A Comparative Analysis of the Roles of von Willebrand Factor and ADAMTS13 in Hepatocellular Carcinoma: A Bioinformatics and Microarray-Based Study
Abstract
1. Introduction
2. Materials and Methods
2.1. The Expression Profile Analysis of VWF and ADAMTS13
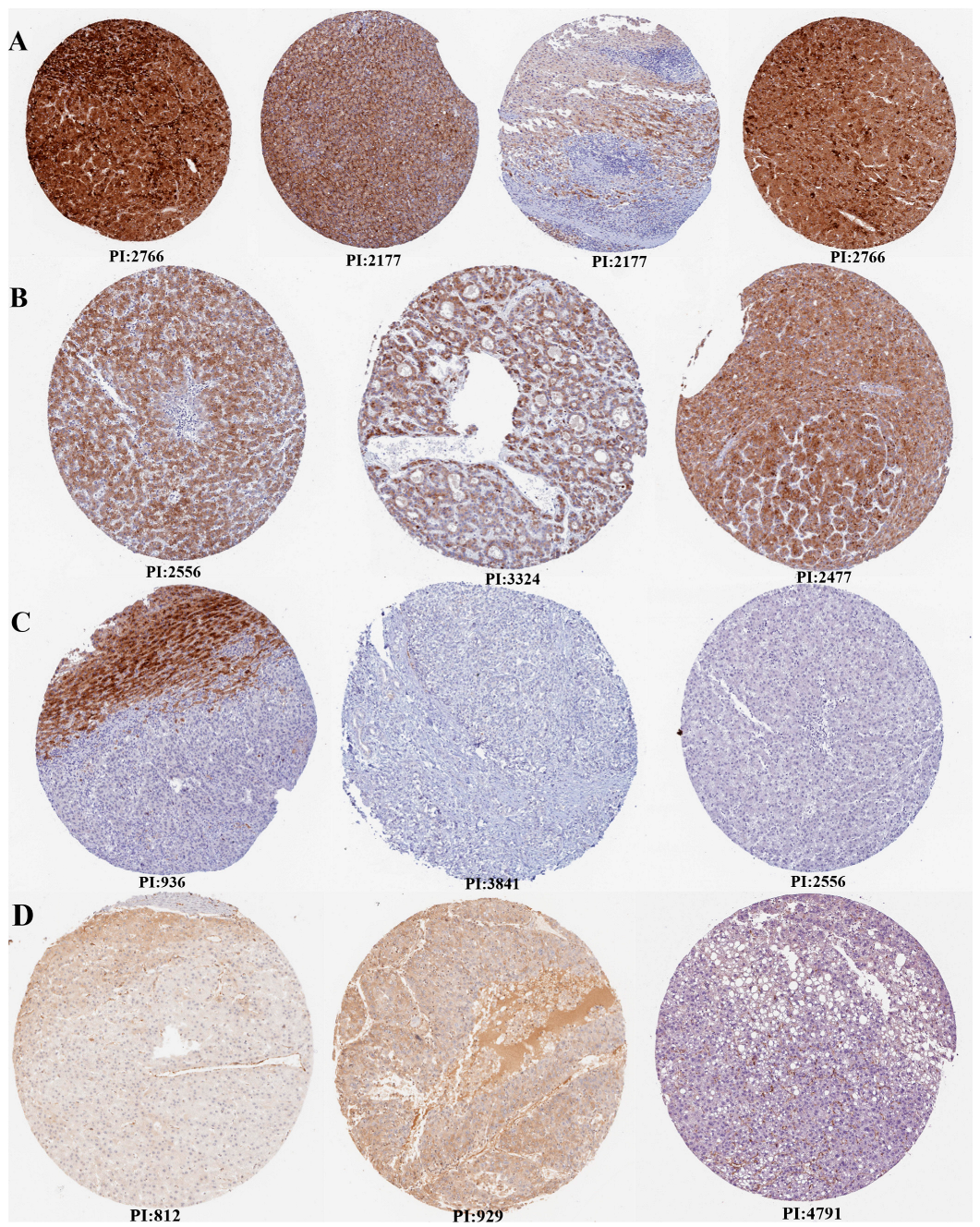
2.2. Human Protein Atlas (HPA) Analysis
2.3. The Methylation Status Analysis of VWF and ADAMTS13
2.4. Immune Infiltration Analysis of VWF and ADAMTS13
