NAC Gene Family in Lagerstroemia indica: Genome-Wide Identification, Characterization, Expression Analysis, and Key Regulators Involved in Anthocyanin Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sequence Analysis of TFs in LiNAC
2.3. Sequence Analysis of L. indica NACs
2.4. Phylogenetic Analysis
2.5. Weighted Correlation Network Analysis (WGCNA) and Construction of Protein Interaction Networks
2.6. qRT-PCR Analysis of 7 LiNAC Genes
3. Result
3.1. Identification of NACs in L. indica
3.2. Analysis of Physical and Chemical Properties of L. indica NACs
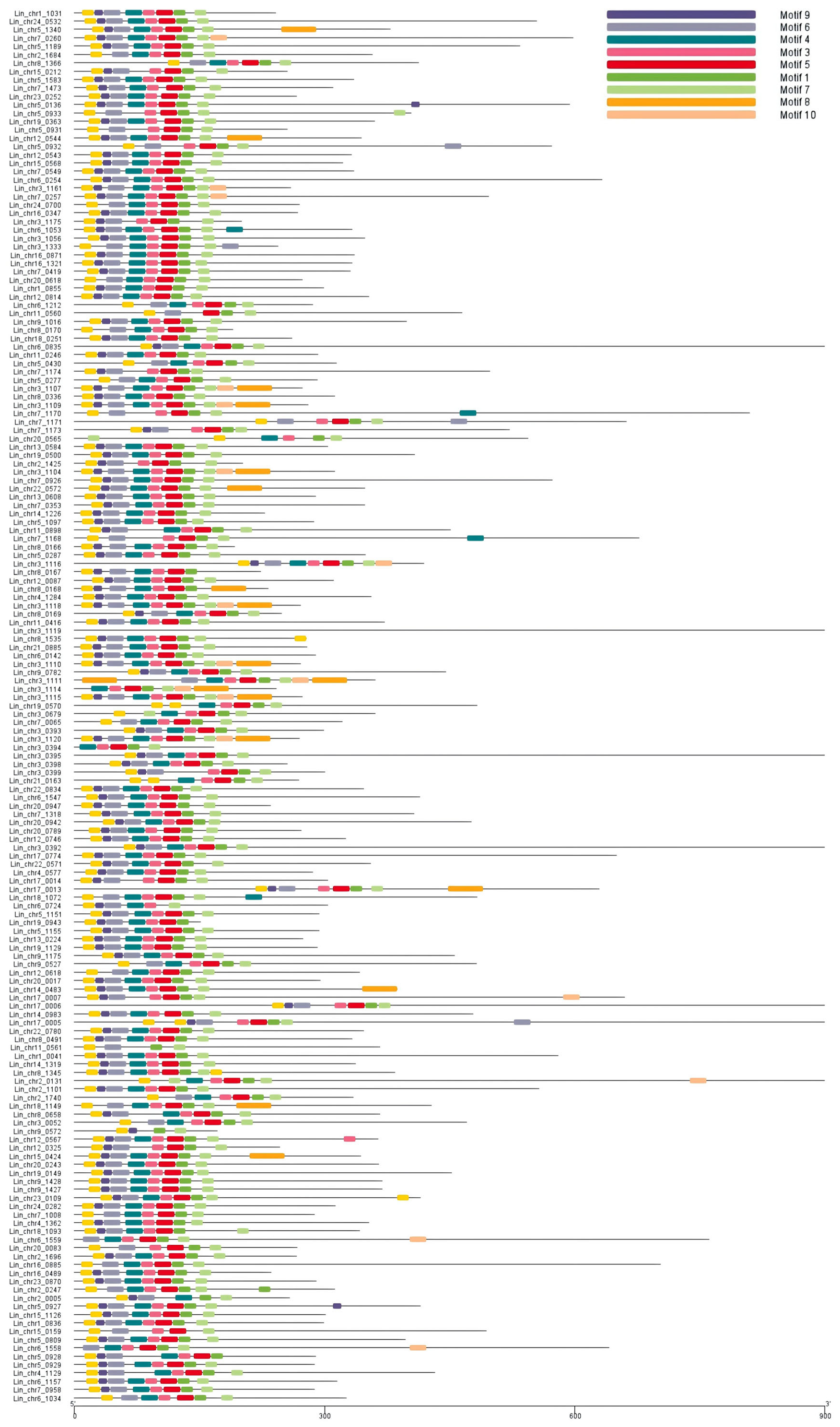
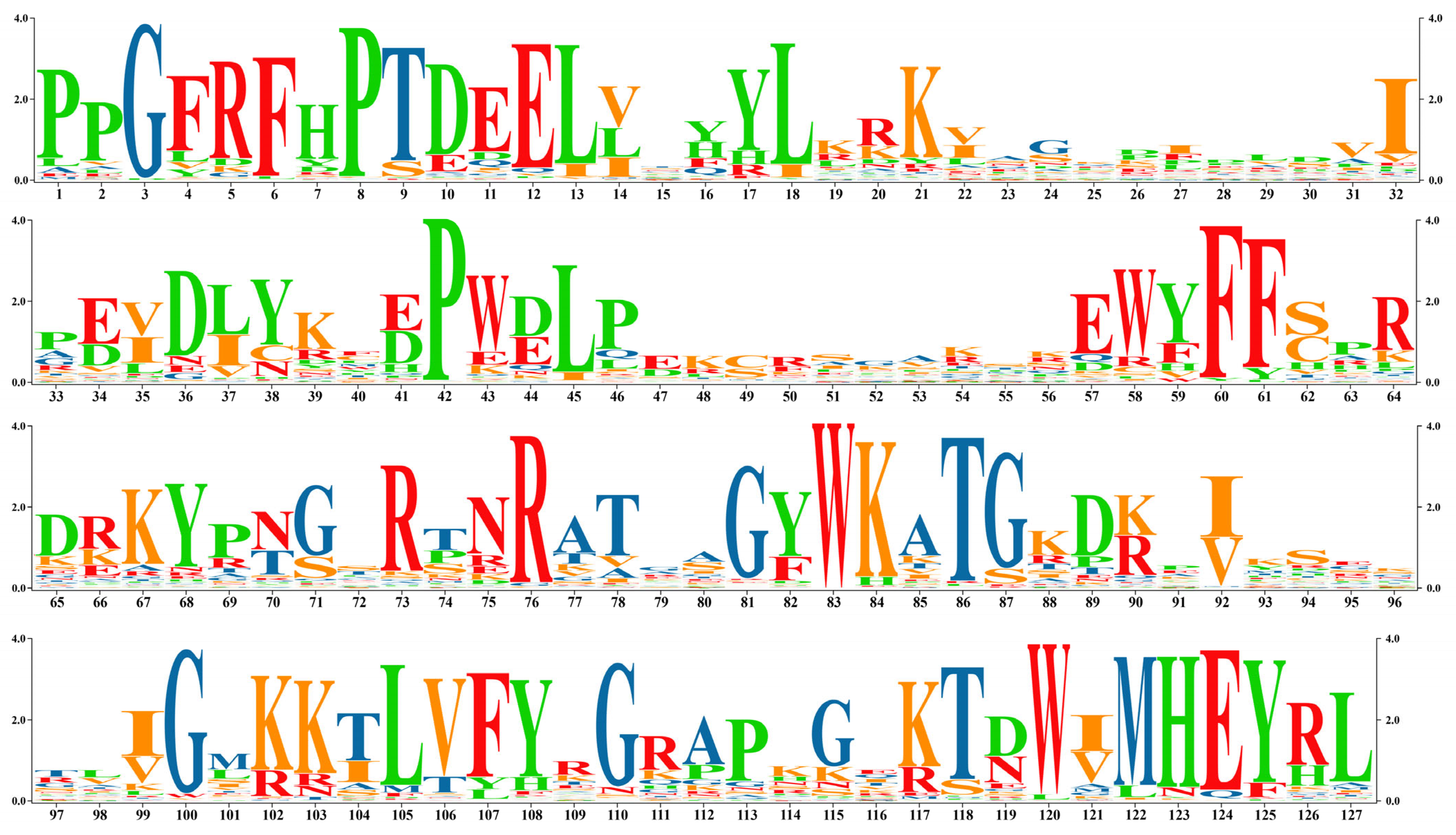
3.3. Conserved Domain Analysis of NAC in L. indica
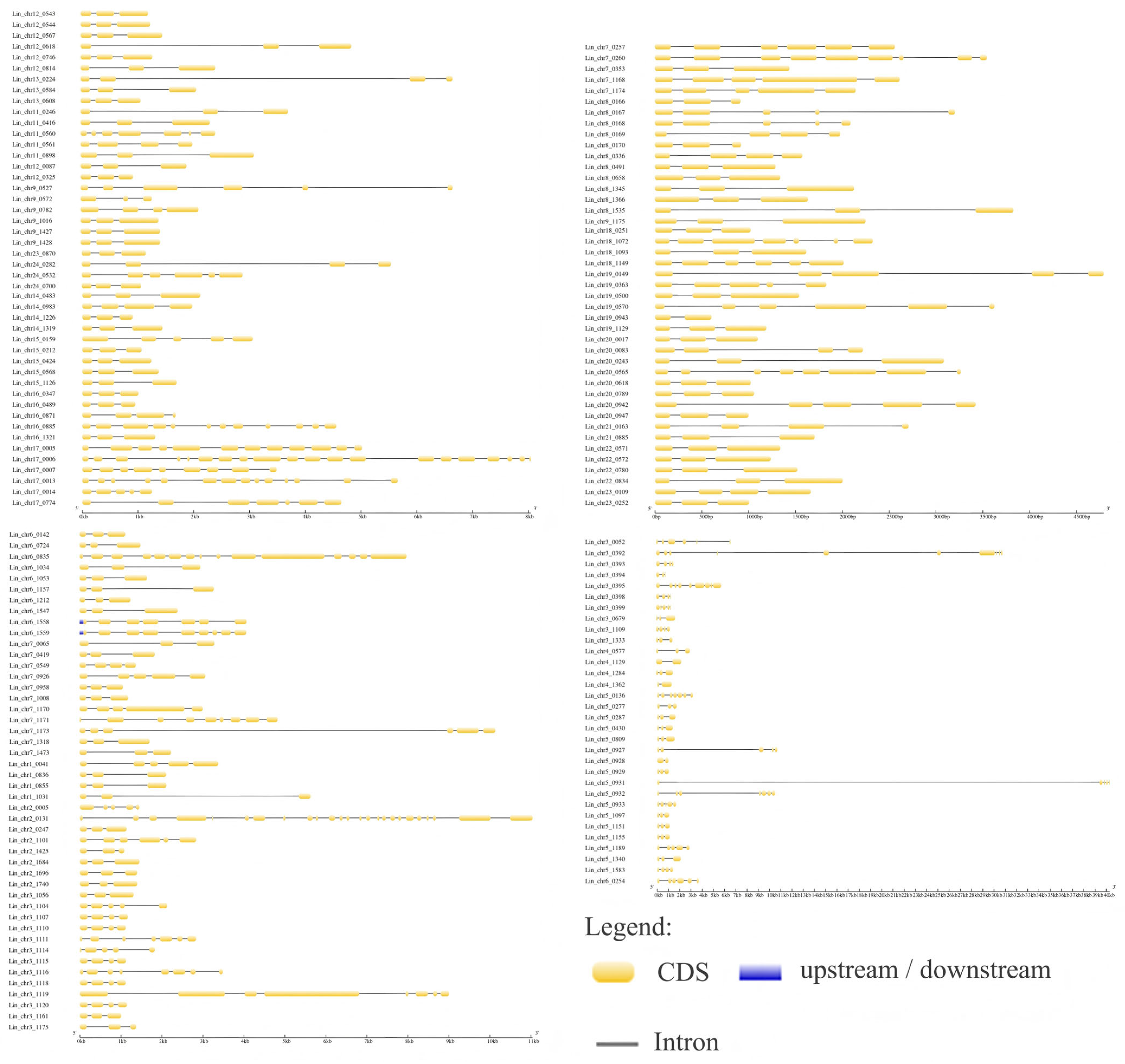
3.4. GSDS Analysis of L. indica NAC Structure
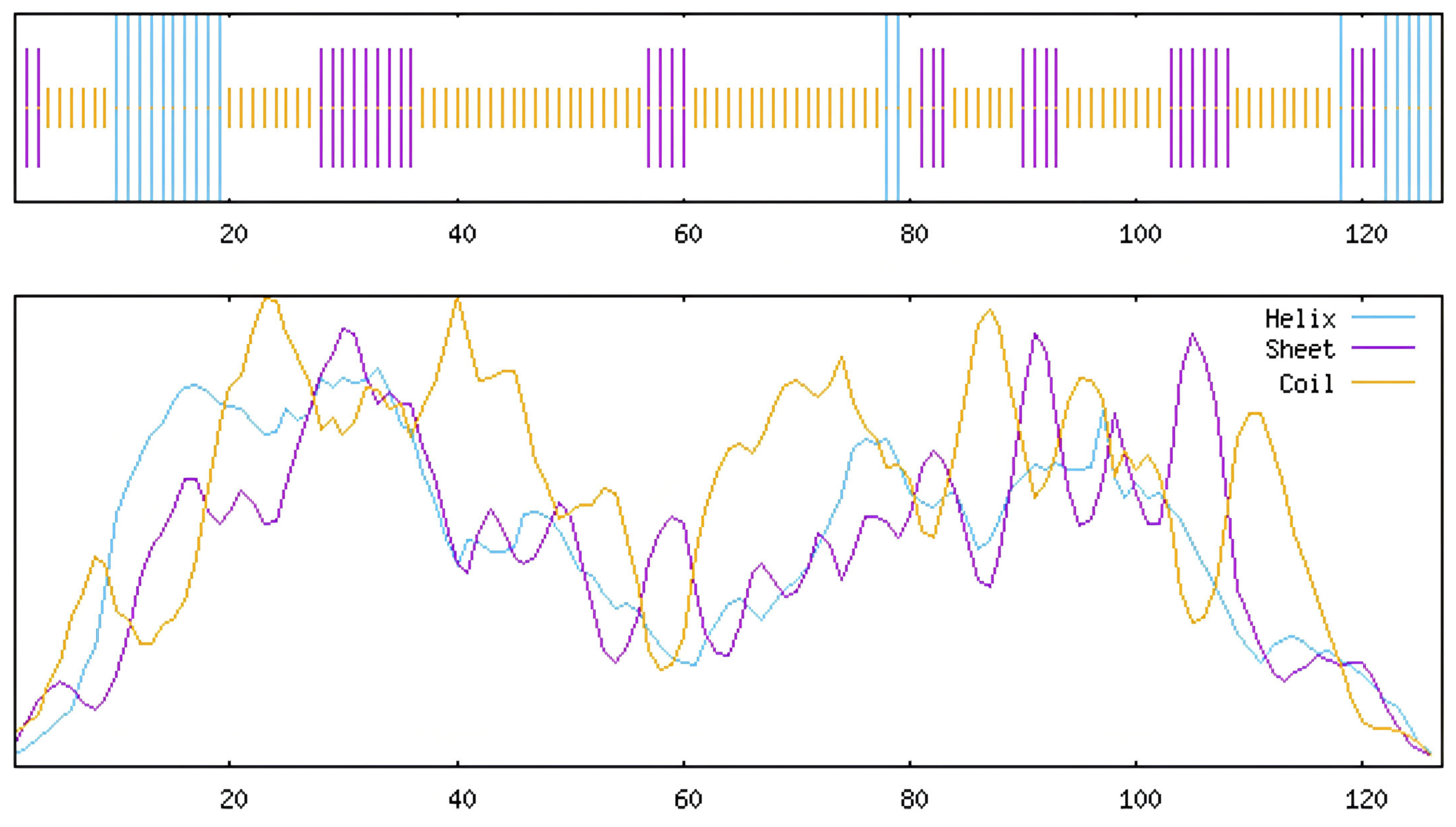
3.5. Prediction of Subcellular Localization of L. indica NACs
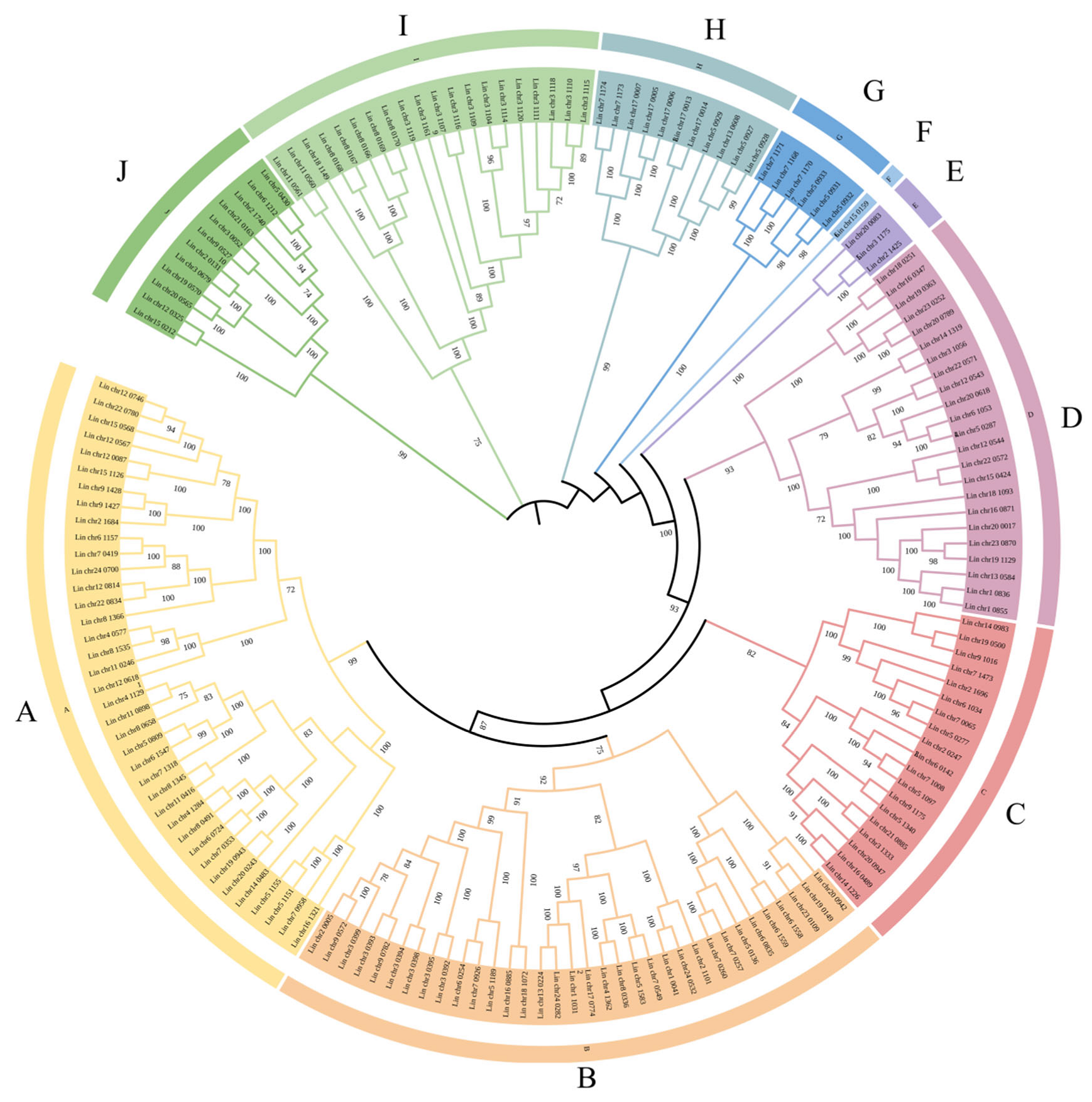
3.6. Constructing a Phylogenetic Tree of L. indica NACs
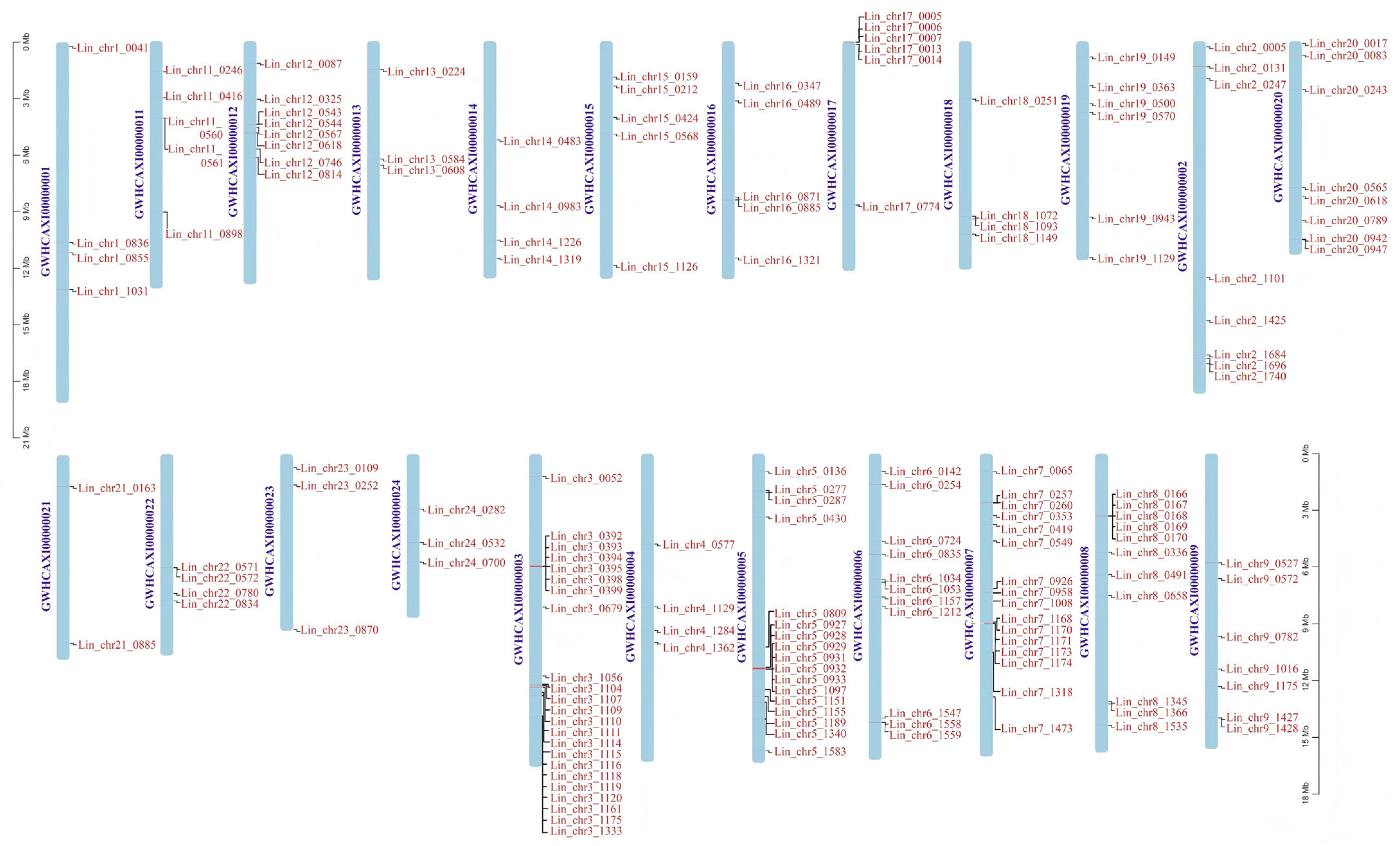
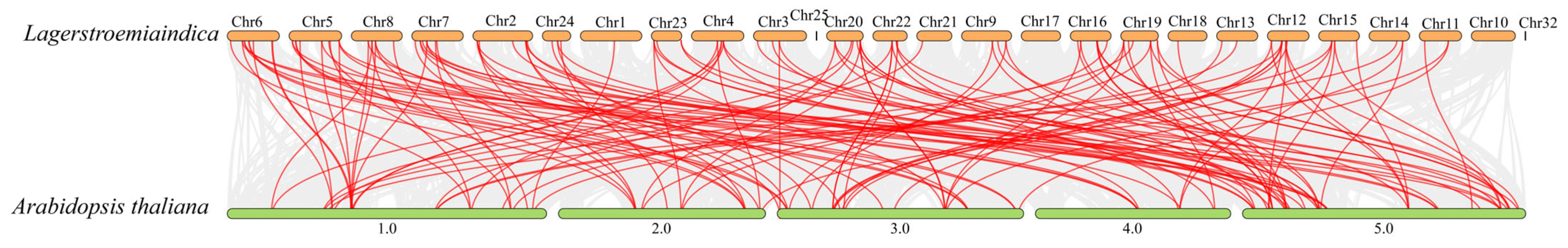
3.7. Chromosomal Localization and Collinearity Analysis of L. indica NACs
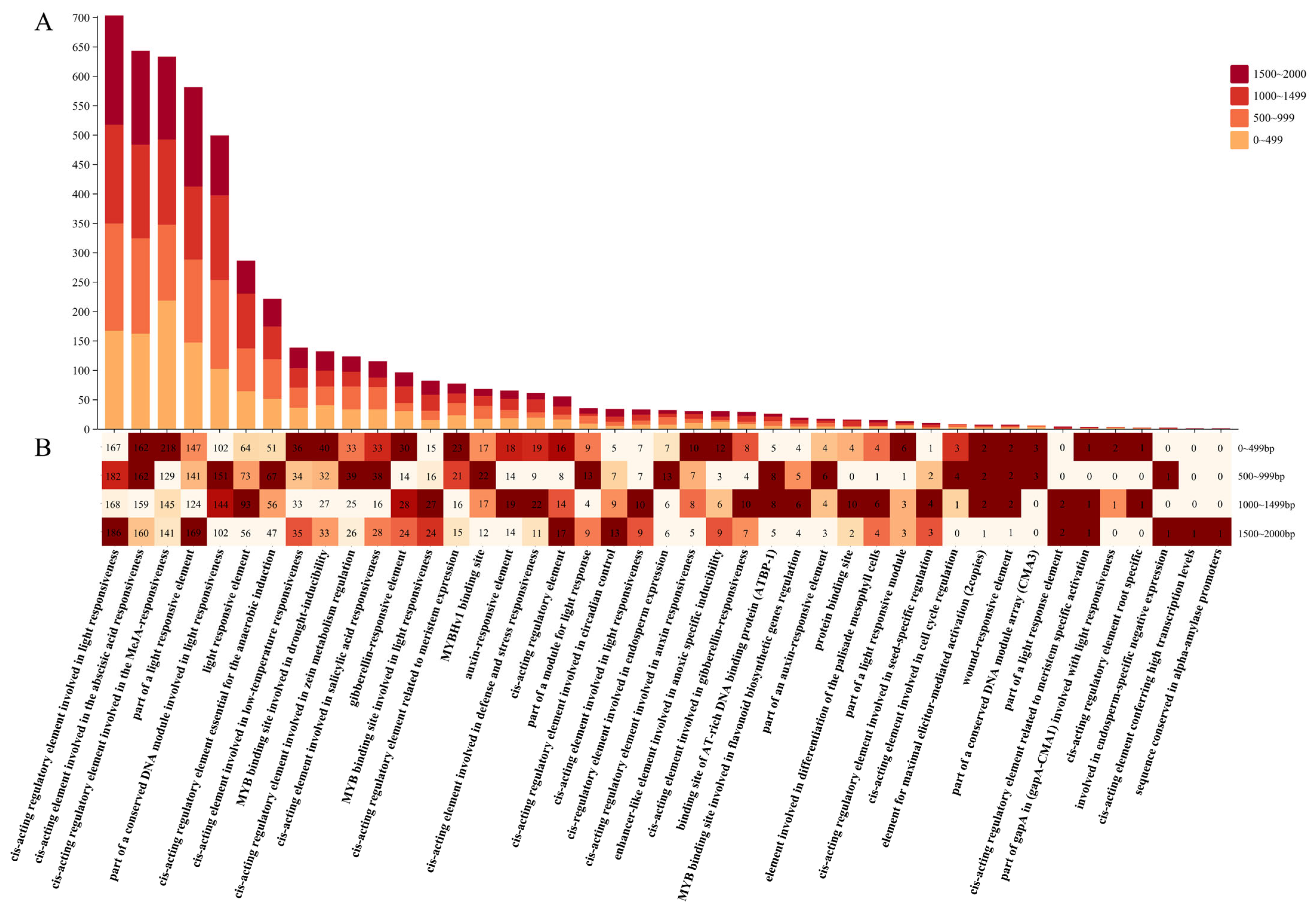
3.8. Analysis of NAC Promoter Components in L. indica
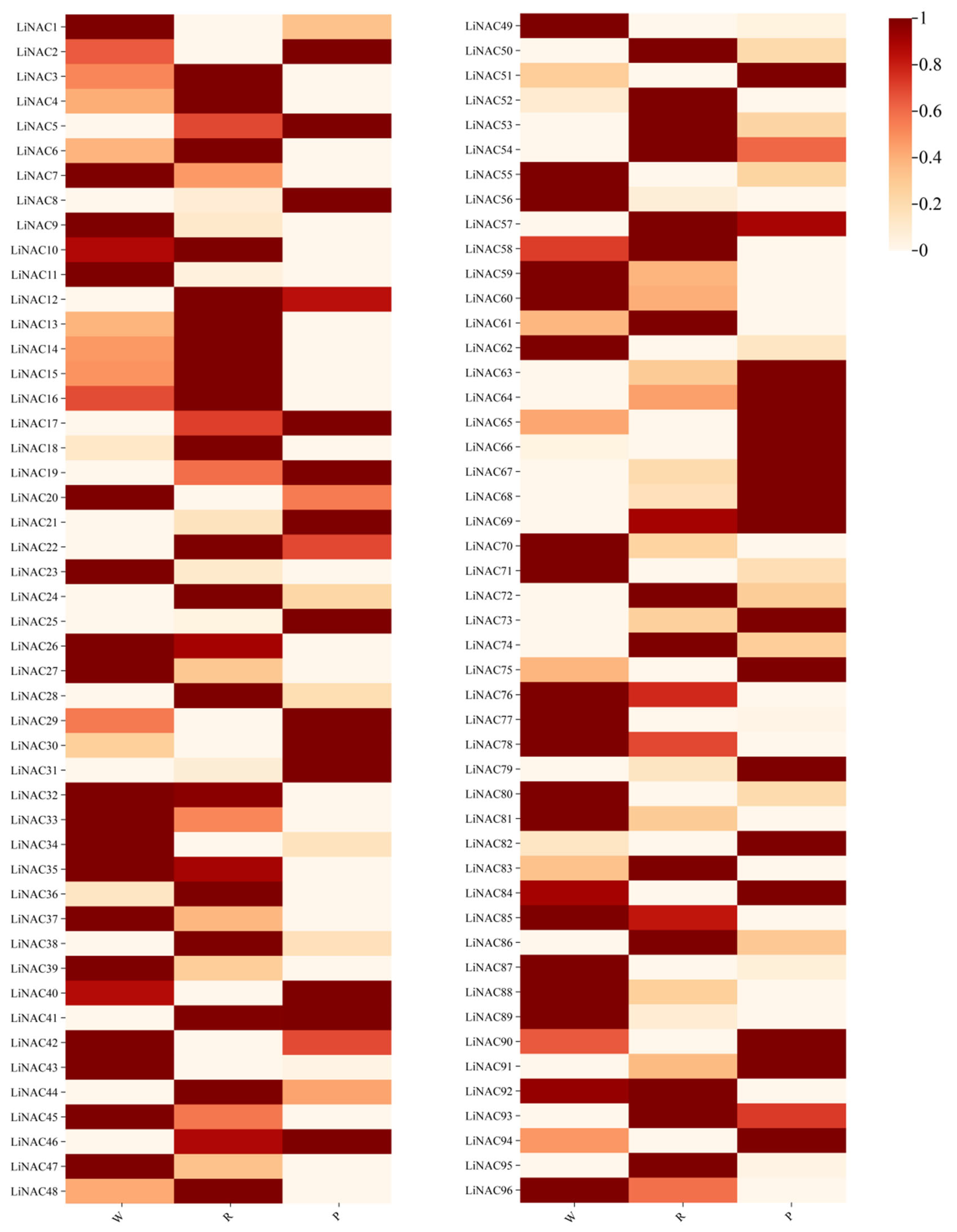
3.9. Expression Analysis of LiNACs
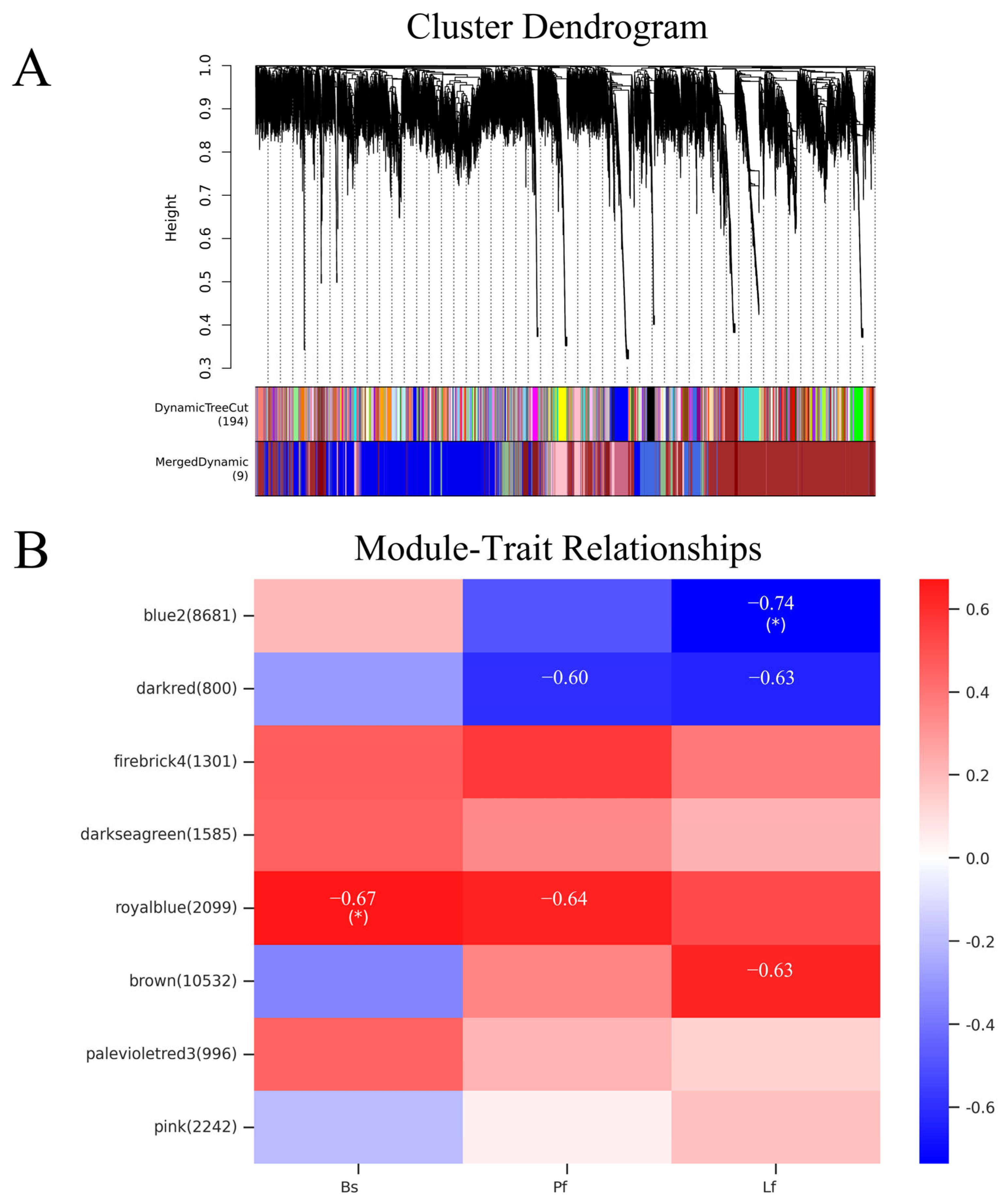
3.10. WGCNA of LiNACs
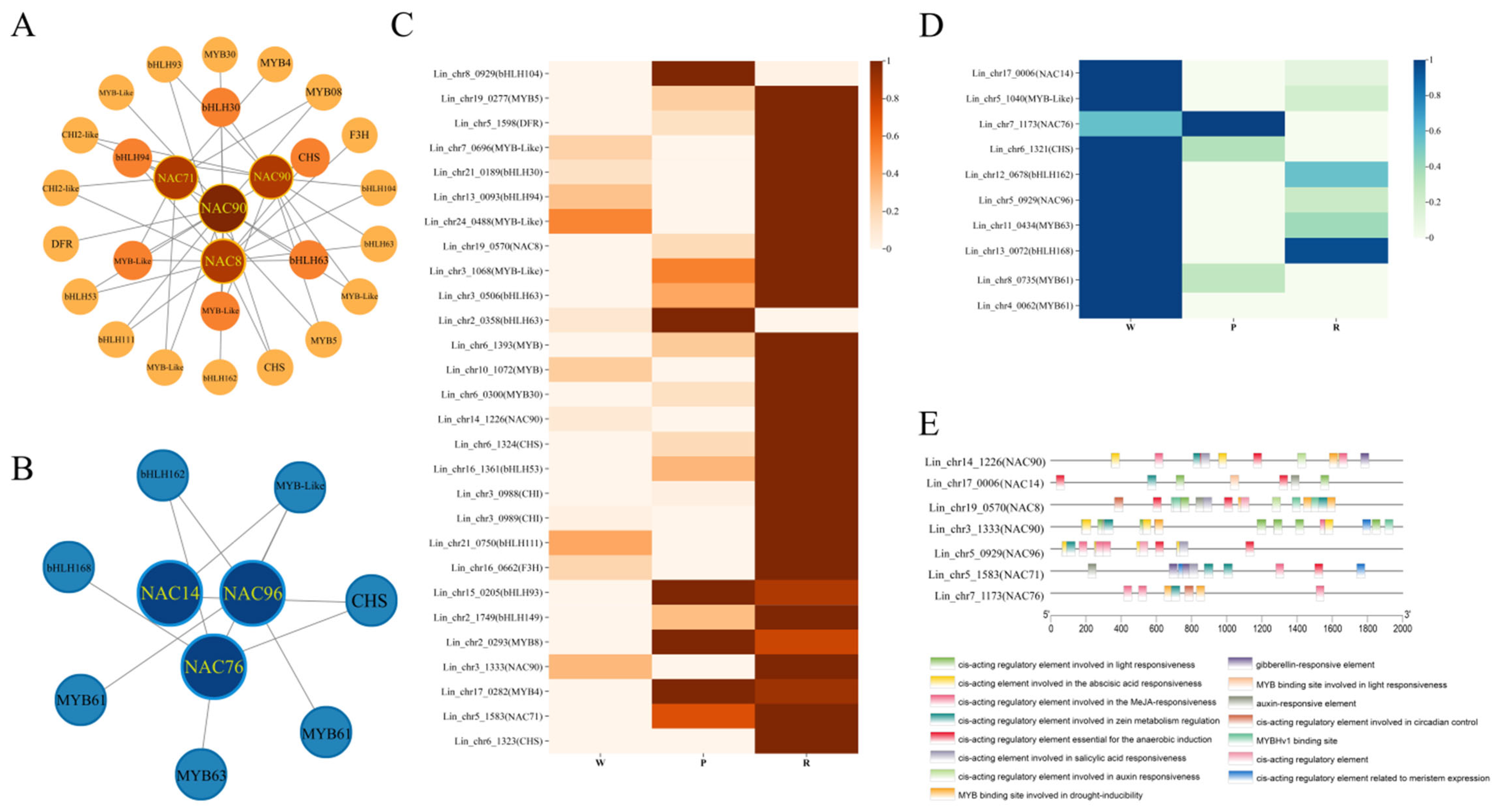
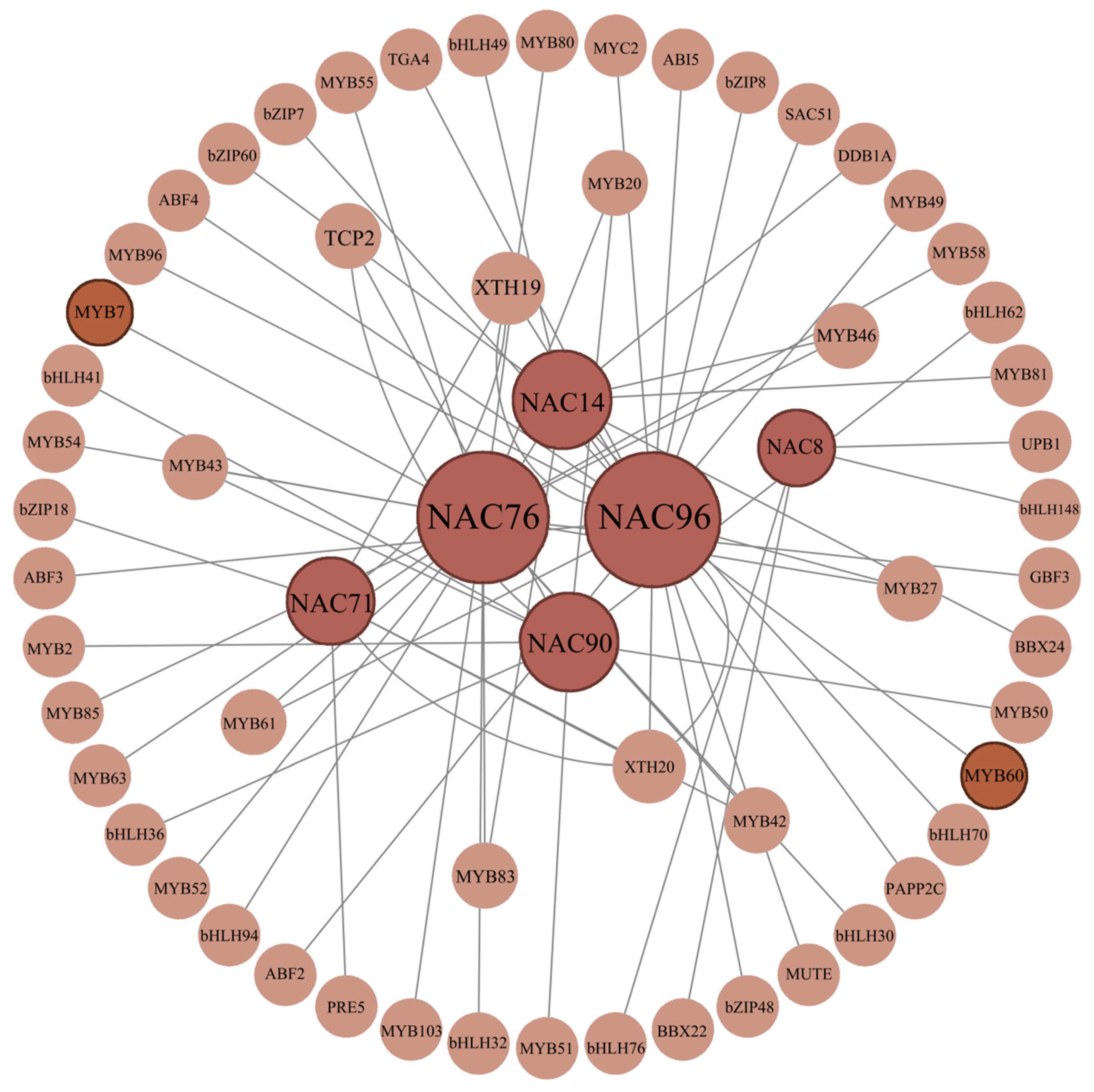
3.11. Gene Co-Expression Network Analysis and Protein Interaction Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hong, J.C. General Aspects of Plant Transcription Factor Families. In Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2016; pp. 35–56. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Keddie, J.; Adam, L.; Ratcliffe, J.; Samaha, R.R.; Creelman, R.; Pilgrim, M.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Riaño-Pachón, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An Integrative Plant Transcription Factor Database. BMC Bioinform. 2007, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, G.L.; Marques, C.S.; Costa, M.D.B.L.; Reis, P.A.B.; Alves, M.S.; Carvalho, C.M.; Fietto, L.G.; Fontes, E.P.B. Complete Inventory of Soybean NAC Transcription Factors: Sequence Conservation and Expression Analysis Uncover Their Distinct Roles in Stress Response. Gene 2009, 444, 10–23. [Google Scholar] [CrossRef]
