lncRNAs as Biomarkers of Hepatocellular Carcinoma Risk and Liver Damage in Advanced Chronic Hepatitis C
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALT | Alanine Aminotransferase |
| AST | Aspartate Aminotransferase |
| AFP | alpha-fetoprotein |
| AUC | area under curve |
| B-TILs | tumor-infiltrating B cells |
| cDNA | complementary DNA |
| ceRNA | competing endogenous RNAs |
| CHC | chronic hepatitis C |
| circRNA | circular RNA |
| CT | computed tomography |
| DAA | direct-acting antiviral |
| HCC | hepatocellular carcinoma |
| HCV | hepatitis C virus |
| miRNA | microRNA |
| MRI | magnetic resonance imaging |
| RCF | relative centrifugal force |
| ROC | receiver operating characteristic |
| RQ | relative quantification |
| RT-qPCR | real-time quantitative reverse transcription PCR |
References
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Fraser, P. No-nonsense functions for long noncoding RNAs. Cell 2011, 145, 178–181. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Chen, W.; Hu, X.; Li, J.; Liu, C. Regulatory roles of long noncoding RNAs implicated in cancer hallmarks. Int. J. Cancer 2020, 146, 906–916. [Google Scholar] [CrossRef]
- Wei, G.H.; Wang, X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3850–3856. [Google Scholar] [PubMed]
- Zhang, F.; Zhang, L.; Zhang, C. Long noncoding RNAs and tumorigenesis: Genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016, 37, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Mohankumar, S.; Patel, T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Funct. Genom. 2016, 15, 249–256. [Google Scholar] [CrossRef]
- Zeng, Z.; Dong, J.; Li, Y.; Dong, Z.; Liu, Z.; Huang, J.; Wang, Y.; Zhen, Y.; Lu, Y. The expression level and clinical significance of lncRNA X91348 in hepatocellular carcinoma. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3067–3071. [Google Scholar] [CrossRef]
- Klingenberg, M.; Groß, M.; Goyal, A.; Polycarpou-Schwarz, M.; Miersch, T.; Ernst, A.S.; Leupold, J.; Patil, N.; Warnken, U.; Allgayer, H.; et al. The Long Noncoding RNA Cancer Susceptibility 9 and RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein L Form a Complex and Coregulate Genes Linked to AKT Signaling. Hepatology 2018, 68, 1817–1832. [Google Scholar] [CrossRef]
- Zuo, X.; Chen, Z.; Gao, W.; Zhang, Y.; Wang, J.; Wang, J.; Cao, M.; Cai, J.; Wu, J.; Wang, X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020, 13, 5. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R Soc. Lond. B Biol. Sci. 2014, 369, 20130502. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front. Genet. 2013, 4, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Chen, K.K.; Zhang, J.; Xiao, B.; Huang, Z.; Ju, C.; Sun, J.; Zhang, F.; Lv, X.B.; Huang, G. The decade of exosomal long RNA species: An emerging cancer antagonist. Mol. Cancer 2018, 17, 75. [Google Scholar] [CrossRef]
- Cholongitas, E.; Senzolo, M.; Standish, R.; Marelli, L.; Quaglia, A.; Patch, D.; Dhillon, A.P.; Burroughs, A.K. A systematic review of the quality of liver biopsy specimens. Am. J. Clin. Pathol. 2006, 125, 710–721. [Google Scholar] [CrossRef]
- Chindamo, M.C.; Nunes-Pannain, V.L.; Araújo-Neto, J.M.; Moraes-Coelho, H.S.; Luiz, R.R.; Villela-Nogueira, C.A.; Perez, R.M. Intermediate fibrosis staging in hepatitis C: A problem not overcome by optimal samples or pathologists’ expertise. Ann. Hepatol. 2015, 14, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Clavien, P.A.; Lesurtel, M.; Bossuyt, P.M.; Gores, G.J.; Langer, B.; Perrier, A.; OLT for HCC Consensus Group. Recommendations for liver transplantation for hepatocellular carcinoma: An international consensus conference report. Lancet Oncol. 2012, 13, e11–e22. [Google Scholar] [CrossRef]
- Silva, M.A.; Hegab, B.; Hyde, C.; Guo, B.; Buckels, J.A.; Mirza, D.F. Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: A systematic review and meta-analysis. Gut 2008, 57, 1592–1596. [Google Scholar] [CrossRef]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef] [PubMed]
- De Stefano, F.; Chacon, E.; Turcios, L.; Marti, F.; Gedaly, R. Novel biomarkers in hepatocellular carcinoma. Dig. Liver Dis. 2018, 50, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Mo, Z.; Hu, Z.; Zhang, L.; Qin, S.; Qin, X.; Li, S. Diagnostic value of fibrinogen to prealbumin ratio and gamma-glutamyl transpeptidase to platelet ratio in the progression of AFP-negative hepatocellular carcinoma. Cancer Cell Int. 2020, 20, 77. [Google Scholar] [CrossRef]
- Devarbhavi, H.; Asrani, S.K.; Arab, J.P.; Nartey, Y.A.; Pose, E.; Kamath, P.S. Global burden of liver disease: 2023 update. J. Hepatol. 2023, 79, 516–537. [Google Scholar] [CrossRef]
- Jeong, S.W.; Jang, J.Y.; Chung, R.T. Hepatitis C virus and hepatocarcinogenesis. Clin. Mol. Hepatol. 2012, 18, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Fuchs, B.C.; Bardeesy, N.; Baumert, T.F.; Chung, R.T. Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma. J. Hepatol. 2014, 61, S79–S90. [Google Scholar] [CrossRef]
- Alberti, A.; Chemello, L.; Benvegnù, L. Natural history of hepatitis C. J. Hepatol. 1999, 31 (Suppl. 1), 17–24. [Google Scholar] [CrossRef]
- Ikeda, K.; Saitoh, S.; Suzuki, Y.; Kobayashi, M.; Tsubota, A.; Koida, I.; Arase, Y.; Fukuda, M.; Chayama, K.; Murashima, N.; et al. Disease progression and hepatocellular carcinogenesis in patients with chronic viral hepatitis: A prospective observation of 2215 patients. J. Hepatol. 1998, 28, 930–938. [Google Scholar] [CrossRef]
- World Health Organization. Hepatitis-Number of Persons Living with Chronic Hepatitis. Available online: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/hepatitis---number-of-persons-living-with-chronic-hepatitis (accessed on 25 April 2025).
