Trials 2003, 4, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Sutera, V.A.; Lovett, S.T. The role of replication initiation control in promoting survival of replication fork damage. Mol. Microbiol. 2006, 60, 229–239. [Google Scholar] [CrossRef]
- Canadian Cancer Statistics Advisory in collaboration with, the Canadian Cancer Society, Statistics Canada and the, Public Health Agency of Canada. Canadian Cancer Statistics: A 2022 Special Report on Cancer Prevalence. 2022. Available online: https://cdn.cancer.ca/-/media/files/research/cancer-statistics/2022-statistics/2022-special-report/2022_prevalence_report_final_en.pdf?rev=7755f9f350e845d58e268a59e3be608e&hash=3F3F30CADD8CAF0049636B5A41EDBB13&_gl=1*21rw8e*_ga*MTI5OTE3MTQ0Ni4xNjQzMzA0MDMz*_ga_23YMKBE2C3*MTY2NzkxMDY0My4zMTcuMS4xNjY3OTEwNjQ2LjU3LjAuMA (accessed on 1 July 2023).
- Bukhari, A.B.; Chan, G.K.; Gamper, A.M. Targeting the DNA Damage Response for Cancer Therapy by Inhibiting the Kinase Wee1. Front. Oncol. 2022, 12, 828684. [Google Scholar] [CrossRef]
- Fu, S.; Yao, S.; Previs, R.A.; Elias, A.D.; Carvajal, R.D.; George, T.J.; Yuan, Y.; Yu, L.; Westin, S.N.; Xing, Y.; et al. Multicenter Phase II Trial of the WEE1 Inhibitor Adavosertib in Refractory Solid Tumors Harboring CCNE1 Amplification. J. Clin. Oncol. 2023, 41, 1725–1734. [Google Scholar] [CrossRef]
- Cuneo, K.C.; Morgan, M.A.; Sahai, V.; Schipper, M.J.; Parsels, L.A.; Parsels, J.D.; Devasia, T.; Al-Hawaray, M.; Cho, C.S.; Nathan, H.; et al. Dose Escalation Trial of the Wee1 Inhibitor Adavosertib (AZD1775) in Combination with Gemcitabine and Radiation for Patients with Locally Advanced Pancreatic Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 2643–2650. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.; Cooney, T.; Yang, X.; Pal, S.; Ermoian, R.; Gajjar, A.; Liu, X.; Prem, K.; Minard, C.G.; Reid, J.M.; et al. Wee1 kinase inhibitor adavosertib with radiation in newly diagnosed diffuse intrinsic pontine glioma: A Children’s Oncology Group phase I consortium study. Neuro-Oncol. Adv. 2022, 4, vdac073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Peng, K.; Liu, Q.; Huang, Q.; Liu, T. Adavosertib and beyond: Biomarkers, drug combination and toxicity of WEE1 inhibitors. Crit. Rev. Oncol. 2023, 193, 104233. [Google Scholar] [CrossRef]

| Strain | ID | Genotype | Source [Ref] |
|---|---|---|---|
| wild type (WT) | SY 17 | h+ leu1–32::hENT1-leu1+ (pJAH29) his7–366::hsv-tk-his7+ (pJAH31) ura4-D18 ade6-M210 | Susan Forsburg [38] |
| cds1∆ | SY 91 | h+ cds1Δ::ura4+ leu1–32::hENT1-leu+ (pJAH29) his7–366::hsv-tk-his7+ (pJAH31) ura4-D18 ade6-M210 | Susan Forsburg [39] |
| chk1∆ | SY 92 | h+ chk1Δ::ura4+ leu1–32::hENT1-leu1+ (pJAH29) his7–366::hsv-tk-his7+ (pJAH31) ura4-D18 ade6-M210 | Susan Forsburg [39] |