- Wang, Y.-X.; Liu, Z.-W.; Wu, Z.-J.; Li, H.; Zhuang, J. Transcriptome-Wide Identification and Expression Analysis of the NAC Gene Family in Tea Plant [Camellia sinensis (L.) O. Kuntze]. PLoS ONE 2016, 11, e0166727. [Google Scholar] [CrossRef]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive Analysis of NAC Domain Transcription Factor Gene Family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef]
- Kikuchi, K.; Ueguchi-Tanaka, M.; Yoshida, K.T.; Nagato, Y.; Matsusoka, M.; Hirano, H.-Y. Molecular Analysis of the NAC Gene Family in Rice. Mol. Gen. Genet. 2000, 262, 1047–1051. [Google Scholar] [CrossRef]
- Duval, M.; Hsieh, T.-F.; Kim, S.Y.; Thomas, T.L. Molecular Characterization of AtNAM: A Member of the Arabidopsis NAC Domain Superfamily. Plant Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef]
- Souer, E.; Van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The No Apical Meristem Gene of Petunia Is Required for Pattern Formation in Embryos and Flowers and Is Expressed at Meristem and Primordia Boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Sablowski, R.W.M.; Meyerowitz, E.M. A Homolog of NO APICAL MERISTEM Is an Immediate Target of the Floral Homeotic Genes. Cell 1998, 92, 93–103. [Google Scholar] [CrossRef]
- He, X.; Mu, R.; Cao, W.; Zhang, Z.; Zhang, J.; Chen, S. AtNAC2, a Transcription Factor Downstream of Ethylene and Auxin Signaling Pathways, Is Involved in Salt Stress Response and Lateral Root Development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC Gene Regulating Senescence Improves Grain Protein, Zinc, and Iron Content in Wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-G.; Kim, S.-Y.; Park, C.-M. A Membrane-Associated NAC Transcription Factor Regulates Salt-Responsive Flowering via FLOWERING LOCUS T in Arabidopsis. Planta 2007, 226, 647–654. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Kim, S.-G.; Park, J.-E.; Park, H.-Y.; Lim, M.-H.; Chua, N.-H.; Park, C.-M. A Membrane-Bound NAC Transcription Factor Regulates Cell Division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef]
