Distinct Transcriptomic Profile Underlying High CO2 and Ethylene-Induced Deastringency in ‘Daebong’ Persimmon Fruit
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, High CO2 and Ethylene Treatments
2.2. RNA Extraction
2.3. cDNA Library Construction, Sequencing, and Quality Control
2.4. De Novo Assembly and Contig Filtering
2.5. Identification of DEGs and Functional Enrichment Analysis
2.6. Firmness, Ethanol Insoluble (EIS), Pectin Content, and Polygalacturonase (PG) Activity
2.7. Ethylene Production and Respiration Rate
2.8. Soluble Tannin Content and Total Phenolics
2.9. Antioxidant Activities
2.10. Color Changes and Overall Sensory Evaluation
2.11. Statistical Analysis of Quality Parameters
3. Results and Discussion
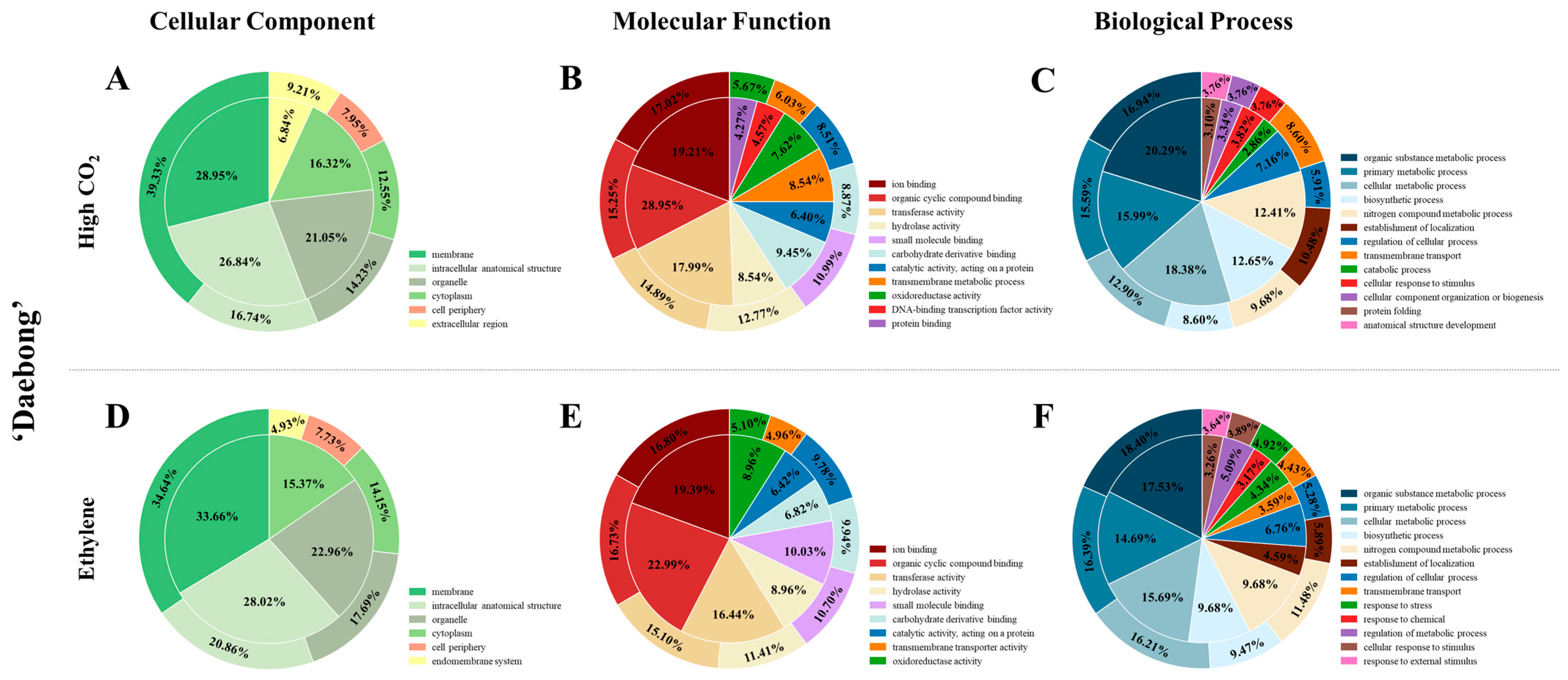
3.1. Assembly, Annotation, and DEGs
