Cross-Activity Analysis of CRISPR/Cas9 Editing in Gene Families of Solanum lycopersicum Detected by Long-Read Sequencing
Abstract
1. Introduction
2. Materials and Methods
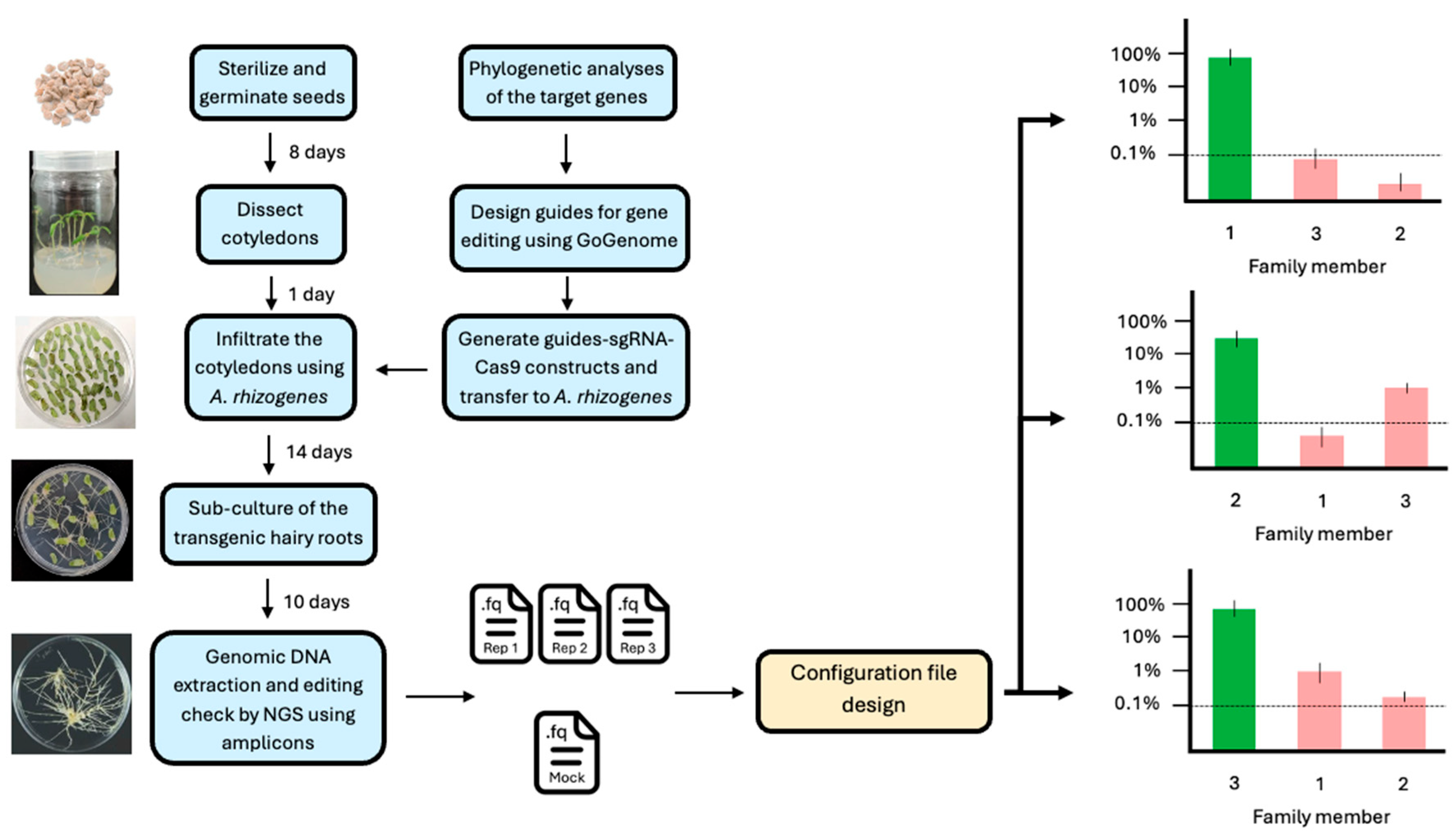
2.1. Plant Material and Generation of Genome-Edited Hairy Roots
2.2. Generation of the Constructs for Genome Editing
2.3. Multiplex PCR NGS Assay
2.4. Setting up CRISPECTOR for Gene Family Analysis by Preparing Configuration Files
- Collect all 20 base pairs’ long subsequences adjacent to any PAM sequence, in both forward and reverse sequences.