- Jensen, M.K.; Hagedorn, P.H.; De Torres-Zabala, M.; Grant, M.R.; Rung, J.H.; Collinge, D.B.; Lyngkjaer, M.F. Transcriptional Regulation by an NAC (NAM–ATAF1,2–CUC2) Transcription Factor Attenuates ABA Signalling for Efficient Basal Defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Zhou, L.-J.; Peng, J.; Chen, C.; Liu, S.; Song, A.; Jiang, J.; Chen, S.; Chen, F. CmNAC25 Targets CmMYB6 to Positively Regulate Anthocyanin Biosynthesis during the Post-Flowering Stage in Chrysanthemum. BMC Biol. 2023, 21, 211. [Google Scholar] [CrossRef]
- Zou, S.-C.; Zhuo, M.-G.; Abbas, F.; Hu, G.-B.; Wang, H.-C.; Huang, X.-M. Transcription Factor LcNAC002 Coregulates Chlorophyll Degradation and Anthocyanin Biosynthesis in Litchi. Plant Physiol. 2023, 192, 1913–1927. [Google Scholar] [CrossRef]
- Jiang, G.; Li, Z.; Song, Y.; Zhu, H.; Lin, S.; Huang, R.; Jiang, Y.; Duan, X. LcNAC13 Physically Interacts with LcR1MYB1 to Coregulate Anthocyanin Biosynthesis-Related Genes during Litchi Fruit Ripening. Biomolecules 2019, 9, 135. [Google Scholar] [CrossRef]
- Geng, M.; Shao, Y.; Zhang, M.; Zheng, X.; Tan, B.; Wang, W.; Zhang, L.; Ye, X.; Li, M.; Li, J.; et al. Overexpression of Peach NAC25 Promotes Anthocyanin Biosynthesis in Poplar Shoots. Fruit Res. 2022, 2, 21. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Wang, P.; Wang, S.; Ma, L.; Li, L.; Yang, R.; Ma, Y.; Wang, Q. Comprehensive Transcriptome Analysis Discovers Novel Candidate Genes Related to Leaf Color in a Lagerstroemia Indica Yellow Leaf Mutant. Genes Genom. 2015, 37, 851–863. [Google Scholar] [CrossRef]
- Al-Snafi, A.E. A Review on Lagerstroemia Indica: A Potential Medicinal Plant. IOSR J. Pharm. 2019, 9, 36–42. [Google Scholar]
- Hyeon, H.N.; Byung, K.C. Anti-Inflammatory Effects of Lagerstroemia Indica and Protection Effects on on Reflux-Esophagitis in Rats. Planta Medica Int. Open 2017, 4, Tu-PO-139. [Google Scholar] [CrossRef]
- Kong, X.-M.; Cui, X.-J.; Chang, M.-F.; Kang, W.-Y. Antioxidant Activity of Lagerstroemia Indica Flower. Nat. Prod. Res. Dev. 2015, 27, 264. [Google Scholar]