3.2. DEGs in the Comparison of High CO2 and Ethylene Treated vs. Control Persimmon Fruit
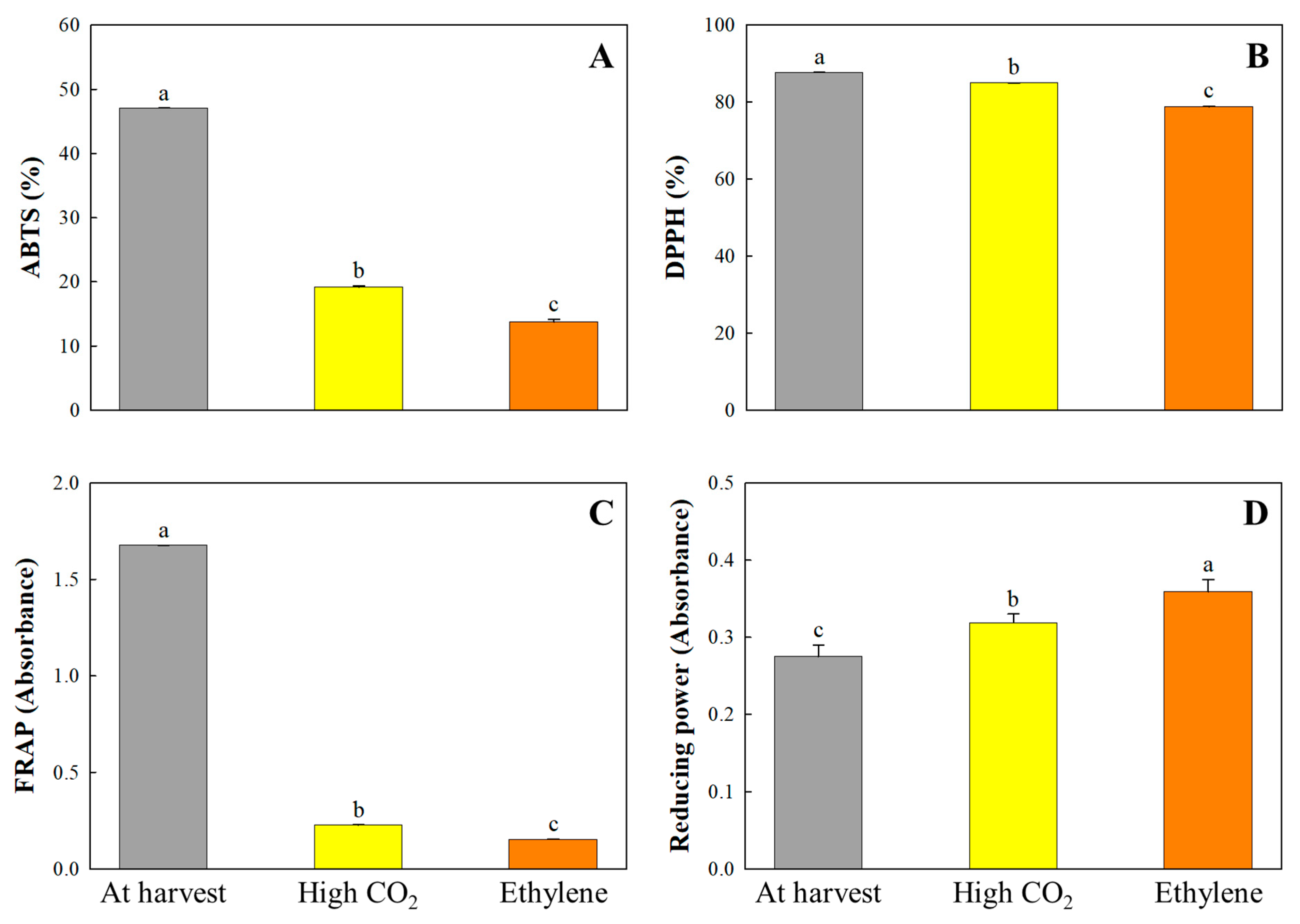
3.3. Soluble Tannin, Total Phenolics, and Genes Related to Deastringency
3.4. Firmness, EIS, Total Pectin, PG Activity, and Related Genes
3.5. Ethylene Production and Respiration Rates and Related Genes
3.6. Antioxidant Activities and Stress-Related Genes
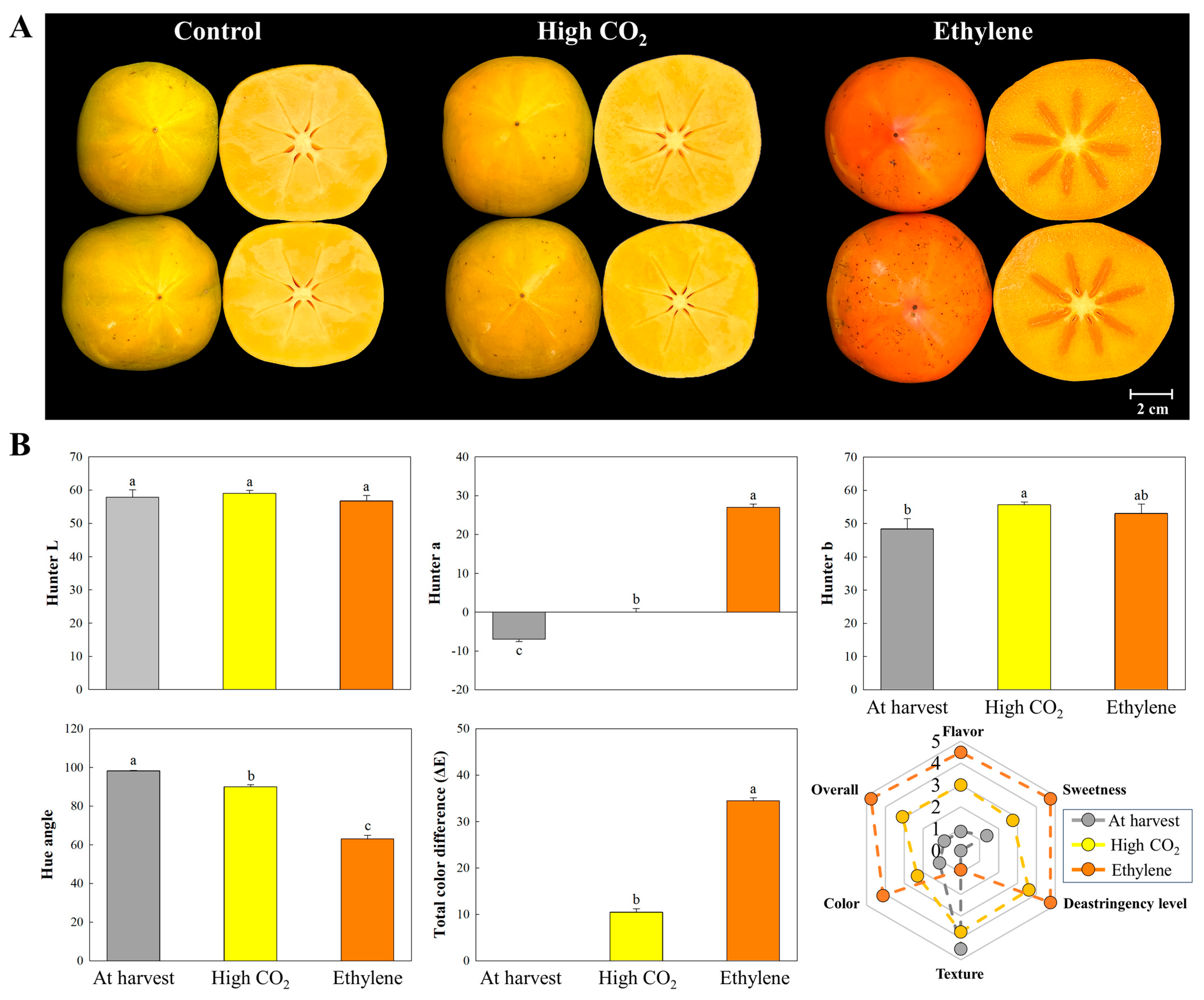
3.7. Color Changes, Overall Sensory Quality, and the Related Genes
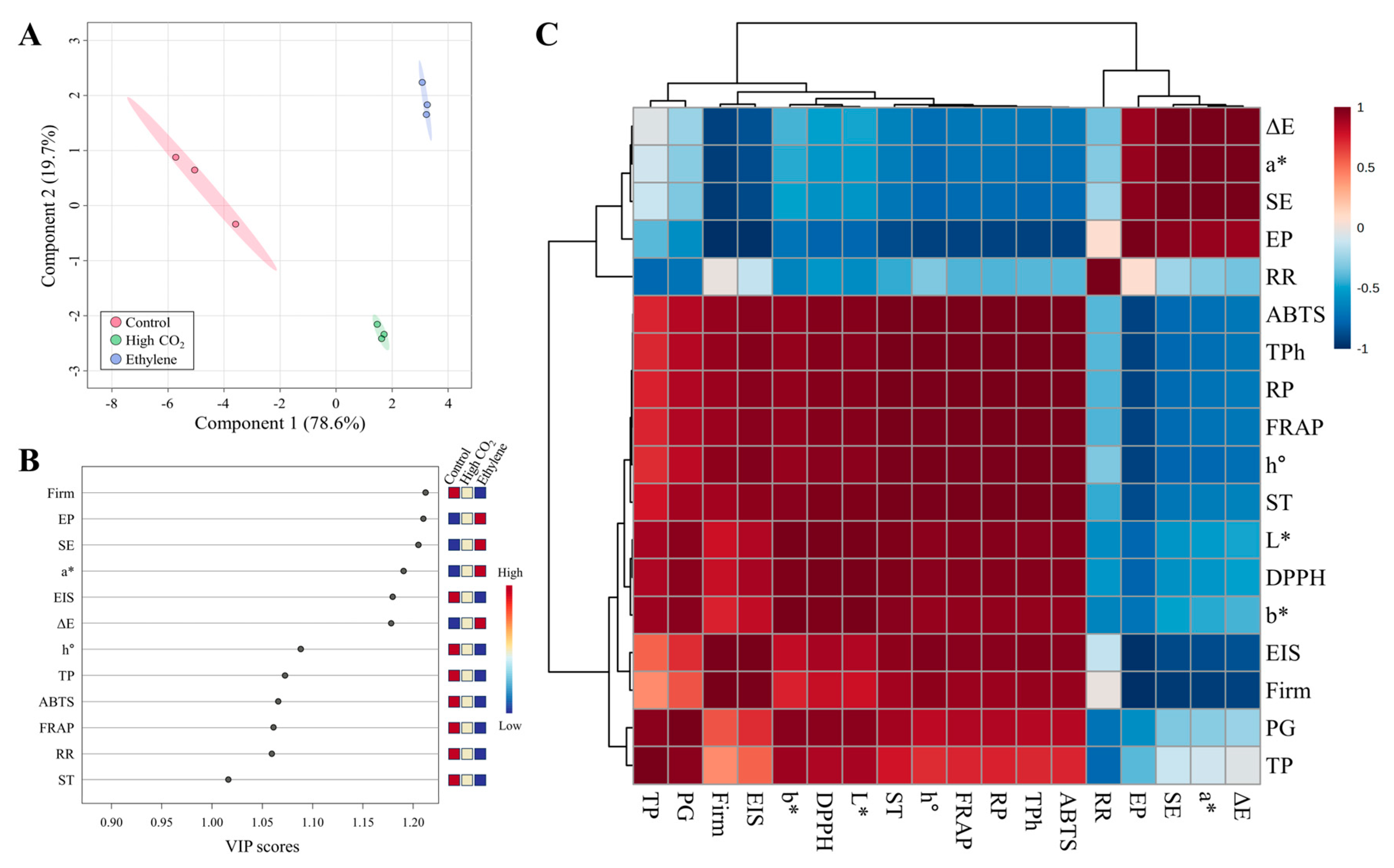
3.8. Partial Least Squares-Discriminant and Correlation Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- FAOSTAT. Food and Agriculture Organization of the United Nations Statistics Division. Available online: http://www.fao.org/home/en (accessed on 1 May 2024).