- Dai, M.; Chen, S.; Wei, X.; Zhu, X.; Lan, F.; Dai, S.; Qin, X. Diagnosis, prognosis and bioinformatics analysis of lncRNAs in hepatocellular carcinoma. Oncotarget 2017, 8, 95799–95809. [Google Scholar] [CrossRef]
- Choi, D.T.; Kum, H.C.; Park, S.; Ohsfeldt, R.L.; Shen, Y.; Parikh, N.D.; Singal, A.G. Hepatocellular Carcinoma Screening Is Associated with Increased Survival of Patients with Cirrhosis. Clin. Gastroenterol. Hepatol. 2019, 17, 976–987.e4. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.-L.; Schirmacher, P.; Vilgrain, V. EASL clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, L.; Gu, J.; Zhang, H.; Yuan, J.; Lian, Q.; Lv, G.; Wang, S.; Wu, Y.; Yang, Y.T.; et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat. Commun. 2017, 8, 14421. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhou, J.K.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Ferrasi, A.C.; Fernandez, G.J.; Grotto, R.M.T.; Silva, G.F.; Goncalves, J.; Costa, M.C.; Enguita, F.J.; Pardini, M.I.M.C. New LncRNAs in Chronic Hepatitis C progression: From fibrosis to hepatocellular carcinoma. Sci. Rep. 2020, 10, 9886. [Google Scholar] [CrossRef]
- Takahashi, K.; Yan, I.K.; Kogure, T.; Haga, H.; Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014, 4, 458–467. [Google Scholar] [CrossRef]
- Gezer, U.; Özgür, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Bedossa, P.; Poynard, T.; The METAVIR Cooperative Study Group. An algorithm for the grading of activity in chronic hepatitis C. Hepatology 1996, 24, 289–293. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−∆∆CT) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mazzara, S.; Rossi, R.L.; Grifantini, R.; Donizetti, S.; Abrignani, S.; Bombaci, M. CombiROC: An interactive web tool for selecting accurate marker combinations of omics data. Sci. Rep. 2017, 7, 45477. [Google Scholar] [CrossRef]
- Zweig, M.H.; Campbell, G. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin. Chem. 1993, 39, 561–577, Erratum in Clin. Chem. 1993, 39, 1589. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ren, X.; Zhang, Q. Incidence, risk factors, and prognosis in patients with primary hepatocellular carcinoma and lung metastasis: A population-based study. Cancer Manag. Res. 2019, 11, 2759–2768. [Google Scholar] [CrossRef] [PubMed]
- Sarveazad, A.; Agah, S.; Babahajian, A.; Amini, N.; Bahardoust, M. Predictors of 5 year survival rate in hepatocellular carcinoma patients. J. Res. Med. Sci. 2019, 24, 86. [Google Scholar] [CrossRef]
- Wang, Z.; Qin, H.; Liu, S.; Sheng, J.; Zhang, X. Precision diagnosis of hepatocellular carcinoma. Chin. Med. J. 2023, 136, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, T.; Zhou, W.; Li, J.; Li, X.; Wang, Q.; Jin, X.; Yin, J.; Chen, L.; Zhang, Y.; et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun. 2020, 11, 1000. [Google Scholar] [CrossRef]
- Lv, E.; Sheng, J.; Yu, C.; Rao, D.; Huang, W. LncRNA influence sequential steps of hepatocellular carcinoma metastasis. Biomed. Pharmacother. 2021, 136, 111224. [Google Scholar] [CrossRef]
- Jiang, N.; Pan, J.; Fang, S.; Zhou, C.; Han, Y.; Chen, J.; Meng, X.; Jin, X.; Gong, Z. Liquid biopsy: Circulating exosomal long noncoding RNAs in cancer. Clin. Chim. Acta 2019, 495, 331–337. [Google Scholar] [CrossRef]
- Bao, H.; Su, H. Long Noncoding RNAs Act as Novel Biomarkers for Hepatocellular Carcinoma: Progress and Prospects. Biomed. Res. Int. 2017, 2017, 6049480. [Google Scholar] [CrossRef]
- Kamel, M.M.; Matboli, M.; Sallam, M.; Montasser, I.F.; Saad, A.S.; El-Tawdi, A.H.F. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl. Res. 2016, 168, 134–145. [Google Scholar] [CrossRef]
- Shehab-Eldeen, S.; Essa, A.; Arafat, E.S.; Sleem, A.S.; Alhosary, A.A.; Darwish, E.; Essa, A.; Al-Omair, O.A.; Al-Khoufi, E.A.; Al Abdulqader, A.K.; et al. Serum LINC00152 and UCA1 in HCV-Induced Hepatocellular Carcinoma: Clinical Significance and Prognostic Value. Biologics 2023, 17, 137–149. [Google Scholar] [CrossRef]