- Yue, Z.; Xu, Y.; Cai, M.; Fan, X.; Pan, H.; Zhang, D.; Zhang, Q. Floral Elegance Meets Medicinal Marvels: Traditional Uses, Phytochemistry, and Pharmacology of the Genus lagerstroemia L. Plants 2024, 13, 3016. [Google Scholar] [CrossRef] [PubMed]
- Zetter, R.; Ferguson, D.K. Lagerstroemia (Lythraceae) Pollen from the Miocene of Eastern China. Grana 2008, 47, 262–271. [Google Scholar]
- Wang, J.F.; Liu, X.H.; Chen, Z.M. Research Progress in Breeding of Lagerstroemia Plant. Acta Hortic. Sin. 2013, 40, 1795. [Google Scholar]
- Li, K.-B. ClustalW-MPI: ClustalW Analysis Using Distributed and Parallel Computing. Bioinformatics 2003, 19, 1585–1586. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Zhou, Y.; Zheng, T.; Cai, M.; Feng, L.; Chi, X.; Shen, P.; Wang, X.; Wan, Z.; Yuan, C.; Zhang, M.; et al. Genome Assembly and Resequencing Analyses Provide New Insights into the Evolution, Domestication and Ornamental Traits of Crape Myrtle. Hortic. Res. 2023, 10, uhad146. [Google Scholar] [CrossRef]
- AtMYB7, a New Player in the Regulation of UV-Sunscreens in Arabidopsis thaliana|Plant and Cell Physiology|Oxford Academic. Available online: https://academic.oup.com/pcp/article-abstract/55/3/507/1938070?login=false (accessed on 1 December 2024).
- Park, J.-S.; Kim, J.-B.; Cho, K.-J.; Cheon, C.-I.; Sung, M.-K.; Choung, M.-G.; Roh, K.-H. Arabidopsis R2R3-MYB Transcription Factor AtMYB60 Functions as a Transcriptional Repressor of Anthocyanin Biosynthesis in Lettuce (Lactuca sativa). Plant Cell Rep. 2008, 27, 985–994. [Google Scholar] [CrossRef]
- Al-Snafi, A.E. Medicinal value of lagerstroemia speciosa: An updated review. Int. J. Curr. Pharm. Res. 2019, 11, 18–26. [Google Scholar] [CrossRef]
- Hong, S.; Wang, J.; Wang, Q.; Zhang, G.; Zhao, Y.; Ma, Q.; Wu, Z.; Ma, J.; Gu, C. Decoding the Formation of Diverse Petal Colors of Lagerstroemia Indica by Integrating the Data from Transcriptome and Metabolome. Front. Plant Sci. 2022, 13, 970023. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Mori, M.; Kondo, T. Blue FLower Color Development by Anthocyanins: From Chemical Structure to Cell Physiology. Nat. Prod. Rep. 2009, 26, 884–915. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of Plant Pigments: Anthocyanins, Betalains and Carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Yoshida, K.; Oyama, K.; Kondo, T. Insight into Chemical Mechanisms of Sepal Color Development and Variation in Hydrangea. Proc. Jpn. Acad. Ser. B 2021, 97, 51–68. [Google Scholar] [CrossRef]