- Select the subsequences with alignment score over some user-defined threshold with respect to . These are determined by the Bio.pairwise2 module (Biopython v1.85).
- Further refine the set of subsequences and keep those with an edit distance under some user-defined threshold with respect to .
Example
3. Results
3.1. On-Target Activity
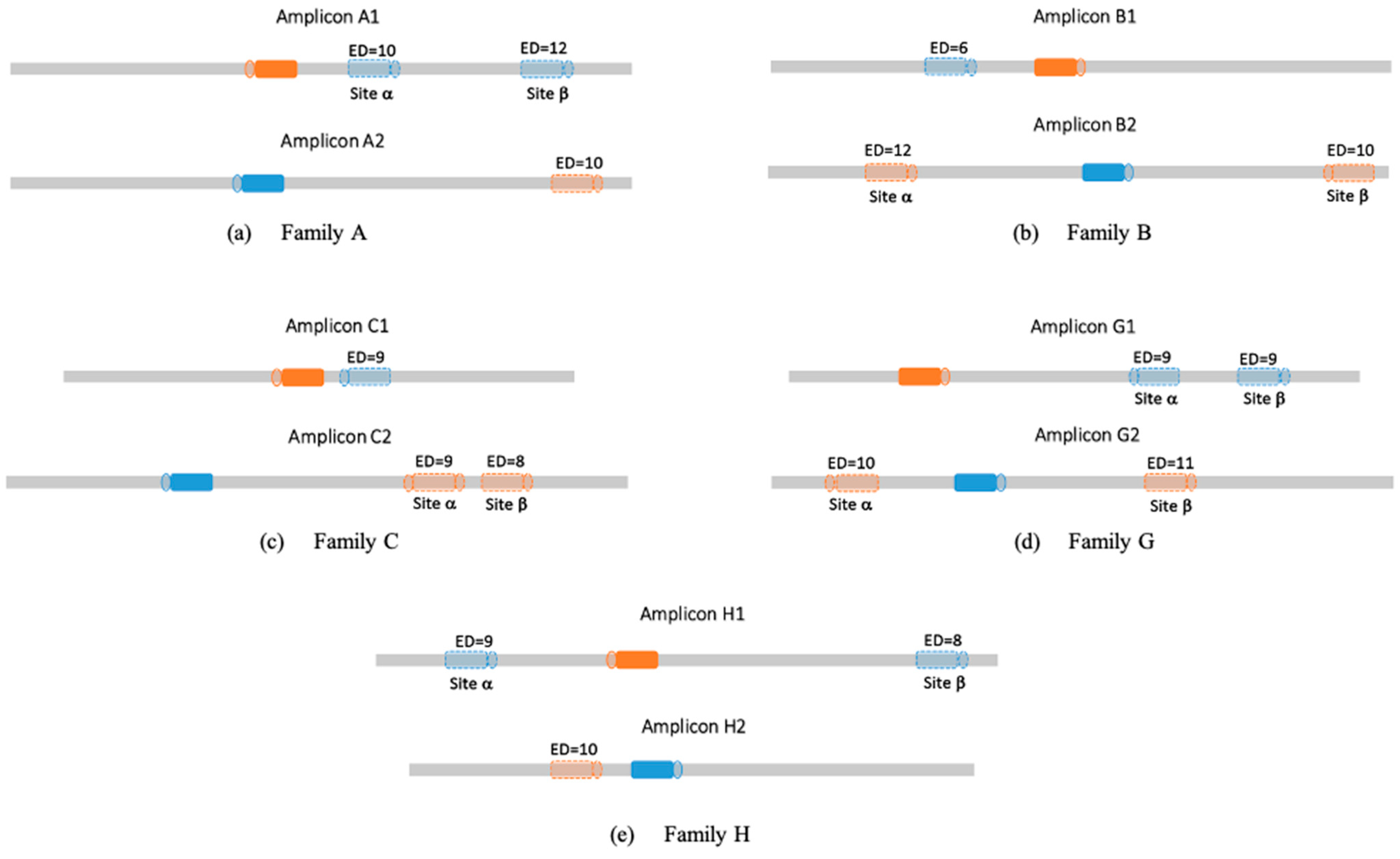
3.2. Off-Target Activity in Families
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Family | Members’ Guide Sequence | Amplicon Length bp | Targeted Gene | Gene ID |
|---|---|---|---|---|
| A | A1: TCTTCATCTCCAGTAAGCCT | 1012 | Ethylene Response Factor D.3 | Solyc01g108240 |
| A2: TCCGGTACGCGAAACAAGGG | 1009 | Ethylene Response Factor D.4 | Solyc10g050970 | |
| B | B1: GCGGCTTCCACGGCACCCAA | 956 | UDP-glycosyltransferase 75C1 | Solyc09g092500 |
| B2: GTAACAACATTTCCAGCACA | 975 | UDP-glycosyltransferase | Solyc12g098590 | |
| C | C1: TGCTAGGAAGACAGTAGCGT | 907 | LOB domain-containing protein 40 | Solyc05g009320 |
| C2: TGAGATTGAGAAGCCCCGCA | 1100 | LOB domain-containing protein 41 | Solyc03g119530 | |
| D | D1: TTCACTGTGCTGCAGCTGGT | 982 | Zinc finger transcription factor 39 | Solyc05g052570 |
| E | E1: GTCATCGTCGTTTTCTGAAG | 972 | C-repeat binding factor 1 | Solyc03g026280 |
| E2: CAATCACTACTCCCCTAATG | 1116 | Dehydration-responsive element-binding transcription factor | Solyc03g124110 | |
| F | F1: GGATGGAATCGAAATCCAGC | 987 | NAC domain-containing protein | Solyc11g017470 |
| F2: ACTGGTGCGGATAAGCCGAT | 1207 | NAC domain-containing protein | Solyc06g060230 | |
| F3: TATCTCTGCAGGAAATGCGC | 1101 | NAC domain-containing protein | Solyc04g009440 | |
| G | G1: GCAAAGGCCAGCAGCAGCTT | 1021 | C2H2-type zinc finger protein | Solyc04g077980 |