- Baek, M.W.; Choi, H.R.; Hwang, I.G.; Tilahun, S.; Jeong, C.S. Prediction of tannin content and quality parameters in astringent persimmons from visible and near-infrared spectroscopy. Front. Plant Sci. 2023, 14, 1260644. [Google Scholar] [CrossRef]
- Park, D.S.; Tilahun, S.; Heo, J.Y.; Jeong, C.S. Quality and expression of ethylene response genes of ‘Daebong’ persimmon fruit during ripening at different temperatures. Postharvest Biol. Technol. 2017, 133, 57–63. [Google Scholar] [CrossRef]
- Joo, O.-S.; Kang, S.-T.; Jeong, C.-H.; Lim, J.-W.; Park, Y.-G.; Cho, K.-M. Manufacturing of the enhances antioxidative wine using a ripe daebong persimmon (Dispyros kaki L.). J. Appl. Biol. Chem. 2011, 54, 126–134. [Google Scholar] [CrossRef]
- Sato, A.; Yamada, M. Persimmon breeding in Japan for pollination-constant non-astringent (PCNA) type with marker-assisted selection. Breed. Sci. 2016, 66, 60–68. [Google Scholar] [CrossRef]
- Zhu, Q.-G.; Xu, Y.; Yang, Y.; Guan, C.-F.; Zhang, Q.-Y.; Huang, J.-W.; Grierson, D.; Chen, K.-S.; Gong, B.-C.; Yin, X.-R. The persimmon (Diospyros oleifera Cheng) genome provides new insights into the inheritance of astringency and ancestral evolution. Hortic. Res. 2019, 6, 138. [Google Scholar] [CrossRef]
- Das, P.R.; Eun, J.-B. Removal of astringency in persimmon fruits (Diospyros kaki) subjected to different freezing temperature treatments. J. Food Sci. Technol. 2021, 58, 3154–3163. [Google Scholar] [CrossRef]
- Chung, H.-S.; Kim, H.-S.; Lee, Y.-G.; Seong, J.-H. Effect of deastringency treatment of intact persimmon fruits on the quality of fresh-cut persimmons. Food Chem. 2015, 166, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, A.E.; Candir, E.; Toplu, C.; Yildiz, E. Effect of hot water treatment on astringency removal in persimmon cultivars. Int. J. Fruit Sci. 2020, 20 (Suppl. S2), S557–S569. [Google Scholar] [CrossRef]
- Cortés, V.; Rodríguez, A.; Blasco, J.; Rey, B.; Besada, C.; Cubero, S.; Salvador, A.; Talens, P.; Aleixos, N. Prediction of the level of astringency in persimmon using visible and near-infrared spectroscopy. J. Food Eng. 2017, 204, 27–37. [Google Scholar] [CrossRef]
- Tanaka, T.; Matsuo, Y.; Saito, Y. Solubility of tannins and preparation of oil-soluble derivatives. J. Oleo Sci. 2018, 67, 1179–1187. [Google Scholar] [CrossRef]
- Eram, M.S.; Ma, K. Decarboxylation of pyruvate to acetaldehyde for ethanol production by hyperthermophiles. Biomolecules 2013, 3, 578–596. [Google Scholar] [CrossRef]
- Waarde, A. Alcoholic fermentation in multicellular organisms. Physiol. Zool. 1991, 64, 895–920. [Google Scholar] [CrossRef]
- Kou, J.; Wei, C.; Zhao, Z.; Guan, J.; Wang, W. Effects of ethylene and 1-methylcyclopropene treatments on physiological changes and ripening-related gene expression of ‘Mopan’ persimmon fruit during storage. Postharvest Biol. Technol. 2020, 166, 111185. [Google Scholar] [CrossRef]
- Shimomura, M. Ripening control with ethylene and ethephon on a rapid-deastringency system in persimmon fruits. Int. Persimmon Symp. 1996, 436, 215–234. [Google Scholar] [CrossRef]
- Kato, K. Astringency removal and ripening in persimmons treated with ethanol and ethylene. Hortic. Sci. 1990, 25, 205–207. [Google Scholar] [CrossRef]
- He, Y.; Xue, J.; Li, H.; Han, S.; Jiao, J.; Rao, J. Ethylene response factors regulate ethylene biosynthesis and cell wall modification in persimmon (Diospyros kaki L.) fruit during ripening. Postharvest Biol. Technol. 2020, 168, 111255. [Google Scholar] [CrossRef]
- Park, D.S.; Tilahun, S.; Park, K.C.; Choi, I.Y.; Jeong, C.S. Transcriptome analysis of astringent ‘Cheongdo-Bansi’ persimmon fruit treated with ethylene for removal of astringency. Postharvest Biol. Technol. 2019, 150, 52–59. [Google Scholar] [CrossRef]
- Nishiyama, S.; Onoue, N.; Kono, A.; Sato, A.; Yonemori, K.; Tao, R. Characterization of a gene regulatory network underlying astringency loss in persimmon fruit. Planta 2018, 247, 733–743. [Google Scholar] [CrossRef]
- Ding, Y.; Shen, X.; Ding, Y.; Zhang, P.; Zhu, Q.; Wang, Y.; Zhang, Q.; Luo, Z.; Yang, Y.; Du, X.; et al. A comprehensive transcriptomic and metabolomic map reveals the molecular mechanism of persimmon fruit deastringency upon 40 °C warm water treatment. Postharvest Biol. Technol. 2025, 220, 113313. [Google Scholar] [CrossRef]
- Salvador, A.; Arnal, L.; Besada, C.; Larrea, V.; Quiles, A.; Pérez-Munuera, I. Physiological and structural changes during ripening and deastringency treatment of persimmon fruit cv. ‘Rojo Brillante’. Postharvest Biol. Technol. 2007, 46, 181–188. [Google Scholar] [CrossRef]
- Wu, W.; Wang, M.-M.; Gong, H.; Liu, X.-F.; Guo, D.-L.; Sun, N.-J.; Huang, J.-W.; Zhu, Q.-G.; Chen, K.-S.; Yin, X.-R.; et al. High CO2/hypoxia-induced softening of persimmon fruit is modulated by DkERF8/16 and DkNAC9 complexes. J. Exp. Bot. 2020, 71, 2690–2700. [Google Scholar] [CrossRef]
- Mamrot, J.; Legaie, R.; Ellery, S.J.; Wilson, T.; Seemann, T.; Powell, D.R.; Gardner, D.K.; Walker, D.W.; Temple-Smith, P.; Papenfuss, A.T.; et al. De novo transcriptome assembly for the spiny mouse (Acomys cahirinus). Sci. Rep. 2017, 7, 8996. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Choi, H.R.; Tilahun, S.; Park, D.S.; Lee, Y.M.; Choi, J.H.; Baek, M.W.; Jeong, C.S. Harvest time affects quality and storability of kiwifruit (Actinidia spp.) Cultivars during long-term cool storage. Sci. Hortic. 2019, 256, 108523. [Google Scholar] [CrossRef]
- Seo, M.H.; Tilahun, S.; Park, D.S.; Melaku, A.; Jeong, C.S. Effect of ripening conditions on the quality and storability of muskmelon (Cucumis melo L.) fruits. J. Hortic. Sci. Technol. 2018, 36, 741–755. [Google Scholar] [CrossRef]
- Tewari, R.; Gupta, M.; Ahmad, F.; Rout, P.K.; Misra, L.; Patwardhan, A.; Vasudeva, R. Extraction, quantification and antioxidant activities of flavonoids, polyphenols and pinitol from wild and cultivated Saraca asoca bark using RP-HPLC-PDA-RI method. Ind. Crop. Prod. 2017, 103, 73–80. [Google Scholar] [CrossRef]
- Baek, M.W.; Choi, H.R.; Solomon, T.; Jeong, C.S.; Lee, O.-H.; Tilahun, S. Preharvest Methyl Jasmonate Treatment Increased the Antioxidant Activity and Glucosinolate Contents of Hydroponically Grown Pak Choi. Antioxidants 2021, 10, 131. [Google Scholar] [CrossRef]
- Choi, Y.-E.; Choi, S.-I.; Han, X.; Men, X.; Jang, G.-W.; Kwon, H.-Y.; Kang, S.-R.; Han, J.-S.; Lee, O.-H. Radical scavenging-linked anti-adipogenic activity of aster scaber ethanolic extract and its bioactive compound. Antioxidants 2020, 9, 1290. [Google Scholar] [CrossRef]
- Cervera-Chiner, L.; Vilhena, N.Q.; Larrea, V.; Moraga, G.; Salvador, A. Influence of temperature on ‘Rojo Brillante’ persimmon drying. Quality characteristics and drying kinetics. LWT 2024, 197, 115902. [Google Scholar] [CrossRef]
- Choi, H.R.; Baek, M.W.; Cheol, L.H.; Jeong, C.S.; Tilahun, S. Changes in metabolites and antioxidant activities of green ‘Hayward’ and gold ‘Haegeum’ kiwifruits during ripening with ethylene treatment. Food Chem. 2022, 384, 132490. [Google Scholar] [CrossRef]
- Sarkhosh, A.; Habibi, F.; Brecht, J.K. Alleviating Astringency in Persimmon Fruit for Enhanced Palatability and Consumer Acceptability: HS1483, 5/2024. EDIS 2024, 3, 1–7. [Google Scholar] [CrossRef]
- Arnal, L.; del Río, M.A. Effect of cold storage and removal astringency on quality of persimmon fruit (Diospyros kaki L.) cv. Rojo Brillante. Food Sci. Technol. Int. 2004, 10, 179–185. [Google Scholar] [CrossRef]