- Kong, J.-M.; Chia, L.-S.; Goh, N.-K.; Chia, T.-F.; Brouillard, R. Analysis and Biological Activities of Anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef]
- Mathew, I.E.; Agarwal, P. May the Fittest Protein Evolve: Favoring the Plant-Specific Origin and Expansion of NAC Transcription Factors. BioEssays 2018, 40, 1800018. [Google Scholar] [CrossRef]
- Gu, C.; Shang, L.; Zhang, G.; Wang, Q.; Ma, Q.; Hong, S.; Zhao, Y.; Yang, L. Identification and Expression Analysis of NAC Gene Family in Weeping Trait of Lagerstroemia Indica. Plants 2022, 11, 2168. [Google Scholar] [CrossRef]
- Rauf, M. ORE1 Balances Leaf Senescence against Maintenance by Antagonizing G2-like-Mediated Transcription. EMBO Rep. 2013, 14, 382–388. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic Acid Promotes Degreening via MYC 2/3/4- and ANAC 019/055/072-mediated Regulation of Major Chlorophyll Catabolic Genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Liu, G.; Li, X.; Jin, S.; Liu, X.; Zhu, L.; Nie, Y.; Zhang, X. Overexpression of Rice NAC Gene SNAC1 Improves Drought and Salt Tolerance by Enhancing Root Development and Reducing Transpiration Rate in Transgenic Cotton. PLoS ONE 2014, 9, e86895. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N.; et al. Apple NAC Transcription Factor MdNAC52 Regulates Biosynthesis of Anthocyanin and Proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef] [PubMed]
- Morishita, T.; Kojima, Y.; Maruta, T.; Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Arabidopsis NAC Transcription Factor, ANAC078, Regulates Flavonoid Biosynthesis under High-Light. Plant Cell Physiol. 2009, 50, 2210–2222. [Google Scholar] [CrossRef]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis Transcription Factor ANAC032 Represses Anthocyanin Biosynthesis in Response to High Sucrose and Oxidative and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Liu, W.; Mei, Z.; Yu, L.; Gu, T.; Li, Z.; Zou, Q.; Zhang, S.; Fang, H.; Wang, Y.; Zhang, Z.; et al. The ABA-Induced NAC Transcription Factor MdNAC1 Interacts with a bZIP-Type Transcription Factor to Promote Anthocyanin Synthesis in Red-Fleshed Apples. Hortic. Res. 2023, 10, uhad049. [Google Scholar] [CrossRef]