| G2: ATCGCGTAGCATCGTCATGG | 1112 | C2H2-type zinc finger protein | Solyc12g088390 | |
| H | H1: TTTGGTTGCAGATTGAACAA | 1061 | Auxin Response Factor 9B | Solyc08g008380 |
| H2: CGAGTTTACTACTTTCCACA | 964 | Auxin Response Factor 18 | Solyc01g096070 | |
| I | I1: GAAGCTGTCACCCACGGTGG | 1059 | Zinc finger protein | Solyc01g107170 |
| J | J1: GCTGGGTACTTTTGATACGG | 1126 | Ethylene Response Factor F.4 | Solyc07g053740 |
| J2: TACTTTCGATACTGCGGAGG | 996 | Ethylene Response Factor F.5 | Solyc10g009110 | |
| K | K1: TCCCCAAATCAAGAATCTGC | 1002 | syntaxin-121-like | Solyc10g081850 |
| L | L1: GTTTCCGAATCTCAACGACC | 924 | LOB domain-containing protein 38 | Solyc01g107190 |
| M | M1: GGAACATCCCCAGCAAATGG | 948 | Multidrug resistance protein ABC transporter family protein | Solyc01g079550 |
References
- Brownie, J.; Shawcross, S.; Theaker, J.; Whitcombe, D.; Ferrie, R.; Newton, C.; Little, S. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997, 25, 3235–3241. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Edgar, R. NCBI GEO: Mining tens of millions of expression profiles—Database and tools update. Nucleic Acids Res. 2007, 35, D760–D765. [Google Scholar] [CrossRef]
- Ben-Dor, A.; Chor, B.; Karp, R.; Yakhini, Z. Discovering local structure in gene expression data: The order-preserving submatrix problem. J. Comput. Biol. 2003, 10, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2014, 33, 187–197. [Google Scholar] [CrossRef]
- Purcell, O.; Peccoud, J.; Lu, T.K. Rule-based design of synthetic transcription factors in eukaryotes. ACS Synth. Biol. 2014, 3, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, S.; Yang, X.; Wu, H.; Tang, H.; Yang, L. PlantGF: An analysis and annotation platform for plant gene families. Database 2022, 2022, baab088. [Google Scholar] [CrossRef]