- Tanaka, T.; Takahashi, R.; Kouno, I.; Nonaka, G.-I. Chemical evidence for the de-astringency (insolubilization of tannins) of persimmon fruit. J. Chem. Soc. Perkin Trans. 1994, 1, 3013–3022. [Google Scholar] [CrossRef]
- Zhao, W.; Zheng, M.; Li, X.; Song, K.; Shi, D. Fruit Astringency: Mechanisms, Technologies, and Future Directions. Horticulturae 2025, 11, 699. [Google Scholar] [CrossRef]
- Amorim, C.; Antoniolli, L.R.; Orsi, B.; Kluge, R.A. Advances in metabolism and genetic control of astringency in persimmon (Diospyros kaki Thunb.) fruit: A review. Sci. Hortic. 2023, 308, 111561. [Google Scholar] [CrossRef]
- Tessmer, M.A.; Appezzato-Da-Glória, B.; Kluge, R.A. Astringency in ‘Giombo’ persimmon and its relationship with the harvest time. Rev. Ceres 2016, 63, 646–652. [Google Scholar] [CrossRef]
- Barbosa, M.C.F.; Morais, M.A.S.; Melo, M.F.; Souza, P.A.; Sarmento, J.D.A.; Morais, P.L.D. Maturation and antioxidant activity of ‘Giombo’ and ‘Rama Forte’ persimmons produced in the Brazilian semiarid. Braz. J. Biol. 2024, 84, e276146. [Google Scholar] [CrossRef]
- Denev, P.; Yordanov, A. Total polyphenol, proanthocyanidin and flavonoid content, carbohydrate composition and antioxidant activity of persimmon (Diospyros kaki L.) fruit in relation to cultivar and maturity stage. Bulg. J. Agric. Sci. 2013, 19, 981–988. [Google Scholar]
- Del Bubba, M.; Giordani, E.; Pippucci, L.; Cincinelli, A.; Checchini, L.; Galvan, P. Changes in tannins, ascorbic acid and sugar content in astringent persimmons during on-tree growth and ripening and in response to different postharvest treatments. J. Food Compos. Anal. 2009, 22, 668–677. [Google Scholar] [CrossRef]
- Seymour, G.B.; Østergaard, L.; Chapman, N.H.; Knapp, S.; Martin, C. Fruit development and ripening. Annu. Rev. Plant Biol. 2013, 64, 219–241. [Google Scholar] [CrossRef]
- Giovannoni, J.J. Genetic regulation of fruit development and ripening. Plant Cell 2004, 16 (Suppl. S1), S170–S180. [Google Scholar] [CrossRef]
- Taira, S.; Ono, M.; Matsumoto, N. Reduction of persimmon astringency by complex formation between pectin and tannins. Postharvest Biol. Technol. 1997, 12, 265–271. [Google Scholar] [CrossRef]
- Choi, H.R.; Jeong, M.J.; Baek, M.W.; Choi, J.H.; Lee, H.C.; Jeong, C.S.; Tilahun, S. Transcriptome Analysis of Pre-Storage 1-MCP and High CO2-Treated ‘Madoka’ Peach Fruit Explains the Reduction in Chilling Injury and Improvement of Storage Period by Delaying Ripening. Int. J. Mol. Sci. 2021, 22, 4437. [Google Scholar] [CrossRef]
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Katsube, T.; Tabata, H.; Ohta, Y.; Yamasaki, Y.; Anuurad, E.; Shiwaku, K.; Yamane, Y. Screening for antioxidant activity in edible plant products: Comparison of low-density lipoprotein oxidation assay, DPPH radical scavenging assay, and Folin−Ciocalteu assay. J. Agric. Food Chem. 2004, 52, 2391–2396. [Google Scholar] [CrossRef]


| Treatment | Raw Data | Trimmed Data | Mapped Reads | Mapping Rate (%) | Number of DEGs (Control vs. Treatments) p < 0.05, | ||
|---|---|---|---|---|---|---|---|
| Control (at harvest) | 67,200,370 | 64,955,644 | 43,602,204 | 67.1 | Up | Down | Total |
| High CO2 (1 day) | 76,167,104 | 72,745,056 | 44,372,198 | 61 | 274 | 231 | 505 |
| Ethylene (4 day) | 75,785,722 | 73,072,096 | 47,458,880 | 64.9 | 798 | 968 | 1766 |
| Gene ID | Gene Descriptions | Log2 Fold Change | Gene Functions | |
|---|---|---|---|---|
| High CO2 | Ethylene | |||
| Upregulated | ||||
| LOC127802412 | F-box protein PP2-B15-like | 12.26 | 13.86 | Protein binding |
| AID51426.1 | ethylene response factor 21 | 11.63 | 8.04 | |
| LOC127797932 | flavanone 3-dioxygenase 3 | 10.30 | 6.55 | |
| LOC127803199 | PI-PLC X domain-containing protein At5g67130-like | 9.85 | 13.57 | Posphoric diester hydrolase activity |
| LOC127810275 | F-box protein VBF-like | 9.15 | 7.60 | Potein binding |
| LOC127794125 | 1-aminocyclopropane-1-carboxylate oxidase 1 | 9.11 | 11.64 | |
| LOC127790996 | ethylene-responsive transcription factor ERF115-like | 9.05 | 9.49 | DNA-binding transcription factor activity |
| LOC127798805 | ethylene-responsive transcription factor ABR1-like isoform X2 | 8.16 | 7.20 | DNA-binding transcription factor activity |
| LOC127802859 | lysine histidine transporter-like 8 | 8.14 | 6.93 | |
| LOC127812266 | sucrose synthase 2-like isoform X2 | 8.13 | 5.18 | Sucrose synthase activity |
| LOC127802270 | receptor-like protein kinase 7 | 7.91 | 6.35 | Potein kinase activity |
| PSR89067.1 | Membrane-associated kinase regulator | 7.91 | 8.97 | |
| LOC127803148 | calcium-binding protein CML46 | 7.63 | 4.57 | Calcium ion binding |
| LOC127801080 | E3 ubiquitin-protein ligase ATL31-like | 7.54 | 8.88 | |
| LOC127798535 | E3 ubiquitin-protein ligase RGLG5-like | 7.44 | 9.60 | |
| LOC127795244 | malate synthase, glyoxysomal | 7.17 | 13.89 | Malate synthase activity |
| LOC127808940 | mitogen-activated protein kinase kinase kinase 5-like isoform X1 | 7.12 | 5.93 | ATP binding |
| LOC127797657 | NAC domain-containing protein 1 | 6.72 | 6.49 | DNA binding |
| AID51417.1 | ethylene response factor 12 | 6.33 | 5.72 | |
| LOC127790200 | cationic peroxidase 1-like | 6.17 | 7.09 | Peroxidase activity |
| LOC127787838 | galacturonosyltransferase-like 9 | 6.09 | 6.17 | Glycosyltransferase activity |
| LOC127809143 | cytochrome P450 86B1-like | 5.70 | 11.06 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| PSR99923.1 | 3-isopropylmalate dehydratase large subunit like | 5.67 | 10.58 | |
| AEC11088.1 | MYB transcription factor PA1 | 5.54 | 10.13 | |
| LOC127797399 | NDR1/HIN1-like protein 13 | 5.23 | 5.39 | |
| LOC127809456 | transcription factor MYB108-like | 5.17 | 6.15 | DNA-binding transcription factor activity |
| LOC127808889 | WRKY transcription factor 31 | 5.06 | 8.36 | DNA-binding transcription factor activity |
| LOC127801120 | L-ascorbate oxidase homolog | 4.98 | 4.53 | Oxidoreductase activity |
| LOC127789678 | plant cysteine oxidase 1-like | 4.82 | 6.68 | Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen |
| LOC127790398 | cytochrome P450 76A1-like | 4.40 | 11.89 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| QGV56724.1 | WRKY transcription factor 15 | 4.33 | 3.53 | |
| LOC127801139 | beta-glucosidase 11-like isoform X1 | 4.31 | 6.55 | Beta-glucosidase activity |
| AZL19548.1 | transcription factor WRKY11 | 4.30 | 5.83 | |
| LOC127792859 | flavonol 3-O-glucosyltransferase UGT89B1-like | 4.30 | 4.68 | UDP-glycosyltransferase activity |
| LOC127811263 | UDP-glycosyltransferase 75C1-like | 4.11 | 6.04 | UDP-glycosyltransferase activity |
| LOC127795520 | ethylene-responsive transcription factor WIN1-like | 3.51 | 8.57 | DNA-binding transcription factor activity |
| LOC127800536 | NAC domain-containing protein 2 | 3.24 | 3.20 | DNA binding |
| Downregulated | ||||
| LOC127787223 | pectinesterase inhibitor 9-like | −7.66 | −5.35 | Enzyme inhibitor activity |
| LOC127812875 | cellulose synthase-like protein G3 isoform X4 | −7.45 | −5.02 | Cellulose synthase (UDP-forming) activity |
| LOC127800039 | germin-like protein subfamily 1 member 11 | −7.14 | −6.30 | Manganese ion binding |
| LOC127806638 | wax ester synthase/diacylglycerol acyltransferase 5-like | −6.67 | −12.37 | Long-chain-alcohol O-fatty-acyltransferase activity |
| LOC114274781 | E3 ubiquitin-protein ligase MARCH10 | −6.15 | −7.31 | Zinc ion binding |
| LOC127810041 | beta-xylosidase/alpha-L-arabinofuranosidase 2-like isoform X2 | −6.00 | −8.57 | Alpha-L-arabinofuranosidase activity |