| rad3∆ | SY 93 | h+ rad3∆::ura4+ leu1–32::hENT1-leu1+ (pJAH29) his7–366::hsv-tk-his7+ (pJAH31) ura4-D18 ade6-M210 | Susan Forsburg [39] |
| Calculated IC50 ± SD (n) | |||||
|---|---|---|---|---|---|
| Strain | Thy | 5FU | BrdU | Gem | AraC |
| wild type (WT) | >500 a (5) | 383 ± 458 (5) | 11.9 ± 4.2 (6) | >500 (4) | >500 (4) |
| chk1∆ | >500 a (4) | >500 (4) | 7.7 ± 2.7 (4) | >500 (4) | 37.9 ± 35.8 (4) |
| cds1∆ | 21.6 ± 5.8 (3) | 73.6 ± 30.6 (4) | 15.9 ± 7.1 (4) | 2.2 ± 0.4 (4) | 13.6 ± 9.3 (3) |
| rad3∆ | 14.9 ± 1.1 (4) | 36.1 ± 16.2 (4) | 3.6 ± 1.3 (4) | 1.1 ± 0.1 (4) | 0.15 ± 0.05 (4) |
| Gem | Gem Dose (µM) | Mean Length ± SD (µm) | Number of Cells (n) | p-Value (Untreated Compared to Treated) |
|---|---|---|---|---|
| Wild type | 0 | 6.5 (1.1) | 100 | – |
| 500 | 10.9 (5.5) | 105 | 0.005 | |
| rad3∆ | 0 | 6.3 (0.1) | 301 (3) | – |
| IC50 | 10.7 (0.7) | 312 (3) | 0.01 | |
| 500 | 8.8 (0.5) | 304 (3) | 0.03 |
| BrdU | BrdU Dose (µM) | Mean Length (±SD, µm) | Number of Cells (n) | p Value (Significance) |
|---|---|---|---|---|
| Wild type | 0 | 6.5 (0.9) | 289 | – |
| 5 | 15.9 (1.9) | 317 | 0.05 (*) | |
| 500 | 11.1 (1.1) | 282 | 0.10 (ns) | |
| cds1∆ | 0 | 6.3 (0.4) | 282 | – |
| 5 | 12.5 (2.1) | 224 | 0.05 (*) | |
| 500 | 6.9 (0.3) | 208 | 0.12 (ns) | |
| chk1∆ | 0 | 6.7 (0.5) | 287 | – |
| 5 | 12.3 (0.7) | 269 | 0.02 (*) | |
| 500 | 7.6 (1.3) | 306 | 0.60 (ns) | |
| rad3∆ | 0 | 6.5 (0.4) | 306 | – |
| 5 | 10.1 (0.3) | 292 | <0.0001 (***) | |
| 500 | 8.7 (0.3) | 308 | <0.04 (*) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kagalwala, Z.B.; Chhipa, M.A.; Kianfard, Z.; Karam, E.; Magalage, S.P.; Sabatinos, S.A. Checkpoint-Dependent Sensitivities to Nucleoside Analogues Uncover Specific Patterns of Genomic Instability. Curr. Issues Mol. Biol. 2025, 47, 756. https://doi.org/10.3390/cimb47090756
Kagalwala ZB, Chhipa MA, Kianfard Z, Karam E, Magalage SP, Sabatinos SA. Checkpoint-Dependent Sensitivities to Nucleoside Analogues Uncover Specific Patterns of Genomic Instability. Current Issues in Molecular Biology. 2025; 47(9):756. https://doi.org/10.3390/cimb47090756
Chicago/Turabian StyleKagalwala, Zainab Burhanuddin, Mohammed Ayan Chhipa, Zohreh Kianfard, Essam Karam, Sirasie P. Magalage, and Sarah A. Sabatinos. 2025. "Checkpoint-Dependent Sensitivities to Nucleoside Analogues Uncover Specific Patterns of Genomic Instability" Current Issues in Molecular Biology 47, no. 9: 756. https://doi.org/10.3390/cimb47090756
APA StyleKagalwala, Z. B., Chhipa, M. A., Kianfard, Z., Karam, E., Magalage, S. P., & Sabatinos, S. A. (2025). Checkpoint-Dependent Sensitivities to Nucleoside Analogues Uncover Specific Patterns of Genomic Instability. Current Issues in Molecular Biology, 47(9), 756. https://doi.org/10.3390/cimb47090756