- Samir, A.; Abdeldaim, A.; Mohammed, A.; Ali, A.; Alorabi, M.; Hussein, M.M.; Bakr, Y.M.; Ibrahim, A.M.; Abdelhafiz, A.S. Analysis of four long non-coding RNAs for hepatocellular carcinoma screening and prognosis by the aid of machine learning techniques. Sci. Rep. 2024, 14, 29582. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Tang, J.; Xiang, T.; Lin, J.; Deng, C.; Peng, Y.; Zheng, J.; Hu, G. Genome wide identification of long noncoding RNAs in CCl4 induced liver fibrosis via RNA sequencing. Mol. Med. Rep. 2018, 18, 299–307. [Google Scholar] [CrossRef]
- Xu, J.; Jin, M.; Mu, Z.; Li, Z.; Qi, R.; Han, X.; Jiang, H. Inhibiting melanoma tumor growth: The role of oxidative stress-associated LINC02132 and COPDA1 long non-coding RNAs. Front. Immunol. 2025, 16, 1558292. [Google Scholar] [CrossRef]
- Jing, R.; Liu, S.; Jiang, Y.; Zong, W.; Ju, S.; Cui, M. Determination of serum RP11-731F5.2 as a noninvasive biomarker for gastric cancer diagnosis and prognosis. Pathol. Res. Pract. 2020, 216, 153261. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Hong, W.; Gao, M.; Yi, E.; Zhang, J.; Hao, B.; Liang, C.; Li, X.; Li, C.; Ye, X.; et al. Long Noncoding RNA COPDA1 Promotes Airway Smooth Muscle Cell Proliferation in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2019, 61, 584–596. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, X.; Zhao, F.; Ru, S. Bisphenol S promotes the cell cycle progression and cell proliferation through ERα-cyclin D-CDK4/6-pRb pathway in MCF-7 breast cancer cells. Toxicol. Appl. Pharmacol. 2019, 366, 75–82. [Google Scholar] [CrossRef]
- Strobeck, M.W.; Fribourg, A.F.; Puga, A.; Knudsen, E.S. Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene 2000, 19, 1857–1867. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.W.; Lin, Y.M.; Chang, J.G.; Yeh, K.T.; Chen, R.M.; Tsai, J.J.; Su, W.W.; Hu, R.M. Clinical implications of deregulated CDK4 and Cyclin D1 expression in patients with human hepatocellular carcinoma. Med. Oncol. 2013, 30, 379. [Google Scholar] [CrossRef]
- Rivadeneira, D.B.; Mayhew, C.N.; Thangavel, C.; Sotillo, E.; Reed, C.A.; Graña, X.; Knudsen, E.S. Proliferative suppression by CDK4/6 inhibition: Complex function of the retinoblastoma pathway in liver tissue and hepatoma cells. Gastroenterology 2010, 138, 1920–1930. [Google Scholar] [CrossRef]
- Reed, C.A.; Mayhew, C.N.; McClendon, A.K.; Knudsen, E.S. Unique impact of RB loss on hepatic proliferation: Tumorigenic stresses uncover distinct pathways of cell cycle control. J. Biol. Chem. 2010, 285, 1089–1096. [Google Scholar] [CrossRef]
- Pavlasova, G.; Mraz, M. The regulation and function of CD20: An “enigma” of B-cell biology and targeted therapy. Haematologica 2020, 105, 1494–1506. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, L.; Li, J.; Huang, J.; Xie, J.H.; Menard, L.; Shi, Y.; Zhao, X.; Xie, S.; Zang, W.; et al. Systematic characterization of the tumor microenvironment in Chinese patients with hepatocellular carcinoma highlights intratumoral B cells as a potential immunotherapy target. Oncol. Rep. 2022, 47, 38. [Google Scholar] [CrossRef] [PubMed]
- Cagle, P.; Qi, Q.; Niture, S.; Kumar, D. KCNQ1OT1: An Oncogenic Long Noncoding RNA. Biomolecules 2021, 11, 1602. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhu, Y.; Liu, H.; Liu, Y.; Zhang, X. Long Non-Coding RNA KCNQ1OT1 Promotes Progression of Hepatocellular Carcinoma by miR-148a-3p/IGF1R Axis. Technol. Cancer Res. Treat. 2020, 19, 1533033820980117. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.L.; Li, D.J.; Lv, M.Y.; Pei, Y.J.; Zhang, X.J.; Li, L.; Liu, X.Y.; Fan, A.H. LncRNA KCNQ1OT1 regulates the invasion and migration of hepatocellular carcinoma by acting on S1PR1 through miR-149. Cancer Gene Ther. 2021, 28, 850–863. [Google Scholar] [CrossRef]