- Xu, T.; Yu, L.; Huang, N.; Liu, W.; Fang, Y.; Chen, C.; Jiang, L.; Wang, T.; Zhao, J.; Zhang, Z.; et al. The Regulatory Role of MdNAC14-Like in Anthocyanin Synthesis and Proanthocyanidin Accumulation in Red-Fleshed Apples. Plant Physiol. Biochem. 2023, 204, 108068. [Google Scholar] [CrossRef]
- Dalman, K.; Wind, J.J.; Nemesio-Gorriz, M.; Hammerbacher, A.; Lundén, K.; Ezcurra, I.; Elfstrand, M. Overexpression of PaNAC03, a Stress Induced NAC Gene Family Transcription Factor in Norway Spruce Leads to Reduced Flavonol Biosynthesis and Aberrant Embryo Development. BMC Plant Biol. 2017, 17, 6. [Google Scholar] [CrossRef]
- Wang, J.; Lian, W.; Cao, Y.; Wang, X.; Wang, G.; Qi, C.; Liu, L.; Qin, S.; Yuan, X.; Li, X.; et al. Overexpression of BoNAC019, a NAC Transcription Factor from Brassica oleracea, Negatively Regulates the Dehydration Response and Anthocyanin Biosynthesis in Arabidopsis. Sci. Rep. 2018, 8, 13349. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, H.; Jiang, X.; Wang, P.; Dai, X.; Chen, W.; Gao, L.; Xia, T. A WD40 Repeat Protein from Camellia Sinensis Regulates Anthocyanin and Proanthocyanidin Accumulation through the Formation of MYB–bHLH–WD40 Ternary Complexes. Int. J. Mol. Sci. 2018, 19, 1686. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3-MYB Transcription Factor MYB 6 Promotes Anthocyanin and Proanthocyanidin Biosynthesis but Inhibits Secondary Cell Wall Formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential Regulation of Closely Related R2R3-MYB Transcription Factors Controls Flavonol Accumulation in Different Parts of the Arabidopsis thaliana Seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Totsuka, A.; Okamoto, E.; Miyahara, T.; Kouno, T.; Cano, E.A.; Sasaki, N.; Watanabe, A.; Tasaki, K.; Nishihara, M.; Ozeki, Y. Repressed Expression of a Gene for a Basic Helix-Loop-Helix Protein Causes a White Flower Phenotype in Carnation. Breed. Sci. 2018, 68, 139–143. [Google Scholar] [CrossRef] [PubMed]