| LOC127812875 | cellulose synthase-like protein G3 isoform X2 | −5.94 | −7.63 | Cellulose synthase (UDP-forming) activity |
| LOC127808982 | berberine bridge enzyme-like 15 | −5.75 | −7.91 | Oxidoreductase activity |
| LOC127807607 | thaumatin-like protein 1 | −5.51 | −8.67 | |
| LOC127812875 | cellulose synthase-like protein G3 isoform X1 | −5.30 | −6.95 | Cellulose synthase (UDP-forming) activity |
| LOC127796807 | 4-coumarate--CoA ligase-like 1 | −5.12 | −4.94 | Long-chain fatty acid-CoA ligase activity |
| PSR95878.1 | Pre-neck appendage protein | −5.08 | −4.36 | |
| LOC127809129 | boron transporter 2 | −4.51 | −5.50 | Solute:inorganic anion antiporter activity |
| KAI7982608.1 | putative pectinesterase/pectinesterase inhibitor 40 | −4.20 | −6.32 | |
| LOC109009267 | expansin-A15 isoform X1 | −4.07 | −8.39 | |
| LOC127796063 | beta-amyrin 28-monooxygenase-like | −3.96 | −6.55 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127803927 | cytochrome P450 71AU50-like isoform X3 | −3.77 | −6.86 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| PSS00292.1 | Developmental and secondary metabolism regulator veA like | −3.73 | −4.15 | |
| LOC127800637 | serine/threonine-protein kinase WNK9 isoform X1 | −3.72 | −6.83 | Protein serine/threonine kinase activity |
| LOC127805784 | protein STRICTOSIDINE SYNTHASE-LIKE 4-like isoform X2 | −3.64 | −6.73 | Strictosidine synthase activity |
| LOC127813526 | glucomannan 4-beta-mannosyltransferase 2 | −3.61 | −8.01 | Mannan synthase activity |
| LOC127806942 | NADPH-dependent oxidoreductase 2-alkenal reductase-like | −3.59 | −4.54 | Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor |
| LOC127798462 | IAA-amino acid hydrolase ILR1-like 1 | −3.37 | −9.56 | IAA-Ala conjugate hydrolase activity |
| Gene ID | Gene Descriptions | Log2 Fold Change | Gene Functions |
|---|---|---|---|
| Upregulated | |||
| LOC127810272 | peroxidase 5-like | 11.28 | Peroxidase activity |
| LOC127799450 | 22.0 kDa class IV heat shock protein-like | 9.89 | Unfolded protein binding |
| LOC127803768 | 17.5 kDa class I heat shock protein-like | 9.74 | Unfolded protein binding |
| LOC127808179 | ethylene-responsive transcription factor ERF014-like | 9.22 | DNA-binding transcription factor activity |
| LOC127797208 | glutathione S-transferase U17-like isoform X1 | 9.11 | Glutathione transferase activity |
| LOC127796264 | protein TIFY 9-like | 7.34 | |
| LOC127804909 | xyloglucan endotransglucosylase/hydrolase protein 23 | 6.80 | Xyloglucan:xyloglucosyl transferase activity |
| LOC127800824 | aspartic proteinase GIP2 | 6.78 | Aspartic-type endopeptidase activity |
| PSS29393.1 | Mitochondrial fission regulator like | 6.77 | |
| LOC127797673 | xyloglucan endotransglucosylase/hydrolase protein 33 | 6.72 | Xyloglucan:xyloglucosyl transferase activity |
| LOC127795131 | 17.3 kDa class II heat shock protein-like | 6.72 | Unfolded protein binding |
| LOC127808826 | alcohol dehydrogenase 1 | 6.42 | S-(hydroxymethyl)glutathione dehydrogenase [NAD(P)+] activity |
| LOC127790650 | (−)-isopiperitenol/(−)-carveol dehydrogenase, mitochondrial-like | 6.31 | |
| LOC127796562 | class I heat shock protein | 6.29 | Unfolded protein binding |
| LOC127806544 | heat shock 70 kDa protein | 6.26 | Heat shock protein binding |
| LOC127810698 | 2-oxoglutarate-dependent dioxygenase 19-like | 6.12 | |
| LOC127793595 | pectinesterase/pectinesterase inhibitor 41 | 6.11 | Pectinesterase inhibitor activity |
| LOC127788897 | aspartate aminotransferase, mitochondrial-like isoform X2 | 5.75 | Pyridoxal phosphate binding |
| LOC127791465 | cytochrome P450 72A397-like | 5.68 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127791460 | glucose-6-phosphate/phosphate translocator 2, chloroplastic-like | 5.66 | Triose-phosphate transmembrane transporter activity |
| KAI8032162.1 | Peroxidase 51 | 5.62 | |
| LOC127792124 | 1-aminocyclopropane-1-carboxylate oxidase | 5.44 | |
| LOC127797235 | F-box protein SKIP28 | 5.09 | Protein binding |
| LOC127787406 | wall-associated receptor kinase-like 20 | 4.99 | Protein serine/threonine kinase activity |
| LOC127790698 | cytosolic sulfotransferase 12-like | 4.90 | Sulfotransferase activity |
| LOC127806329 | cucumber peeling cupredoxin-like | 4.87 | Electron transfer activity |
| LOC127800236 | universal stress protein A-like protein | 4.83 | |
| LOC127799949 | alcohol dehydrogenase 3 | 4.71 | S-(hydroxymethyl)glutathione dehydrogenase [NAD(P)+] activity |
| LOC127805737 | jasmonate-induced oxygenase 1-like isoform X1 | 4.66 | |
| ASL69240.1 | ethylene response factor 26 | 4.55 | |
| LOC127793838 | protein TIFY 10A-like | 4.45 | |
| LOC127794477 | wall-associated receptor kinase-like 10 isoform X5 | 4.39 | Polysaccharide binding |
| LOC127789995 | NADPH-dependent aldo-keto reductase, chloroplastic-like | 4.22 | Aldose reductase (NADPH) activity |
| LOC127801110 | WRKY transcription factor 46 | 4.11 | DNA-binding transcription factor activity |
| LOC127801209 | expansin-like A2 | 4.09 | |
| LOC127799270 | glutathione S-transferase | 4.06 | Glutathione transferase activity |
| LOC127793268 | endoplasmic reticulum oxidoreductin-1-like | 4.00 | Thiol oxidase activity |
| LOC127792628 | epidermis-specific secreted glycoprotein EP1-like | 3.66 | |
| LOC127789222 | putative receptor protein kinase ZmPK1 | 3.66 | Protein serine/threonine kinase activity |
| LOC127813273 | WRKY transcription factor WRKY24 | 3.63 | DNA-binding transcription factor activity |
| Q8S932.1 | 1-aminocyclopropane-1-carboxylate oxidase | 3.62 | |
| LOC127808277 | putative 12-oxophytodienoate reductase 11 isoform X1 | 3.18 | Oxidoreductase activity |
| Downregulated | |||
| LOC127795923 | pectate lyase 18 | −8.67 | Pectate lyase activity |
| LOC127799002 | putative beta-D-xylosidase | −8.10 | Xylan 1,4-beta-xylosidase activity |
| KAF5729599.1 | Expansin A4 ALPHA 1.6 EXPA4 | −8.08 | |
| LOC127792836 | nucleobase-ascorbate transporter 4 | −7.20 | Transmembrane transporter activity |
| LOC127787055 | monothiol glutaredoxin-S9-like | −7.01 | |
| LOC127792213 | cytochrome P450 724B1-like | −6.77 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127795951 | GDSL esterase/lipase 1-like isoform X5 | −5.46 | Hydrolase activity, acting on ester bonds |
| LOC127793870 | AP2-like ethylene-responsive transcription factor At1g16060 | −5.33 | DNA-binding transcription factor activity |
| LOC127800698 | aspartyl protease AED3 | −4.97 | Aspartic-type endopeptidase activity |
| LOC127803129 | serine carboxypeptidase-like 25 | −4.90 | Serine-type carboxypeptidase activity |
| PSS05082.1 | Phenylalanine ammonia-lyase | −4.82 | |
| LOC127812252 | stemmadenine O-acetyltransferase-like | −4.73 | |
| LOC127795350 | aspartyl protease AED3-like | −4.61 | Aspartic-type endopeptidase activity |
| LOC127813223 | nudix hydrolase 8-like | −4.60 | NADH pyrophosphatase activity |
| LOC127798278 | peroxidase P7-like | −4.51 | Peroxidase activity |
| LOC127790066 | pyrophosphate-energized vacuolar membrane proton pump-like | −4.40 | Diphosphate hydrolysis-driven proton transmembrane transporter activity |
| LOC127786991 | GDSL esterase/lipase At1g54790-like | −4.33 | Hydrolase activity, acting on ester bonds |
| LOC127793738 | beta-amyrin 28-monooxygenase-like | −4.18 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127795833 | cytochrome P450 736A117-like isoform X1 | −3.96 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127809955 | glycosyl hydrolase 5 family protein-like | −3.94 | Hydrolase activity, hydrolyzing O-glycosyl compounds |