- Sterck, L.; Rombauts, S.; Vandepoele, K.; Rouzé, P.; Van de Peer, Y. How many genes are there in plants (... and why are they there)? Curr. Opin. Plant Biol. 2007, 10, 199–203. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Michael, T.P.; Jupe, F.; Bemm, F.; Motley, S.T.; Sandoval, J.P.; Lanz, C.; Loudet, O.; Weigel, D.; Ecker, J.R. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat. Commun. 2018, 9, 541. [Google Scholar] [CrossRef]
- Clement, K.; Rees, H.; Canver, M.C.; Gehrke, J.M.; Farouni, R.; Hsu, J.Y.; Cole, M.A.; Liu, D.R.; Joung, J.K.; Bauer, D.E.; et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019, 37, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Conant, D.; Hsiau, T.; Rossi, N.; Oki, J.; Maures, T.; Waite, K.; Yang, J.; Joshi, S.; Kelso, R.; Holden, K.; et al. Inference of CRISPR edits from Sanger trace data. CRISPR J. 2022, 5, 123–130. [Google Scholar] [CrossRef]
- Chuai, G.; Ma, H.; Yan, J.; Chen, M.; Hong, N.; Xue, D.; Zhou, C.; Zhu, C.; Chen, K.; Duan, B.; et al. DeepCRISPR: Optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018, 19, 80. [Google Scholar] [CrossRef]
- Amit, I.; Iancu, O.; Levy-Jurgenson, A.; Kurgan, G.; McNeill, M.S.; Rettig, G.R.; Allen, D.; Breier, D.; Ben Haim, N.; Wang, Y.; et al. CRISPECTOR provides accurate estimation of genome editing translocation and off-target activity from comparative NGS data. Nat. Commun. 2021, 12, 3042. [Google Scholar] [CrossRef] [PubMed]
- Assa, G.; Kalter, N.; Rosenberg, M.; Beck, A.; Markovich, O.; Gontmakher, T.; Hendel, A.; Yakhini, Z. Quantifying allele-specific CRISPR editing activity with CRISPECTOR2.0. Nucleic Acids Res. 2024, 52, e78. [Google Scholar] [CrossRef]