- López-Pérez, M.; Aguirre-Garrido, F.; Herrera-Zúñiga, L.; Fernández, F.J. Gene as a Dynamical Notion: An Extensive and Integrative Vision. Redefining the Gene Concept, from Traditional to Genic-Interaction, as a New Dynamical Version. Biosystems 2023, 234, 105060. [Google Scholar] [CrossRef] [PubMed]
- Pagano, A.; Gualtieri, C.; Mutti, G.; Raveane, A.; Sincinelli, F.; Semino, O.; Balestrazzi, A.; Macovei, A. Identification and Characterization of SOG1 (Suppressor of Gamma Response 1) Homologues in Plants Using Data Mining Resources and Gene Expression Profiling. Genes 2022, 13, 667. [Google Scholar] [CrossRef]
- Asahina, M.; Azuma, K.; Pitaksaringkarn, W.; Yamazaki, T.; Mitsuda, N.; Ohme-Takagi, M.; Yamaguchi, S.; Kamiya, Y.; Okada, K.; Nishimura, T.; et al. Spatially Selective Hormonal Control of RAP2.6L and ANAC071 Transcription Factors Involved in Tissue Reunion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 16128–16132. [Google Scholar] [CrossRef]
- Aharoni, A.; Cai, J.; Panda, S.; Kazachkova, Y.; Amzallag, E.; Meir, S.; Rogachev, I. A NAC Triad Mediates Plant Immunity by Negatively Regulating N-Hydroxy Pipecolic Acid Biosynthesis. Nat. Commun. 2024, 15, 7212. [Google Scholar]
- Moro, L. Postharvest Auxin and Methyl Jasmonate Effect on Anthocyanin Biosynthesis in Red Raspberry (Rubus idaeus L.). J. Plant Growth Regul. 2017, 36, 773–782. [Google Scholar] [CrossRef]
- Dong, N.; Lin, H. Contribution of Phenylpropanoid Metabolism to Plant Development and Plant–Environment Interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Clayton-Cuch, D.; Yu, L.; Shirley, N.; Bradley, D.; Bulone, V.; Böttcher, C. Auxin Treatment Enhances Anthocyanin Production in the Non-Climacteric Sweet Cherry (Prunus avium L.). Int. J. Mol. Sci. 2021, 22, 10760. [Google Scholar] [CrossRef]
- Rudell, D.R.; Mattheis, J.P.; Fan, X.; Fellman, J.K. Methyl Jasmonate Enhances Anthocyanin Accumulation and Modifies Production of Phenolics and Pigments in ‘Fuji’ Apples. J. Am. Soc. Hortic. Sci. 2002, 127, 435–441. [Google Scholar] [CrossRef]
- Zhang, D.; Sun, L.; Xi, D.; Li, X.; Gao, L.; Miao, L.; Luo, Y.; Tian, M.; Zhu, H. Methyl Jasmonate-induced bHLH42 Mediates Tissue-specific Accumulation of Anthocyanins via Regulating Flavonoid Metabolism-related Pathways in Caitai. Physiol. Plant. 2024, 176, e14434. [Google Scholar] [CrossRef] [PubMed]