- Zhou, W.; Li, H.; Shang, S.; Liu, F. lncRNA KCNQ1OT1 reverses the effect of sevoflurane on hepatocellular carcinoma progression via regulating the miR-29a-3p/CBX3 axis. Braz. J. Med. Biol. Res. 2021, 54, e10213. [Google Scholar] [CrossRef]
- Hu, H.; Yang, L.; Li, L.; Zeng, C. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ ABCC1 axis. Biochem. Biophys. Res. Commun. 2018, 503, 2400–2406. [Google Scholar] [CrossRef]
- Li, C.; Miao, R.; Zhang, J.; Qu, K.; Liu, C. Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular carcinoma by functioning as a competing endogenous RNA of miR-504. Int. J. Oncol. 2018, 52, 1603–1612. [Google Scholar] [CrossRef]
- Panzitt, K.; Tschernatsch, M.M.; Guelly, C.; Moustafa, T.; Stradner, M.; Strohmaier, H.M.; Buck, C.R.; Denk, H.; Schroeder, R.; Trauner, M.; et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007, 132, 330–342. [Google Scholar] [CrossRef]
- Xie, H.; Ma, H.; Zhou, D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed. Res. Int. 2013, 2013, 136106. [Google Scholar] [CrossRef]
- Huang, J.; Zheng, Y.; Xiao, X.; Liu, C.; Lin, J.; Zheng, S.; Yang, B.; Ou, Q. A Circulating Long Noncoding RNA Panel Serves as a Diagnostic Marker for Hepatocellular Carcinoma. Dis. Markers 2020, 2020, 5417598. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.; Mahfouz, H.; Salama, A.; Medhat, E. Long Non-Coding HULC and miRNA-372 as Diagnostic Biomarkers in Hepatocellular Carcinoma. Rep. Biochem. Mol. Biol. 2020, 9, 230–240. [Google Scholar] [CrossRef]
- Gaber, D.A.; Shaker, O.; Younis, A.T.; El-Kassas, M. LncRNA HULC and miR-122 Expression Pattern in HCC-Related HCV Egyptian Patients. Genes 2022, 13, 1669. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Esmaeili, M.; Taheri, M.; Samsami, M. Highly upregulated in liver cancer (HULC): An update on its role in carcinogenesis. J. Cell. Physiol. 2020, 235, 9071–9079. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zheng, H.; Chan, M.T.; Wu, W.K. HULC: An oncogenic long non-coding RNA in human cancer. J. Cell. Mol. Med. 2017, 21, 410–417. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, F.; Zhao, M.; Yang, Z.; Li, J.; Zhang, S.; Zhang, W.; Ye, L.; Zhang, X. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J. Biol. Chem. 2017, 292, 15395–15407. [Google Scholar] [CrossRef]
- Han, Y.; Ma, Z. LncRNA highly upregulated in liver cancer regulates imatinib resistance in chronic myeloid leukemia via the miR-150-5p/MCL1 axis. Anticancer Drugs 2021, 32, 427–436. [Google Scholar] [CrossRef]
- Guan, L.; Wang, F.; Wang, M.; Han, S.; Cui, Z.; Xi, S.; Xu, H.; Li, S. Downregulation of HULC Induces Ferroptosis in Hepatocellular Carcinoma via Targeting of the miR-3200-5p/ATF4 Axis. Oxidative Med. Cell. Longev. 2022, 2022, 9613095. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Du, Y.; Kong, G.; You, X.; Zhang, S.; Zhang, T.; Gao, Y.; Ye, L.; Zhang, X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J. Biol. Chem. 2012, 287, 26302–26311. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, J.; Luo, F. Serum tumor markers for detection of hepatocellular carcinoma. World J. Gastroenterol. 2006, 12, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.F.; Weng, J.; Xu, G.X.; Chen, C.M.; Jia, C.K. Combination of serum tumor markers dickkopf-1, DCP and AFP for the diagnosis of primary hepatocellular carcinoma. Asian Pac. J. Trop. Med. 2017, 10, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Agopian, V.G.; Harlander-Locke, M.P.; Markovic, D.; Zarrinpar, A.; Kaldas, F.M.; Cheng, E.Y.; Yersiz, H.; Farmer, D.G.; Hiatt, J.R.; Busuttil, R.W. Evaluation of Patients with Hepatocellular Carcinomas That Do Not Produce α-Fetoprotein. JAMA Surg. 2017, 152, 55–64. [Google Scholar] [CrossRef]
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and Other Tools) Resource [Internet]; Food and Drug Administration: Silver Spring, MD, USA; National Institutes of Health: Bethesda, MD, USA, 2016; Glossary. Available online: https://www.ncbi.nlm.nih.gov/books/NBK338448/ (accessed on 10 March 2025).