| AXQ60485.1 | acetaldehyde dehydrogenase | −3.75 | |
| LOC127787809 | UPF0481 protein At3g47200-like isoform X1 | −3.60 | |
| LOC127793571 | 7-deoxyloganetic acid glucosyltransferase-like | −3.48 | UDP-glycosyltransferase activity |
| LOC127791330 | NAC domain-containing protein 72-like | −3.44 | DNA binding |
| GFZ19603.1 | glycosyl hydrolase family protein | −3.26 | |
| LOC127799077 | xyloglucan O-acetyltransferase 4-like | −3.23 | O-acetyltransferase activity |
| Gene ID | Gene Descriptions | Log2 Fold Change | Gene Functions |
|---|---|---|---|
| Upregulated | |||
| GFS33599.1 | laccase 5 | 14.88 | |
| LOC127812189 | stemmadenine O-acetyltransferase-like | 14.03 | |
| KAH9668398.1 | Endonuclease | 12.98 | |
| LOC127792808 | 9-cis-epoxycarotenoid dioxygenase NCED2, chloroplastic | 12.97 | Carotenoid dioxygenase activity |
| OVA11305.1 | Cyclin PHO80-like | 12.67 | |
| LOC127812613 | metalloendoproteinase 4-MMP-like | 11.50 | Metalloendopeptidase activity |
| LOC127803872 | indole-3-pyruvate monooxygenase YUCCA7 | 11.09 | N,N-dimethylaniline monooxygenase activity |
| LOC127792678 | glutathione S-transferase parC isoform X2 | 11.05 | Glutathione transferase activity |
| LOC127795217 | EIN3-binding F-box protein 1-like | 10.87 | Protein binding |
| BAB89348.1 | 1-aminocyclopropane-1-carboxylate synthase | 10.67 | |
| KAH9793099.1 | Vascular-related protein 1 | 10.60 | |
| LOC127787815 | polygalacturonase-like isoform X2 | 10.25 | Polygalacturonase activity |
| LOC127792636 | (S)-N-methylcoclaurine 3′-hydroxylase isozyme 2 | 9.97 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127792270 | transcription factor MYB101 | 9.47 | |
| LOC127805642 | GABA transporter 1 | 9.45 | |
| LOC127810232 | bidirectional sugar transporter N3-like | 9.15 | Sugar transmembrane transporter activity |
| PSR98642.1 | FMN-dependent NADH-azoreductase | 9.07 | |
| LOC127814200 | EG45-like domain containing protein isoform X2 | 9.00 | |
| LOC127796202 | 7-deoxyloganetic acid glucosyltransferase-like | 8.94 | UDP-glycosyltransferase activity |
| LOC127805921 | NAC domain-containing protein 1-like | 8.88 | DNA binding |
| LOC127813338 | salutaridine reductase-like isoform X2 | 8.76 | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor |
| LOC127789396 | hexose carrier protein HEX6-like isoform X1 | 8.71 | Monosaccharide transmembrane transporter activity |
| LOC127805642 | GABA transporter 1 | 8.67 | |
| AOR05828.1 | xyloglucan endotransglycosylase/hydrolase 10 | 8.64 | |
| LOC127808774 | alcohol acyltransferase 9 | 8.59 | Acyltransferase activity, transferring groups other than amino-acyl groups |
| LOC114318315 | laccase-15-like isoform X1 | 8.49 | Hydroquinone:oxygen oxidoreductase activity |
| LOC127795412 | GDSL esterase/lipase 5-like | 8.30 | Hydrolase activity, acting on ester bonds |
| LOC127793042 | myb-related protein 305-like | 8.25 | Sequence-specific DNA binding |
| LOC127790046 | very-long-chain aldehyde decarbonylase CER3-like isoform X1 | 8.02 | Oxidoreductase activity |
| PSS24712.1 | Axin-1 like | 7.97 | |
| LOC127790278 | cytochrome P450 98A2 | 7.94 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127801145 | trehalose-phosphate phosphatase H isoform X1 | 7.89 | Trehalose-phosphatase activity |
| LOC127805724 | protein SMALL AUXIN UPREGULATED RNA 12-like | 7.84 | |
| LOC127807158 | tetrahydroberberine oxidase-like | 7.73 | Oxidoreductase activity |
| AID51423.1 | ethylene response factor 18 | 7.64 | |
| LOC122583038 | 22.0 kDa heat shock protein | 7.54 | |
| LOC127787260 | acidic endochitinase-like | 7.44 | Chitinase activity |
| LOC127803070 | rhamnogalacturonate lyase B isoform X3 | 7.43 | Carbohydrate binding |
| LOC127796343 | AP2/ERF and B3 domain-containing transcription factor RAV1-like | 7.34 | DNA-binding transcription factor activity |
| GFY95051.1 | cupredoxin superfamily protein | 7.08 | |
| LOC127796343 | AP2/ERF and B3 domain-containing transcription factor RAV1-like | 7.01 | DNA-binding transcription factor activity |
| LOC127807942 | monooxygenase 2-like | 6.97 | FAD binding |
| LOC127791424 | glycosyltransferase At3g07620 | 6.90 | Glycosyltransferase activity |
| LOC127802236 | proline-rich receptor-like protein kinase PERK7 | 6.87 | Protein kinase activity |
| LOC8279466 | cytosolic sulfotransferase 15 | 6.75 | Sulfotransferase activity |
| LOC127795644 | 23 kDa jasmonate-induced protein-like | 6.72 | |
| LOC127797898 | 11-beta-hydroxysteroid dehydrogenase A-like | 6.70 | Oxidoreductase activity |
| LOC127789041 | UDP-glycosyltransferase 73C3-like | 6.69 | UDP-glycosyltransferase activity |
| LOC127797462 | peroxidase 4-like | 6.66 | Peroxidase activity |
| LOC127797581 | cytochrome P450 86A1-like | 6.61 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127805525 | putative polyol transporter 1 | 6.60 | Carbohydrate:proton symporter activity |
| LOC127786959 | (+)-neomenthol dehydrogenase-like | 6.59 | |
| LOC127787695 | respiratory burst oxidase homolog protein A-like | 6.58 | NAD(P)H oxidase H2O2-forming activity |
| LOC127791004 | mitochondrial phosphate carrier protein 3, mitochondrial-like | 6.22 | Phosphate transmembrane transporter activity |
| PSS31544.1 | Rhamnogalacturonate lyase | 5.99 | |
| LOC127796209 | galacturonosyltransferase-like 9 | 5.90 | Glycosyltransferase activity |
| AOR05829.1 | xyloglucan endotransglycosylase/hydrolase 11 | 5.88 | |
| LOC127797011 | classical arabinogalactan protein 5-like | 5.86 | |
| BAB89349.1 | 1-aminocyclopropane-1-carboxylate synthase | 5.81 | |
| LOC127796404 | trehalose-phosphate phosphatase 2 isoform X3 | 5.79 | Trehalose-phosphatase activity |
| LOC127808654 | adenylate-forming reductase 03009 | 5.77 | |
| LOC127809972 | polygalacturonase At1g48100-like | 5.60 | Polygalacturonase activity |
| LOC127807241 | ethylene-responsive transcription factor RAP2-1-like | 5.54 | DNA-binding transcription factor activity |
| LOC127799926 | CASP-like protein 1 | 5.54 | |
| LOC127798463 | transcription factor MYB48 | 5.48 | Sequence-specific DNA binding |
| LOC127805880 | inositol oxygenase 1-like isoform X2 | 5.47 | Inositol oxygenase activity |
| LOC127803057 | elongation of fatty acids protein 3-like | 5.38 | Fatty acid elongase activity |
| LOC127803147 | methyltransferase PMT19 isoform X1 | 5.38 | Methyltransferase activity |
| LOC127786954 | (+)-neomenthol dehydrogenase-like | 5.38 | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor |
| LOC127797620 | UDP-glucosyltransferase 29-like | 5.38 | UDP-glycosyltransferase activity |
| AGA15800.1 | ethylene response factor 9 | 5.27 | |
| LOC127809456 | transcription factor MYB108-like | 5.25 | DNA-binding transcription factor activity |
| LOC127809221 | ethylene-responsive transcription factor ERF008-like | 5.24 | DNA-binding transcription factor activity |
| LOC127811484 | tabersonine 16-hydroxylase 2-like | 5.17 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127813020 | cytochrome P450 711A1 isoform X1 | 5.16 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127786644 | cytochrome P450 CYP72A219-like | 5.14 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127792028 | xyloglucan endotransglucosylase protein 1-like | 5.13 | Hydrolase activity, hydrolyzing O-glycosyl compounds |
| LOC127804981 | L-type lectin-domain containing receptor kinase IX.1-like isoform X2 | 5.12 | ATP binding |
| LOC127789757 | protein kinase At2g41970 | 5.10 | Protein tyrosine kinase activity |
| ANO39898.1 | ethylene response factor 24 | 5.10 | |