- Ron, M.; Kajala, K.; Pauluzzi, G.; Wang, D.; Reynoso, M.A.; Zumstein, K.; Garcha, J.; Winte, S.; Masson, H.; Inagaki, S.; et al. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 2014, 166, 455–469. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Kandzia, R.; Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef]
- Murray, M.; Thompson, W. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Guo, C.; Ma, X.; Gao, F.; Guo, Y. Off-target effects in CRISPR/Cas9 gene editing. Front. Bioeng. Biotechnol. 2023, 11, 1143157. [Google Scholar] [CrossRef]
- Jacobs, T.B.; Zhang, N.; Patel, D.; Martin, G.B. Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol. 2017, 174, 2023–2037. [Google Scholar] [CrossRef] [PubMed]

 ) indicates values lower than 0.3%. Note variability of activity levels.
) indicates values lower than 0.3%. Note variability of activity levels.
 ) indicates values lower than 0.3%. Note variability of activity levels.
) indicates values lower than 0.3%. Note variability of activity levels.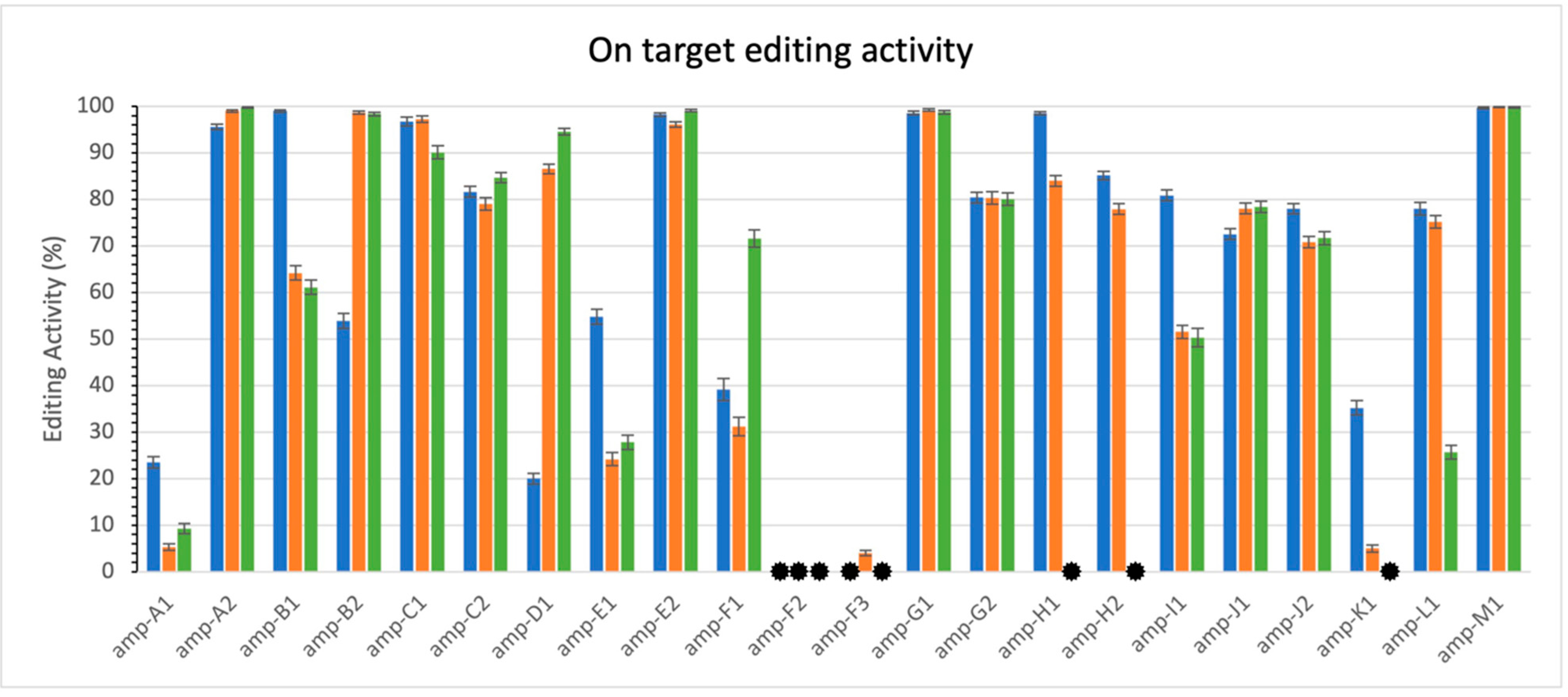
 ) indicates values lower than 0.01%. This figure represents the results from all three replicates. The dashed line marks the 0.1% threshold, a standard benchmark in CRISPR off-target analysis.
) indicates values lower than 0.01%. This figure represents the results from all three replicates. The dashed line marks the 0.1% threshold, a standard benchmark in CRISPR off-target analysis.
 ) indicates values lower than 0.01%. This figure represents the results from all three replicates. The dashed line marks the 0.1% threshold, a standard benchmark in CRISPR off-target analysis.
) indicates values lower than 0.01%. This figure represents the results from all three replicates. The dashed line marks the 0.1% threshold, a standard benchmark in CRISPR off-target analysis.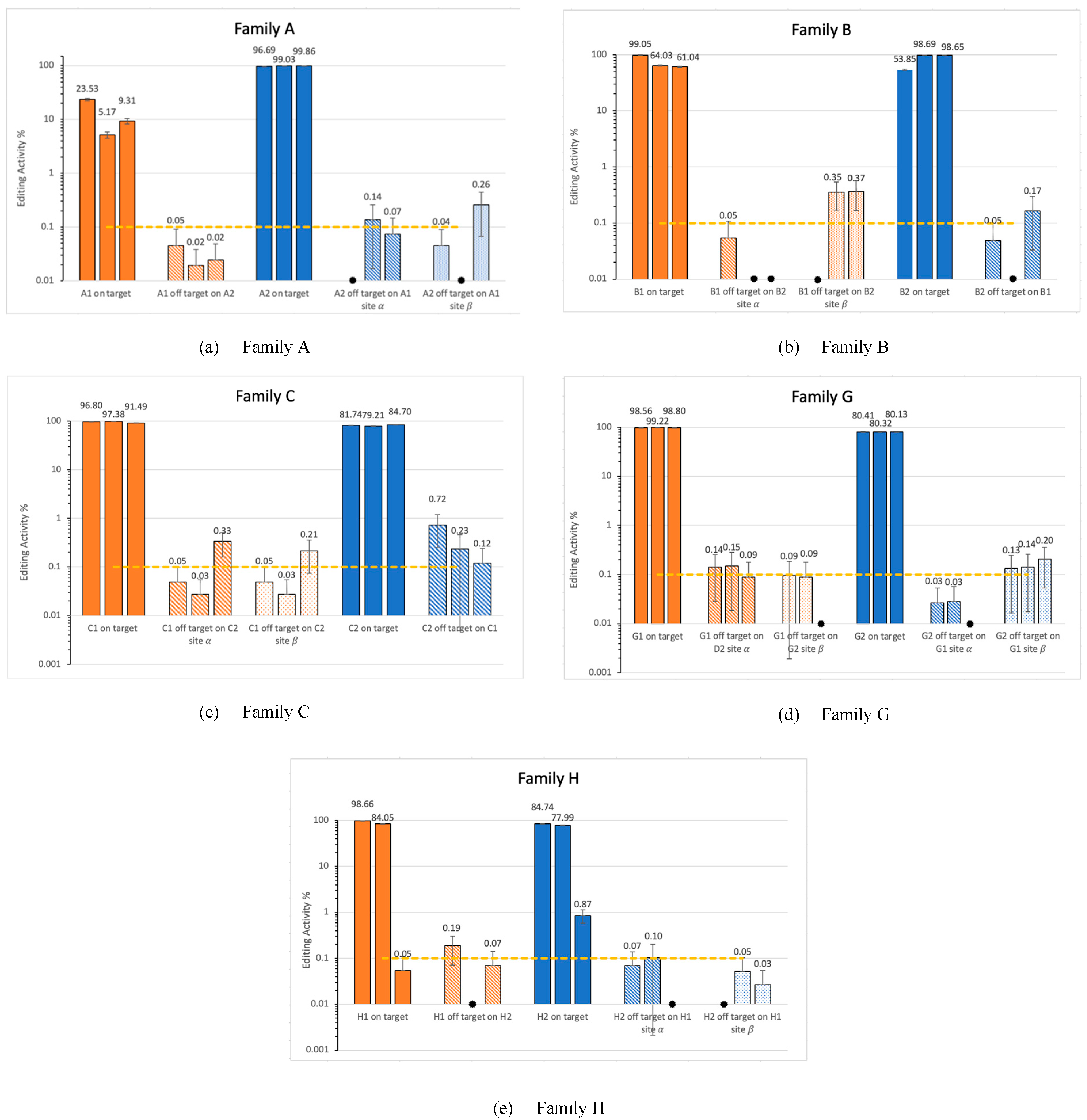