- Ou, F.S.; Michiels, S.; Shyr, Y.; Adjei, A.A.; Oberg, A.L. Biomarker Discovery and Validation: Statistical Considerations. J. Thorac. Oncol. 2021, 16, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Li, Y.; Shao, S.; Guo, B.; Zhang, M.; Zheng, L.; Zhang, K.; Zhou, F.; Zhang, L.; Chen, C.; et al. Protective effect of hepatocyte-enriched lncRNA-Mir122hg by promoting hepatocyte proliferation in acute liver injury. Exp. Mol. Med. 2022, 54, 2022–2035. [Google Scholar] [CrossRef]
- Joshi, M.; Oltean, M.; Patil, P.B.; Hallberg, D.; Kleman, M.; Holgersson, J.; Olausson, M.; Sumitran-Holgersson, S. Chemokine-mediated robust augmentation of liver engraftment: A novel approach. Stem Cells Transl. Med. 2015, 4, 21–30. [Google Scholar] [CrossRef]
- Dhir, A.; Dhir, S.; Proudfoot, N.J.; Jopling, C.L. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat. Struct. Mol. Biol. 2015, 22, 319–327. [Google Scholar] [CrossRef]
- Wu, J.; Gao, L.; Chen, H.; Zhou, X.; Lu, X.; Mao, Z. LINC02535 promotes cell growth in poorly differentiated gastric cancer. J. Clin. Lab. Anal. 2021, 35, e23877. [Google Scholar] [CrossRef]
- Wen, D.; Huang, Z.; Li, Z.; Tang, X.; Wen, X.; Liu, J.; Li, M. LINC02535 co-functions with PCBP2 to regulate DNA damage repair in cervical cancer by stabilizing RRM1 mRNA. J. Cell Physiol. 2020, 235, 7592–7603. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, J.; Zhang, W.; Wang, A.; Jiao, M.; Cai, X.; Zhu, J.; Liu, Z.; Huang, J.A. LINC02535/miR-30a-5p/GALNT3 axis contributes to lung adenocarcinoma progression via the NF-κ B signaling pathway. Cell Cycle 2022, 21, 2455–2470. [Google Scholar] [CrossRef]
- Assal, R.A.; Elemam, N.M.; Mekky, R.Y.; Attia, A.A.; Soliman, A.H.; Gomaa, A.I.; Efthimiadou, E.K.; Braoudaki, M.; Fahmy, S.A.; Youness, R.A. A Novel Epigenetic Strategy to Concurrently Block Immune Checkpoints PD-1/PD-L1 and CD155/TIGIT in Hepatocellular Carcinoma. Transl. Oncol. 