| LOC127811155 | trans-cinnamate 4-monooxygenase | 5.10 | Trans-cinnamate 4-monooxygenase activity |
| LOC127798547 | UDP-glycosyltransferase 88A1-like | 5.06 | UDP-glycosyltransferase activity |
| EOY16099.1 | Structural constituent of ribosome, putative | 5.03 | |
| LOC127808940 | mitogen-activated protein kinase kinase kinase 5-like isoform X1 | 4.97 | ATP binding |
| LOC127797621 | beta-D-glucosyl crocetin beta-1,6-glucosyltransferase-like isoform X8 | 4.95 | UDP-glycosyltransferase activity |
| LOC127806048 | putative GTP diphosphokinase RSH1, chloroplastic | 4.85 | |
| LOC127809200 | D-galacturonate reductase-like | 4.81 | Aldose reductase (NADPH) activity |
| LOC127795794 | pleiotropic drug resistance protein 2-like | 4.79 | ATP binding |
| LOC127788535 | ethylene-overproduction protein 1 | 4.79 | Protein binding |
| AVR54525.1 | MYB transcription factor | 4.78 | |
| LOC127808842 | cytochrome P450 81Q32-like | 4.72 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127798669 | transcription factor MYB73-like | 4.71 | DNA-binding transcription factor activity, RNA polymerase II-specific |
| LOC127802382 | acyl-CoA-binding domain-containing protein 5-like | 4.60 | Protein binding |
| LOC127801844 | cytokinin riboside 5′-monophosphate phosphoribohydrolase LOG1-like isoform X3 | 4.60 | Hydrolase activity, hydrolyzing N-glycosyl compounds |
| PSS05082.1 | Phenylalanine ammonia-lyase | 4.59 | |
| LOC127804247 | protein MIZU-KUSSEI 1-like | 4.53 | |
| LOC114265924 | myb-binding protein 1A-like protein | 4.50 | Transcription factor binding |
| LOC127805731 | UDP-glycosyltransferase 74F2-like | 4.47 | UDP-glycosyltransferase activity |
| AHE13906.1 | xyloglucan endotransglucosylase/hydrolase 9 | 4.43 | |
| LOC127788427 | glycosyltransferase family 92 protein RCOM_0530710 | 4.32 | |
| LOC127806120 | galactinol—sucrose galactosyltransferase 6 | 4.30 | |
| LOC127796158 | glucuronoxylan glucuronosyltransferase IRX7 | 4.29 | Glycosyltransferase activity |
| LOC127810184 | transcription factor BHLH089-like | 4.26 | DNA-binding transcription factor activity |
| LOC127792240 | fasciclin-like arabinogalactan protein 9 | 4.25 | |
| PSR84714.1 | Galacturonosyltransferase 12 | 4.23 | |
| LOC127807192 | beta-galactosidase 16-like | 4.22 | Beta-galactosidase activity |
| LOC127790738 | protein ORANGE-LIKE, chloroplastic isoform X1 | 4.17 | |
| LOC127802040 | flavin-containing monooxygenase FMO GS-OX5-like isoform X2 | 4.16 | N,N-dimethylaniline monooxygenase activity |
| KAJ4723831.1 | Caffeic acid O-methyltransferase | 4.14 | |
| PSS04611.1 | FAD synthase | 4.09 | |
| LOC127802877 | gallate 1-beta-glucosyltransferase 84A24 | 4.08 | UDP-glycosyltransferase activity |
| LOC127796482 | LOB domain-containing protein 15 | 4.06 | |
| LOC127801122 | axial regulator YABBY 1 | 4.04 | |
| PSR93351.1 | Pectinesterase inhibitor domain protein | 3.98 | |
| PSR91886.1 | Chaperone protein like | 3.95 | |
| LOC127800265 | lipase-like isoform X1 | 3.94 | |
| LOC127794167 | dammarenediol II synthase-like | 3.94 | Beta-amyrin synthase activity |
| PSS02795.1 | Plant intracellular Ras-group-related LRR protein | 3.92 | |
| LOC127814076 | auxin-responsive protein SAUR36-like | 3.91 | |
| LOC127797571 | ethylene-responsive transcription factor ERF113-like | 3.90 | DNA-binding transcription factor activity |
| LOC127799406 | polygalacturonase | 3.88 | Polygalacturonase activity |
| LOC127807635 | aldehyde oxidase GLOX | 3.83 | |
| LOC127793192 | scarecrow-like protein 27 | 3.82 | DNA-binding transcription factor activity |
| LOC127807986 | thioredoxin-like protein CXXS1 | 3.76 | |
| LOC127799531 | sugar transporter ERD6-like 16 | 3.75 | Sugar transmembrane transporter activity |
| LOC127805879 | zinc finger A20 and AN1 domain-containing stress-associated protein 5-like | 3.75 | Zinc ion binding |
| LOC127793707 | oxygen-evolving enhancer protein 3, chloroplastic | 3.72 | Electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity |
| GFS45640.1 | thioredoxin superfamily protein | 3.70 | |
| BAH89267.1 | putative leucoanthocyanidin reductase | 3.69 | |
| LOC127813769 | cytokinin dehydrogenase 7 | 3.69 | Cytokinin dehydrogenase activity |
| AYW01721.1 | persimmon protein ERF25 | 3.67 | |
| PSR93233.1 | Acetyl-coenzyme A synthetase | 3.62 | |
| LOC127802856 | (+)-neomenthol dehydrogenase-like | 3.54 | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor |
| LOC127802400 | NAC domain-containing protein 104-like | 3.53 | DNA binding |
| LOC127813180 | ethylene-responsive transcription factor ERF061 | 3.52 | DNA-binding transcription factor activity |
| LOC127798691 | EIN3-binding F-box protein 1-like | 3.49 | Protein binding |
| LOC127810431 | glucan endo-1,3-beta-glucosidase-like | 3.48 | Hydrolase activity, hydrolyzing O-glycosyl compounds |
| LOC127811516 | fructokinase-4 | 3.47 | Fructokinase activity |
| LOC127797006 | cyclin-dependent protein kinase inhibitor SMR13-like | 3.45 | |
| LOC127797433 | ATP-dependent 6-phosphofructokinase 4, chloroplastic isoform X1 | 3.44 | 6-phosphofructokinase activity |
| LOC127804052 | beta-D-xylosidase 6 | 3.43 | Alpha-L-arabinofuranosidase activity |
| LOC127799002 | putative beta-D-xylosidase | 3.43 | Alpha-L-arabinofuranosidase activity |
| LOC127797597 | pyruvate decarboxylase 1-like isoform X2 | 3.42 | Carboxy-lyase activity |
| LOC127790127 | ethylene-responsive transcription factor RAP2-7-like isoform X2 | 3.39 | DNA-binding transcription factor activity |
| LOC127809307 | chloroplast protein FOR GROWTH AND FERTILITY 2-like | 3.35 | |
| LOC127808885 | thylakoidal processing peptidase 2, chloroplastic isoform X3 | 3.29 | Serine-type endopeptidase activity |
| LOC127807957 | protein MULTIPLE CHLOROPLAST DIVISION SITE 1 isoform X2 | 3.26 | |
| Downregulated | |||
| LOC127809111 | alpha-galactosidase 1-like | −12.79 | Hydrolase activity, hydrolyzing O-glycosyl compounds |
| AIL29216.1 | fatty acid hydroperoxide lyase | −8.61 | |
| LOC127803467 | protein CELLULOSE SYNTHASE INTERACTIVE 1 | −8.19 | Microtubule binding |
| LOC127791627 | receptor-like protein kinase At5g24010 | −7.90 | Transmembrane receptor protein tyrosine kinase activity |
| LOC127796215 | 2-methylene-furan-3-one reductase | −7.70 | Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor |
| KAF3627153.1 | putative caffeoyl-CoA O-methyltransferase 5-like | −7.63 | |
| GFY98677.1 | stress response NST1-like protein | −7.57 | |
| PSR89435.1 | Callose synthase | −7.34 | |
| LOC127790844 | dirigent protein 22-like | −7.34 | |
| LOC127797089 | nucleobase-ascorbate transporter 12 | −7.28 | Transmembrane transporter activity |
| LOC127813927 | pectin acetylesterase 8-like isoform X1 | −7.16 | Pectin acetylesterase activity |
| LOC127788263 | serine/threonine-protein kinase PBL16 isoform X1 | −6.94 | Protein kinase activity |
| LOC127786732 | monocopper oxidase-like protein SKU5 | −6.91 | Copper ion binding |
| LOC127803982 | allene oxide cyclase, chloroplastic-like | −6.76 | Allene-oxide cyclase activity |
| LOC127808680 | acetyl-CoA carboxylase 1-like | −6.72 | Acetyl-CoA carboxylase activity |
| LOC127799558 | peroxidase 12-like | −6.57 | Peroxidase activity |
| LOC127803039 | xyloglucan galactosyltransferase XLT2-like | −6.56 | Galactosyltransferase activity |
| LOC127789537 | polygalacturonase inhibitor-like | −6.35 | Protein binding |
| LOC127810198 | cell wall/vacuolar inhibitor of fructosidase 2 | −6.25 | Enzyme inhibitor activity |