| Family | Members’ Guide Sequence | Amplicon Length bp |
|---|---|---|
| A | A1: TCTTCATCTCCAGTAAGCCT | 1012 |
| A2: TCCGGTACGCGAAACAAGGG | 1009 | |
| B | B1: GCGGCTTCCACGGCACCCAA | 956 |
| B2: GTAACAACATTTCCAGCACA | 975 | |
| C | C1: TGCTAGGAAGACAGTAGCGT | 907 |
| C2: TGAGATTGAGAAGCCCCGCA | 1100 | |
| G | G1: GCAAAGGCCAGCAGCAGCTT | 1021 |
| G2: ATCGCGTAGCATCGTCATGG | 1112 | |
| H | H1: TTTGGTTGCAGATTGAACAA | 1061 |
| H2: CGAGTTTACTACTTTCCACA | 964 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kutchinsky, O.; Li, D.; Assa, G.; Aharoni, A.; Yakhini, Z. Cross-Activity Analysis of CRISPR/Cas9 Editing in Gene Families of Solanum lycopersicum Detected by Long-Read Sequencing. Curr. Issues Mol. Biol. 2025, 47, 507. https://doi.org/10.3390/cimb47070507
Kutchinsky O, Li D, Assa G, Aharoni A, Yakhini Z. Cross-Activity Analysis of CRISPR/Cas9 Editing in Gene Families of Solanum lycopersicum Detected by Long-Read Sequencing. Current Issues in Molecular Biology. 2025; 47(7):507. https://doi.org/10.3390/cimb47070507
Chicago/Turabian StyleKutchinsky, Ofri, Dongqi Li, Guy Assa, Asaph Aharoni, and Zohar Yakhini. 2025. "Cross-Activity Analysis of CRISPR/Cas9 Editing in Gene Families of Solanum lycopersicum Detected by Long-Read Sequencing" Current Issues in Molecular Biology 47, no. 7: 507. https://doi.org/10.3390/cimb47070507
APA StyleKutchinsky, O., Li, D., Assa, G., Aharoni, A., & Yakhini, Z. (2025). Cross-Activity Analysis of CRISPR/Cas9 Editing in Gene Families of Solanum lycopersicum Detected by Long-Read Sequencing. Current Issues in Molecular Biology, 47(7), 507. https://doi.org/10.3390/cimb47070507