2024, 45, 101961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gao, F.; Yu, X.; Zhang, Q.; Sun, Z.; He, Y.; Guo, W. LINC00261: A burgeoning long noncoding RNA related to cancer. Cancer Cell Int. 2021, 21, 274. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Lei, S.; Zeng, Z.; Zhang, J.; Xue, Y.; Sun, Y.; Lan, J.; Xu, S.; Mao, D.; Guo, B. Linc00261 inhibits metastasis and the WNT signaling pathway of pancreatic cancer by regulating a miR-552-5p/FOXO3 axis. Oncol. Rep. 2020, 43, 930–942. [Google Scholar] [CrossRef]
- Wang, X.; Gao, X.; Tian, J.; Zhang, R.; Qiao, Y.; Hua, X.; Shi, G. LINC00261 inhibits progression of pancreatic cancer by down-regulating miR-23a-3p. Arch. Biochem. Biophys. 2020, 689, 108469. [Google Scholar] [CrossRef]
- Zhai, S.; Xu, Z.; Xie, J.; Zhang, J.; Wang, X.; Peng, C.; Li, H.; Chen, H.; Shen, B.; Deng, X. Epigenetic silencing of LncRNA LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene 2021, 40, 277–291. [Google Scholar] [CrossRef]
- Zhu, J.; Deng, J.; Zhang, L.; Zhao, J.; Zhou, F.; Liu, N.; Cai, R.; Wu, J.; Shu, B.; Qi, S. Reconstruction of lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveals functional lncRNAs in skin cutaneous melanoma. BMC Cancer 2020, 20, 927. [Google Scholar] [CrossRef] [PubMed]
- Zhan, K.; Pan, H.; Zhou, Z.; Tang, W.; Ye, Z.; Huang, S.; Luo, L. Biological role of long non-coding RNA KCNQ1OT1 in cancer progression. Biomed. Pharmacother. 2023, 169, 115876. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Lev, M. Kats and Pier Paolo Pandolfi. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, D.H.; Wu, N.; Xiao, J.H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef]
- Lu, X.-J.; Gao, A.-M.; Ji, L.-J.; Xu, J. Pseudogene in cancer: Real functions and promising signature. J. Med. Genet. 2015, 52, 17–24. [Google Scholar]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.C.; Shi, Z.T.; Xiao, S.F.; Ke, Y.; Tang, H.R.; Wu, T.G.; Guo, Z.T.; Ni, F.; An, S.; Wang, L. Co-expression analysis and ceRNA network reveal eight novel potential lncRNA biomarkers in hepatocellular carcinoma. PeerJ 2019, 7, e8101. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