- Fernandes De Oliveira, A.; Mercenaro, L.; Del Caro, A.; Pretti, L.; Nieddu, G. Distinctive Anthocyanin Accumulation Responses to Temperature and Natural UV Radiation of Two Field-Grown Vitis vinifera L. Cultivars. Molecules 2015, 20, 2061–2080. [Google Scholar] [CrossRef] [PubMed]
- Blancquaert, E.H.; Oberholster, A.; Ricardo-da-Silva, J.M.; Deloire, A.J. Grape Flavonoid Evolution and Composition Under Altered Light and Temperature Conditions in Cabernet Sauvignon (Vitis vinifera L.). Front. Plant Sci. 2019, 10, 1062. [Google Scholar] [CrossRef] [PubMed]
- Alabd, A.; Ahmad, M.; Zhang, X.; Gao, Y.; Peng, L.; Zhang, L.; Ni, J.; Bai, S.; Teng, Y. Light-Responsive Transcription Factor PpWRKY44 Induces Anthocyanin Accumulation by Regulating PpMYB10 Expression in Pear. Hortic. Res. 2022, 9, uhac199. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Z.; Chen, Z.; Wang, J.; Liu, W. NAC Gene Family in Lagerstroemia indica: Genome-Wide Identification, Characterization, Expression Analysis, and Key Regulators Involved in Anthocyanin Biosynthesis. Curr. Issues Mol. Biol. 2025, 47, 542. https://doi.org/10.3390/cimb47070542
Gao Z, Chen Z, Wang J, Liu W. NAC Gene Family in Lagerstroemia indica: Genome-Wide Identification, Characterization, Expression Analysis, and Key Regulators Involved in Anthocyanin Biosynthesis. Current Issues in Molecular Biology. 2025; 47(7):542. https://doi.org/10.3390/cimb47070542
Chicago/Turabian StyleGao, Zilong, Zhuomei Chen, Jinfeng Wang, and Weixin Liu. 2025. "NAC Gene Family in Lagerstroemia indica: Genome-Wide Identification, Characterization, Expression Analysis, and Key Regulators Involved in Anthocyanin Biosynthesis" Current Issues in Molecular Biology 47, no. 7: 542. https://doi.org/10.3390/cimb47070542
APA StyleGao, Z., Chen, Z., Wang, J., & Liu, W. (2025). NAC Gene Family in Lagerstroemia indica: Genome-Wide Identification, Characterization, Expression Analysis, and Key Regulators Involved in Anthocyanin Biosynthesis. Current Issues in Molecular Biology, 47(7), 542. https://doi.org/10.3390/cimb47070542