| LOC127800689 | LIM domain-containing protein WLIM1 | −6.25 | Actin filament binding |
| LOC127799017 | protein NETWORKED 1A-like | −6.20 | Actin filament binding |
| LOC127805837 | cellulose synthase-like protein D3 | −6.11 | Cellulose synthase (UDP-forming) activity |
| LOC127801708 | formin-like protein 1 | −5.99 | Actin filament binding |
| LOC127789271 | cellulose synthase-like protein G3 | −5.88 | Cellulose synthase (UDP-forming) activity |
| PSS04488.1 | Trichohyalin like | −5.82 | |
| GFS34626.1 | thioredoxin M-type 4 | −5.82 | |
| LOC127809708 | lysine-rich arabinogalactan protein 18-like | −5.81 | |
| LOC127787238 | berberine bridge enzyme-like 6 | −5.80 | FAD binding |
| LOC127801919 | reticulon-like protein B2 | −5.79 | |
| LOC127806788 | leucine-rich repeat extensin-like protein 2 | −5.71 | Protein binding |
| PON73688.1 | Tol-Pal system beta propeller repeat-containing protein | −5.70 | |
| LOC127796357 | WAT1-related protein At3g28050-like isoform X1 | −5.62 | Transmembrane transporter activity |
| LOC127814265 | acyl-lipid (9-3)-desaturase-like | −5.61 | Oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water |
| LOC127813587 | xyloglucan galactosyltransferase GT19 | −5.59 | Glycosyltransferase activity |
| LOC127814002 | GDSL esterase/lipase At4g01130 | −5.57 | Hydrolase activity, acting on ester bonds |
| PSS26602.1 | Polyketide cyclase | −5.49 | |
| LOC127795202 | senescence/dehydration-associated protein At4g35985, chloroplastic-like isoform X1 | −5.41 | |
| LOC127786623 | scopoletin glucosyltransferase-like | −5.37 | UDP-glycosyltransferase activity |
| PSS07274.1 | Protein FAM133B like | −5.33 | |
| LOC127799436 | momilactone A synthase-like | −5.26 | |
| LOC127804145 | callose synthase 3-like | −5.24 | 1,3-beta-D-glucan synthase activity |
| LOC127800990 | germin-like protein 5-1 | −5.21 | Manganese ion binding |
| LOC127795475 | xyloglucan glycosyltransferase 5 | −5.18 | Glycosyltransferase activity |
| LOC127802055 | glycosyltransferase BC10 | −5.15 | Glycosyltransferase activity |
| LOC127799885 | protein trichome birefringence-like 6 | −5.13 | O-acetyltransferase activity |
| LOC127798217 | pectinesterase/pectinesterase inhibitor 34 | −5.12 | Pectinesterase inhibitor activity |
| PSS21721.1 | ATP phosphoribosyltransferase regulatory subunit like | −5.11 | |
| LOC127792829 | flavonol 3-O-glucosyltransferase UGT89B1-like | −5.11 | UDP-glycosyltransferase activity |
| LOC127795273 | trimethyltridecatetraene synthase-like | −5.03 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127792573 | aconitate hydratase, cytoplasmic isoform X2 | −5.02 | Aconitate hydratase activity |
| LOC127810619 | U-box domain-containing protein 6-like | −4.92 | Ubiquitin-protein transferase activity |
| LOC127788941 | RING-H2 finger protein ATL66-like | −4.92 | |
| KAG5222891.1 | myotubularin-related protein | −4.85 | |
| LOC127798906 | protein REDUCED WALL ACETYLATION 2 | −4.83 | Acetyltransferase activity |
| PSS21630.1 | Glucan endo-1,3-beta-glucosidase | −4.63 | |
| LOC114306531 | rootletin-like isoform X3 | −4.61 | |
| LOC127807864 | cellulose synthase A catalytic subunit 2 [UDP-forming]-like isoform X2 | −4.60 | Cellulose synthase (UDP-forming) activity |
| LOC127788688 | tetrapyrrole-binding protein, chloroplastic | −4.52 | Tetrapyrrole binding |
| LOC127810642 | ubiquinol oxidase, mitochondrial-like | −4.50 | Alternative oxidase activity |
| LOC127788265 | mitochondrial uncoupling protein 1 | −4.47 | Oxidative phosphorylation uncoupler activity |
| LOC127808097 | protein LIKE COV 1-like isoform X2 | −4.43 | |
| LOC127793879 | cellulose synthase A catalytic subunit 7 [UDP-forming] | −4.42 | Cellulose synthase (UDP-forming) activity |
| LOC127791234 | aldehyde oxidase GLOX | −4.41 | |
| LOC127814001 | rho GDP-dissociation inhibitor 1-like | −4.40 | Rho GDP-dissociation inhibitor activity |
| LOC127814067 | zerumbone synthase isoform X2 | −4.38 | Oxidoreductase activity |
| LOC127808689 | L-ascorbate oxidase homolog | −4.32 | Oxidoreductase activity |
| LOC127792648 | polyol transporter 4 | −4.26 | Carbohydrate transmembrane transporter activity |
| GFZ03086.1 | 1,3-beta-glucan synthase component | −4.24 | |
| LOC127807082 | putative HVA22-like protein g | −4.16 | |
| PSS30372.1 | Protein rolling stone like | −4.15 | |
| LOC127797069 | 14 kDa proline-rich protein DC2.15-like | −4.14 | |
| KAI8014341.1 | Armadillo repeat-containing protein 6 | −4.13 | |
| XP_023882653.1 | protein WVD2-like 7 isoform X1 | −4.02 | |
| LOC127795124 | cytochrome P450 736A117-like | −3.98 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127795780 | protein trichome birefringence-like 5 | −3.96 | O-acetyltransferase activity |
| LOC127796448 | microtubule-associated protein 70-2 | −3.92 | Microtubule binding |
| LOC127790491 | dirigent protein 22-like | −3.91 | |
| LOC127803088 | lysine-rich arabinogalactan protein 18 | −3.84 | |
| LOC127789071 | protein WVD2-like 7 isoform X2 | −3.81 | |
| LOC127800498 | linoleate 9S-lipoxygenase 5 isoform X2 | −3.81 | Metal ion binding |
| LOC127791405 | peroxidase 3-like | −3.79 | Peroxidase activity |
| LOC118056996 | cytochrome P450 71D10-like | −3.79 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| LOC127793976 | jasmonate ZIM domain-containing protein 1-like | −3.66 | |
| LOC127803184 | WAT1-related protein At4g15540-like | −3.65 | Transmembrane transporter activity |
| LOC127810929 | fasciclin-like arabinogalactan protein 7 | −3.60 | |
| GFS42279.1 | hydroxyproline-rich glycoprotein family protein | −3.58 | |
| LOC127787381 | glutathione synthetase, chloroplastic | −3.55 | Glutathione synthase activity |
| PSS33484.1 | hydroxyproline-rich glycoprotein family protein | −3.53 | |
| LOC127812477 | cinnamoyl-CoA reductase 1-like | −3.49 | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor |
| LOC127795300 | protein trichome birefringence-like 14 | −3.46 | Acetyltransferase activity |
| LOC127800790 | protein WVD2-like 3 | −3.45 | Microtubule binding |
| AOR05817.1 | pectinesterase 3 | −3.43 | |
| LOC127813752 | arabinosyltransferase XEG113 isoform X4 | −3.41 | Arabinosyltransferase activity |
| LOC127797224 | UDP-glucose 6-dehydrogenase 1-like | −3.32 | UDP-glucose 6-dehydrogenase activity |
| LOC127808089 | alcohol dehydrogenase-like 1 | −3.31 | S-(hydroxymethyl)glutathione dehydrogenase [NAD(P)+] activity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, M.W.; Chang, S.M.; Park, D.; Tilahun, S.; Jeong, C.S. Distinct Transcriptomic Profile Underlying High CO2 and Ethylene-Induced Deastringency in ‘Daebong’ Persimmon Fruit. Curr. Issues Mol. Biol. 2025, 47, 689. https://doi.org/10.3390/cimb47090689
Baek MW, Chang SM, Park D, Tilahun S, Jeong CS. Distinct Transcriptomic Profile Underlying High CO2 and Ethylene-Induced Deastringency in ‘Daebong’ Persimmon Fruit. Current Issues in Molecular Biology. 2025; 47(9):689. https://doi.org/10.3390/cimb47090689
Chicago/Turabian StyleBaek, Min Woo, Se Min Chang, DoSu Park, Shimeles Tilahun, and Cheon Soon Jeong. 2025. "Distinct Transcriptomic Profile Underlying High CO2 and Ethylene-Induced Deastringency in ‘Daebong’ Persimmon Fruit" Current Issues in Molecular Biology 47, no. 9: 689. https://doi.org/10.3390/cimb47090689
APA StyleBaek, M. W., Chang, S. M., Park, D., Tilahun, S., & Jeong, C. S. (2025). Distinct Transcriptomic Profile Underlying High CO2 and Ethylene-Induced Deastringency in ‘Daebong’ Persimmon Fruit. Current Issues in Molecular Biology, 47(9), 689. https://doi.org/10.3390/cimb47090689