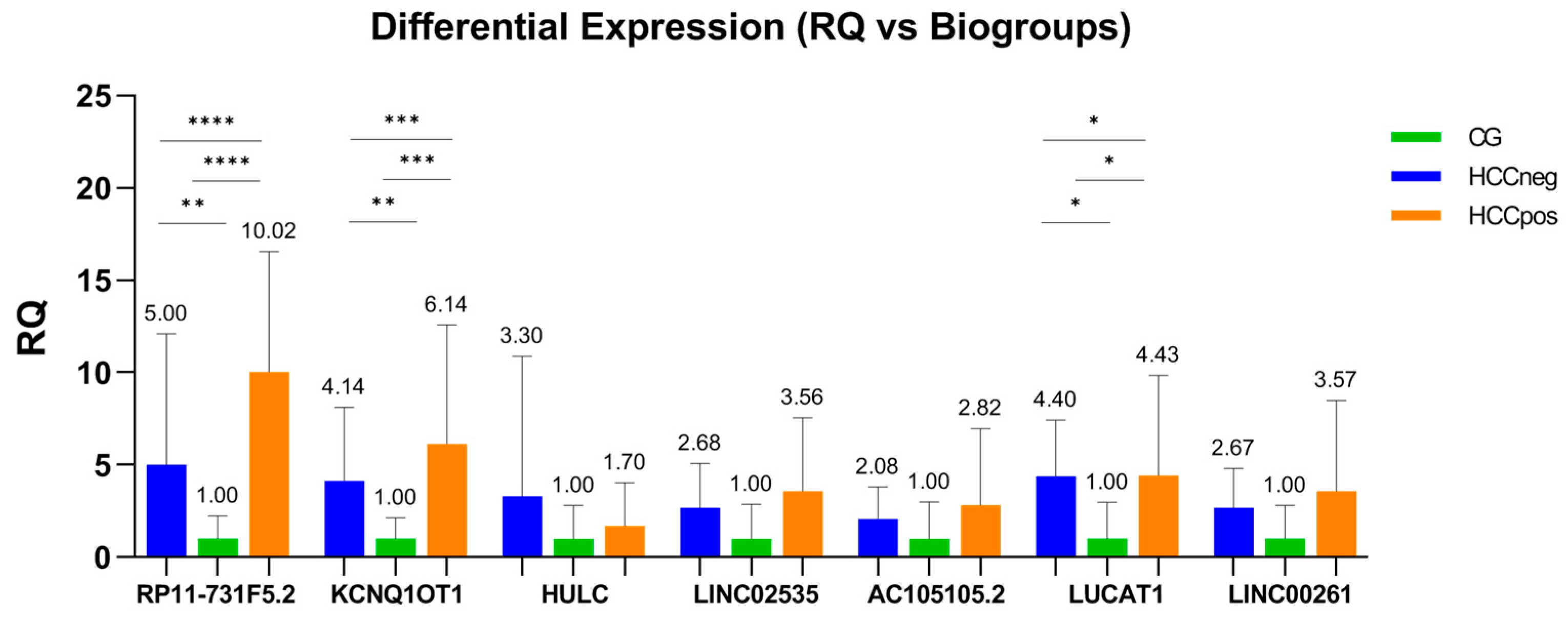
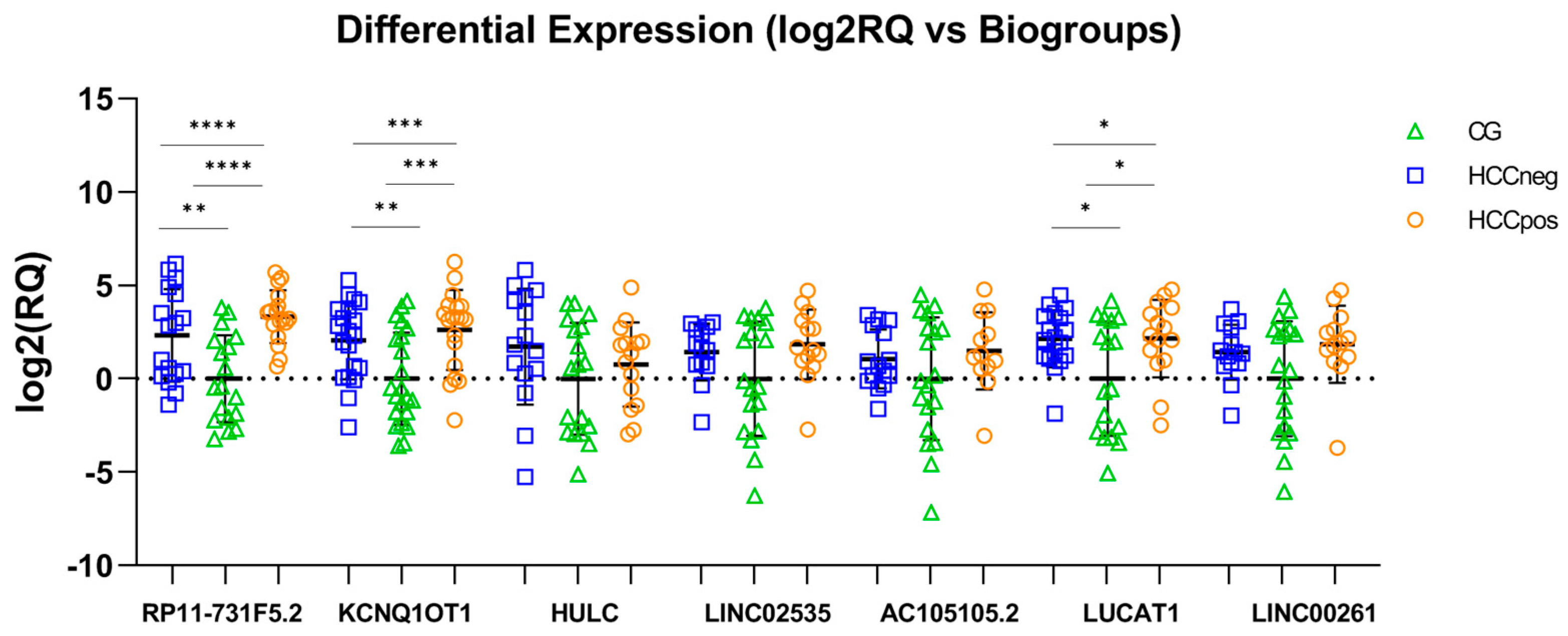
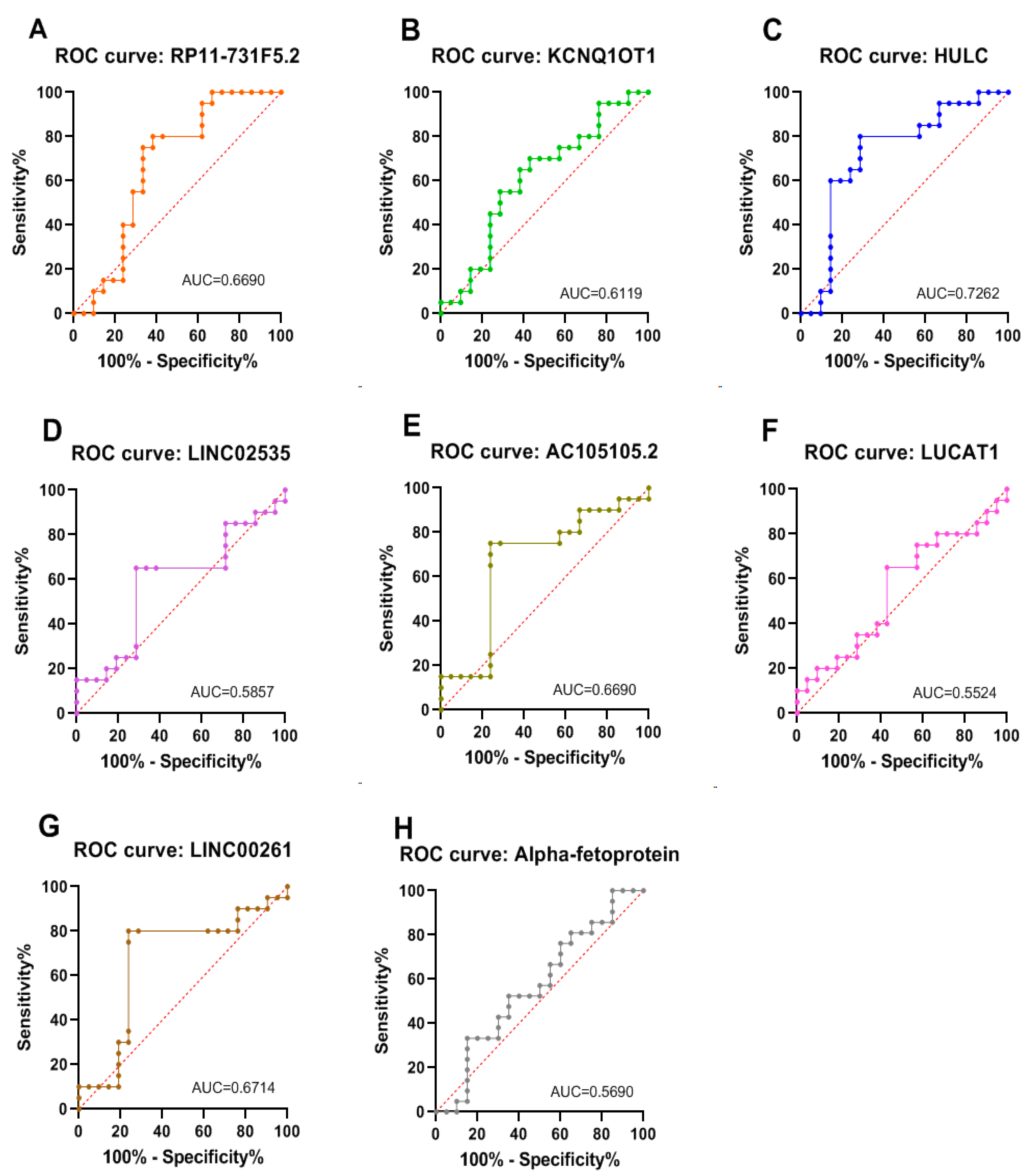
| Variables | BioGroups | |||
|---|---|---|---|---|
| Control Group n = 22 | HCCneg n = 21 | HCCpos n = 20 | p-Value † | |
| Age (years) | 57.2 ± 7.5 | 56 ± 9.0 | 58.9 ± 6.7 | 0.999 |
| Sex | ||||
| Male | 12 (54.5) | 12 (57%) | 15 (75%) | 0.339 |
| Female | 10 (45.5) | 9 (43%) | 5 (25%) | |
| BMI (Kg/m2) | 25.6 ± 7.4 | 28.2 ± 5.8 | 27.2 ± 4.6 | |
| HCV Genotype | ||||
| 1 * | - | 17 (81%) | 14 (70%) | 0.484 |
| Not 1 ** | - | 4 (19%) | 6 (30%) | |
| Fibrosis Grade # | ||||
| F3 | - | 3 (14%) | 3 (15%) | 1.000 |
| F4 | - | 18 (86%) | 17 (85%) | |
| AFP (ng/mL) | ||||
| Median | - | 7.2 (2.7–74.2) | 9.6 (1.3–591) | 0.457 |
| Mean | - | 19 ± 19.8 | 66 ± 140.7 | |
| HCC Diagnosis## | ||||
| Median | - | - | 24 (8–89) | |
| Mean | - | - | 35.5 ± 25.9 | |
| Target | Ensembl ID | Primers (5′ to 3′) | Amplicon (bp) |
|---|---|---|---|
| RP11-731F5.2 | ENSG00000253364 | F-TTCAGTCTTTGCAGCGTGGAG | 121 |
| R-CCTGTTTTGGCGCGGTA | |||
| KCNQ1OT1 | ENSG00000269821 | F-TGCAGAAGACAGGACACTGG | 125 |
| R-CTTTGGTGGGAAAGGACAGA | |||
| HULC | ENST00000503668 | F-ACTCTGAAGTAAAGGCCGGA | 95 |
| R-GCCAGGAAACTTCTTGCTTGT | |||
| LINC02535 | ENST00000455071 | F-AAGGAGCTCTGTTCTCCAGG | 102 |
| R-GCCTCTATGTAGGGCGCTTT | |||
| AC105105.2 | ENSG00000267391 | F-CCCGTGATGCTTCTTTTCTC | 150 |
| R-CCATTGTCACACTCCACAGC | |||
| LINC00261 | ENSG00000259974 | F-TCAGATTGCTCCTGGACACTT | 91 |
| R-GGACCATTGCCTCTTGATTAG | |||
| LUCAT1 | ENSG00000248323 | F-GCTCGGATTGCCTTAGACAG | 114 |
| R-GGGTGAGCTTCTTGTGAGGA | |||
| β-ACTIN | ENSG00000075624 | F-AGAGCCTCGCCTTTGCCGATCC | 103 |
| R-CACATGCCGGAGCCGTTGTCG |
| RP11-731F5 | KCNQ1OT1 | HULC | LINC02535 | AC105105.2 | LUCAT1 | LINC00261 | AFP | |
|---|---|---|---|---|---|---|---|---|
| RP11-731F5 | - | 0.70 | 0.18 | 0.50 | 0.58 | 0.41 | 0.46 | −0.13 |
| KCNQ1OT1 | 0.48 | - | 0.40 | 0.57 | 0.66 | 0.59 | 0.57 | −0.18 |
| HULC | 0.37 | 0.76 | - | 0.32 | 0.36 | 0.32 | 0.26 | −0.01 |
| LINC02535 | 0.37 | 0.66 | 0.33 | - | 0.91 | 0.64 | 0.93 | −0.36 |
| AC105105.2 | 0.46 | 0.65 | 0.32 | 0.89 | - | 0.53 | 0.88 | −0.40 |
| LUCAT1 | 0.63 | 0.59 | 0.43 | 0.75 | 0.76 | - | 0.62 | 0.04 |
| LINC00261 | 0.33 | 0.60 | 0.24 | 0.91 | 0.88 | 0.80 | - | −0.38 |
| AFP | −0.09 | 0.07 | −0.002 | 0.34 | 0.39 | 0.23 | 0.28 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, D.B.; Fernandez, G.J.; Silva, L.T.; Silva, G.F.; Lima, E.O.; Galvani, A.F.; Pereira, G.L.; Ferrasi, A.C. lncRNAs as Biomarkers of Hepatocellular Carcinoma Risk and Liver Damage in Advanced Chronic Hepatitis C. Curr. Issues Mol. Biol. 2025, 47, 348. https://doi.org/10.3390/cimb47050348
dos Santos DB, Fernandez GJ, Silva LT, Silva GF, Lima EO, Galvani AF, Pereira GL, Ferrasi AC. lncRNAs as Biomarkers of Hepatocellular Carcinoma Risk and Liver Damage in Advanced Chronic Hepatitis C. Current Issues in Molecular Biology. 2025; 47(5):348. https://doi.org/10.3390/cimb47050348
Chicago/Turabian Styledos Santos, Driéle B., Geysson J. Fernandez, Letícia T. Silva, Giovanni F. Silva, Estela O. Lima, Aline F. Galvani, Guilherme L. Pereira, and Adriana C. Ferrasi. 2025. "lncRNAs as Biomarkers of Hepatocellular Carcinoma Risk and Liver Damage in Advanced Chronic Hepatitis C" Current Issues in Molecular Biology 47, no. 5: 348. https://doi.org/10.3390/cimb47050348
APA Styledos Santos, D. B., Fernandez, G. J., Silva, L. T., Silva, G. F., Lima, E. O., Galvani, A. F., Pereira, G. L., & Ferrasi, A. C. (2025). lncRNAs as Biomarkers of Hepatocellular Carcinoma Risk and Liver Damage in Advanced Chronic Hepatitis C. Current Issues in Molecular Biology, 47(5), 348. https://doi.org/10.3390/cimb47050348