2.5. The Survival Analysis of VWF and ADAMTS13
2.6. The Correlation, Targetgram, and Gene Signature Analysis of VWF and ADAMTS13
2.7. The Gene–Gene Interaction
2.8. MicroRNA Target Analysis
2.9. Enrichment Analysis of ADAMTS13 and VWF (Enrichr-KG)
2.10. Association of HCC-Associated Long Non-Coding RNAs (LncRNAs) with VWF and ADAMTS13
2.11. Detection of Differentially Expressed Genes (DEGs)
2.12. Statistical Analysis Methods
3. Results
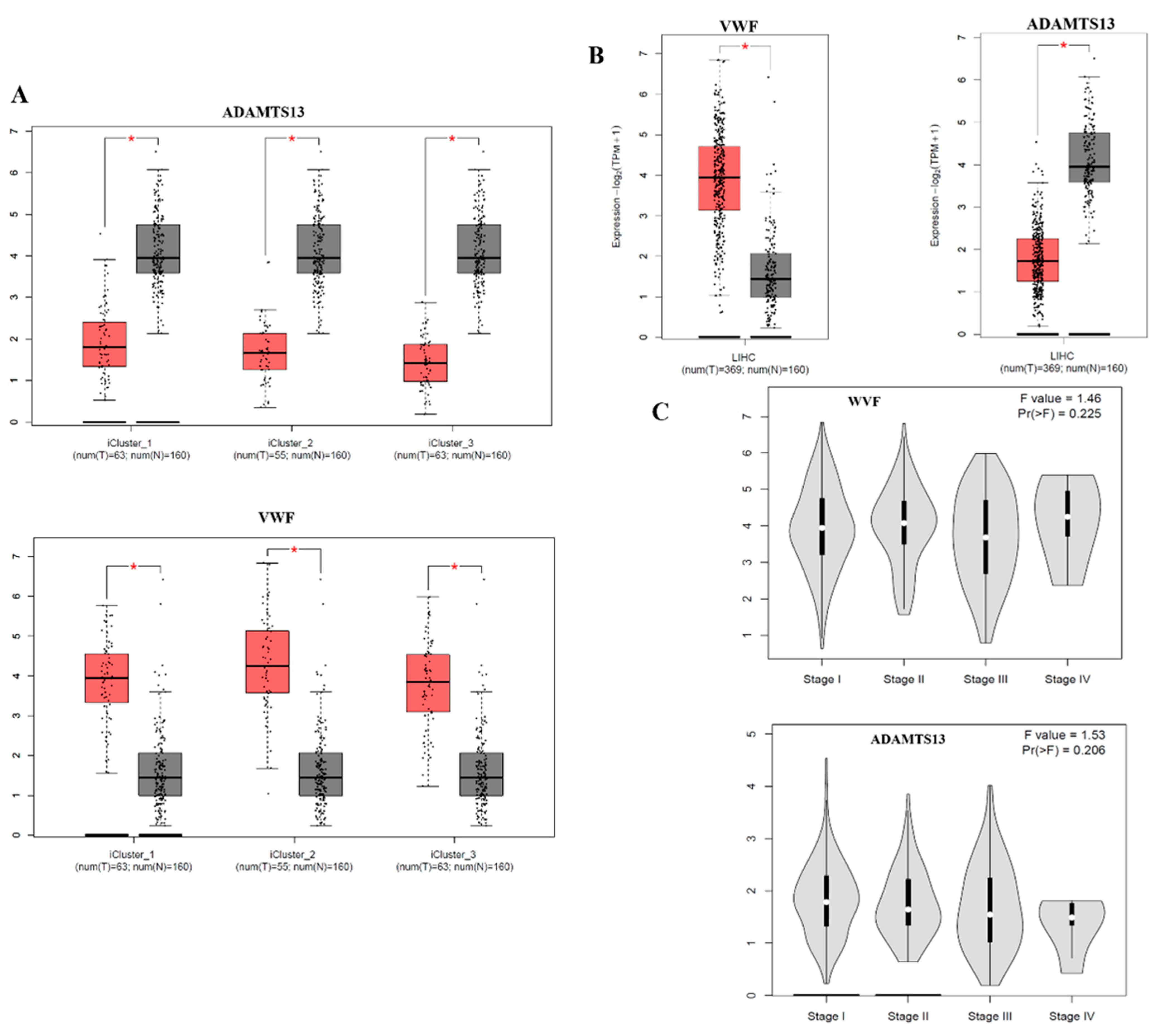
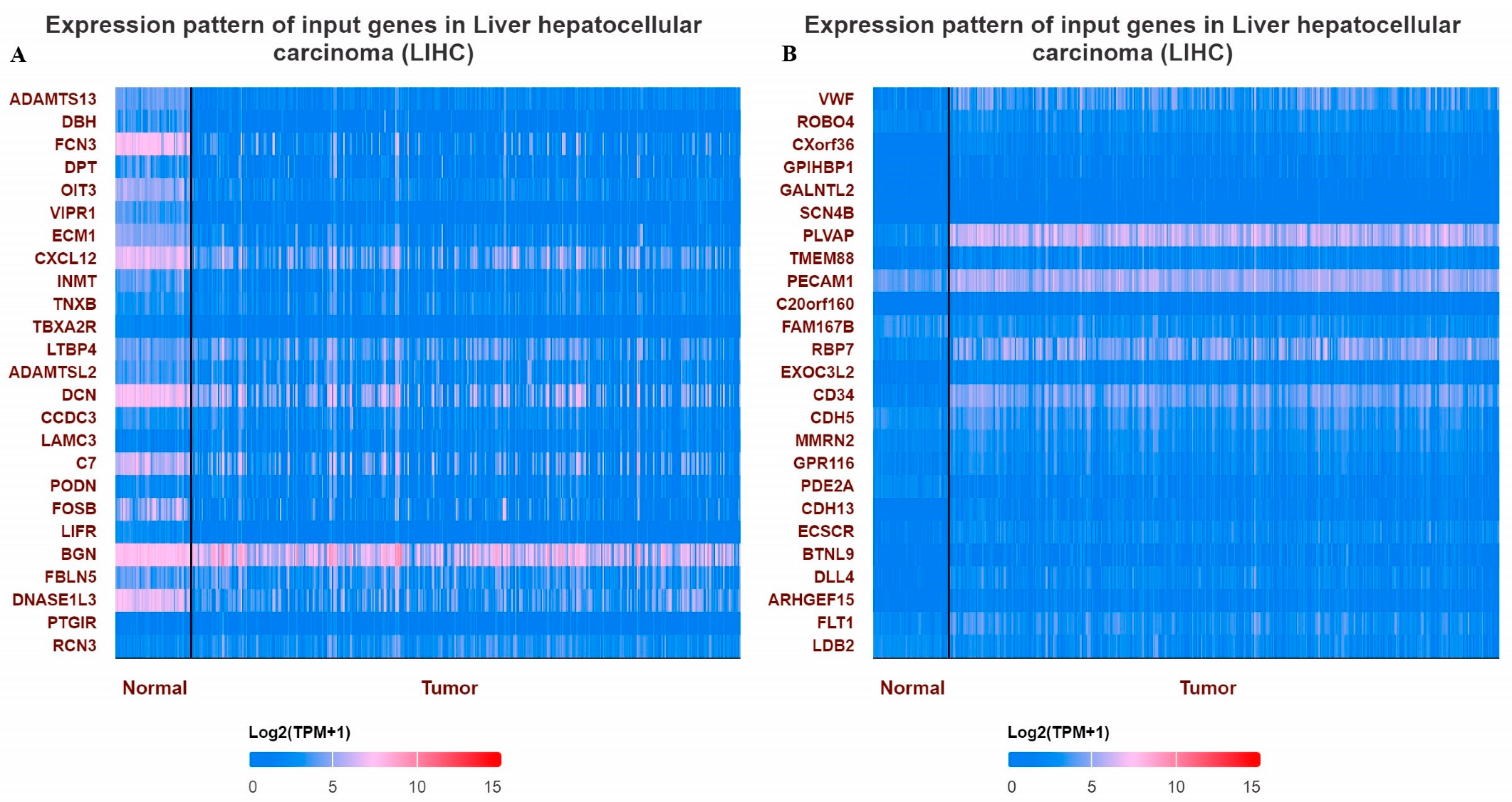
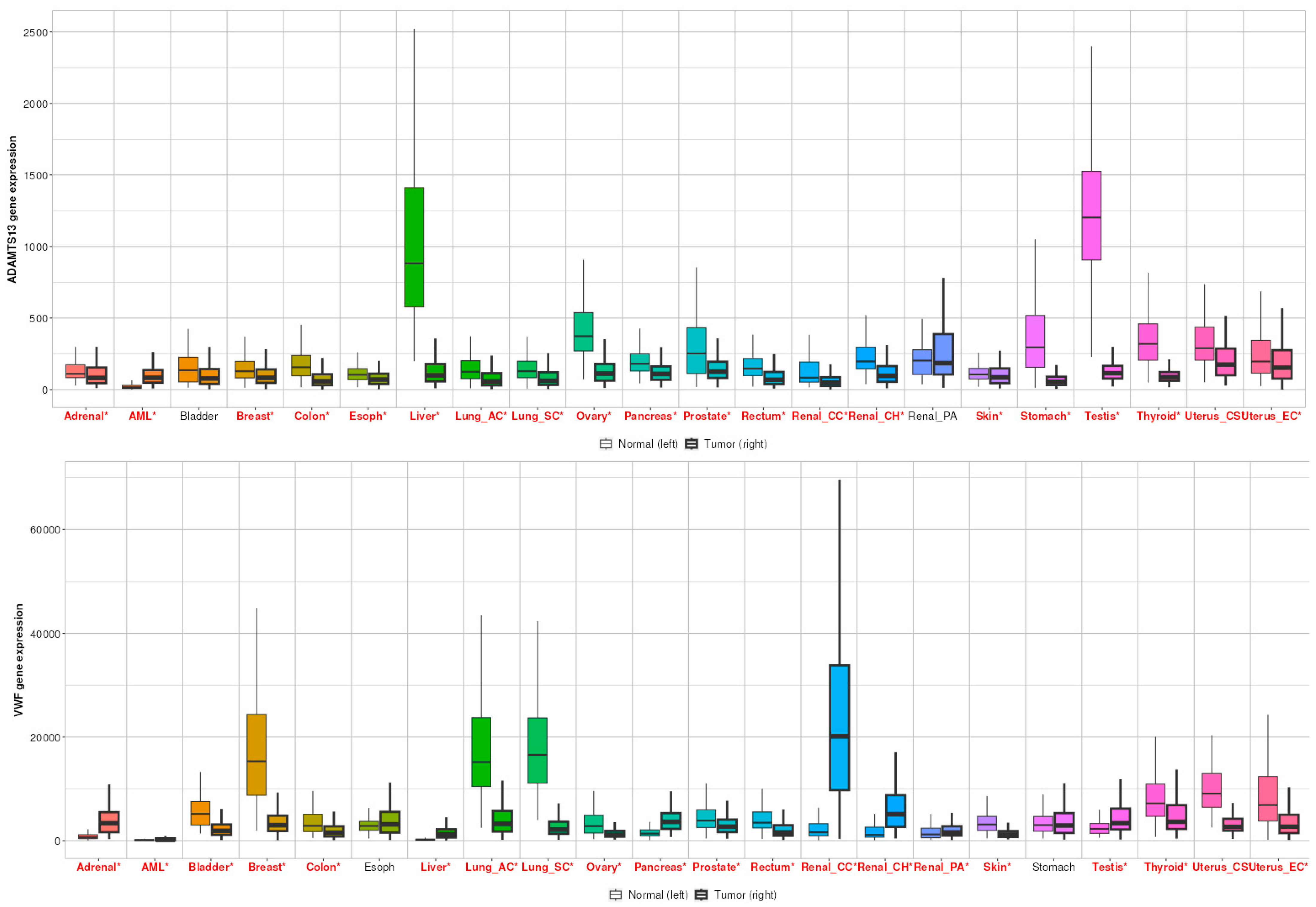
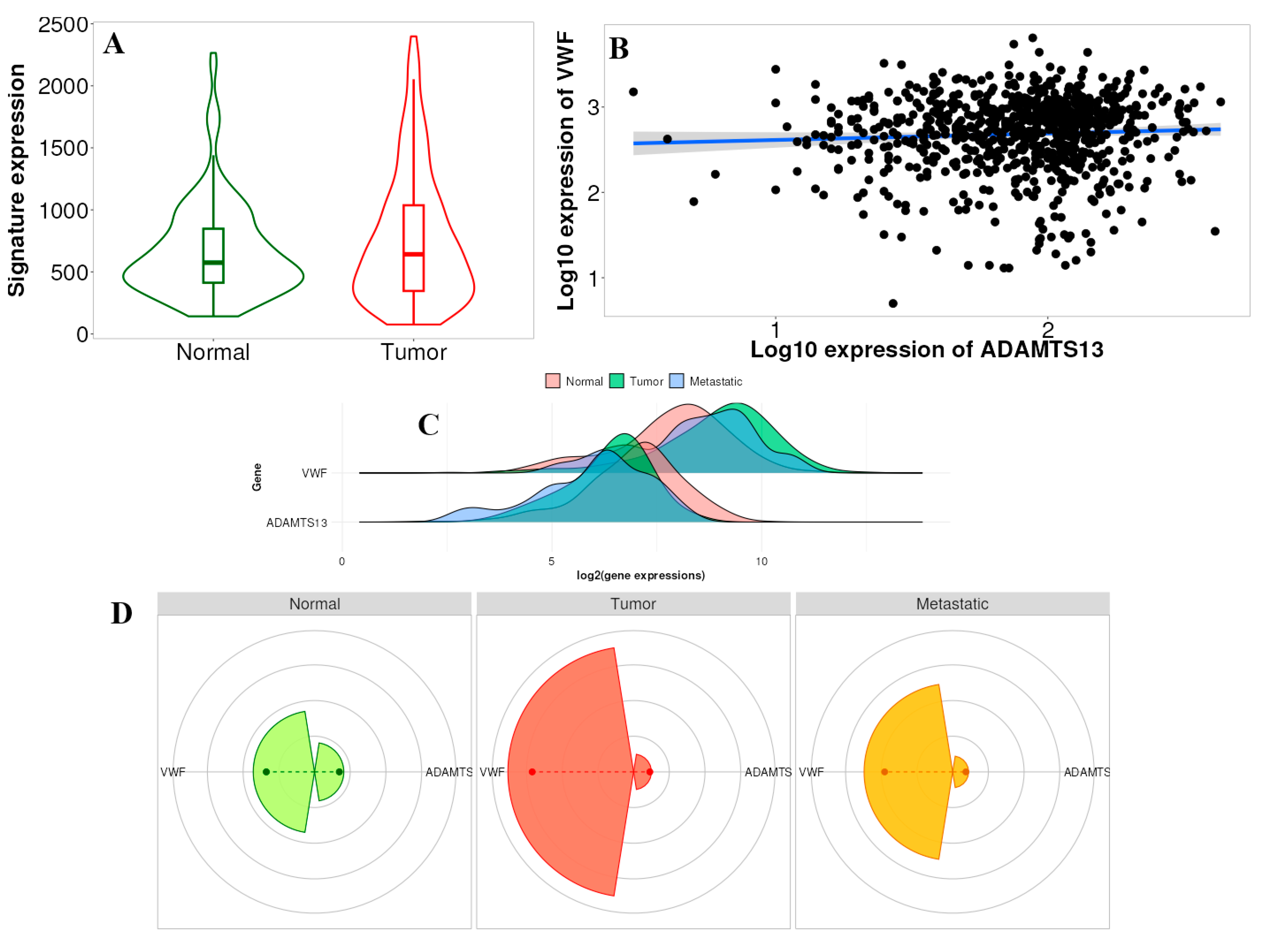
3.1. Expression Profile of VWF and ADAMTS13
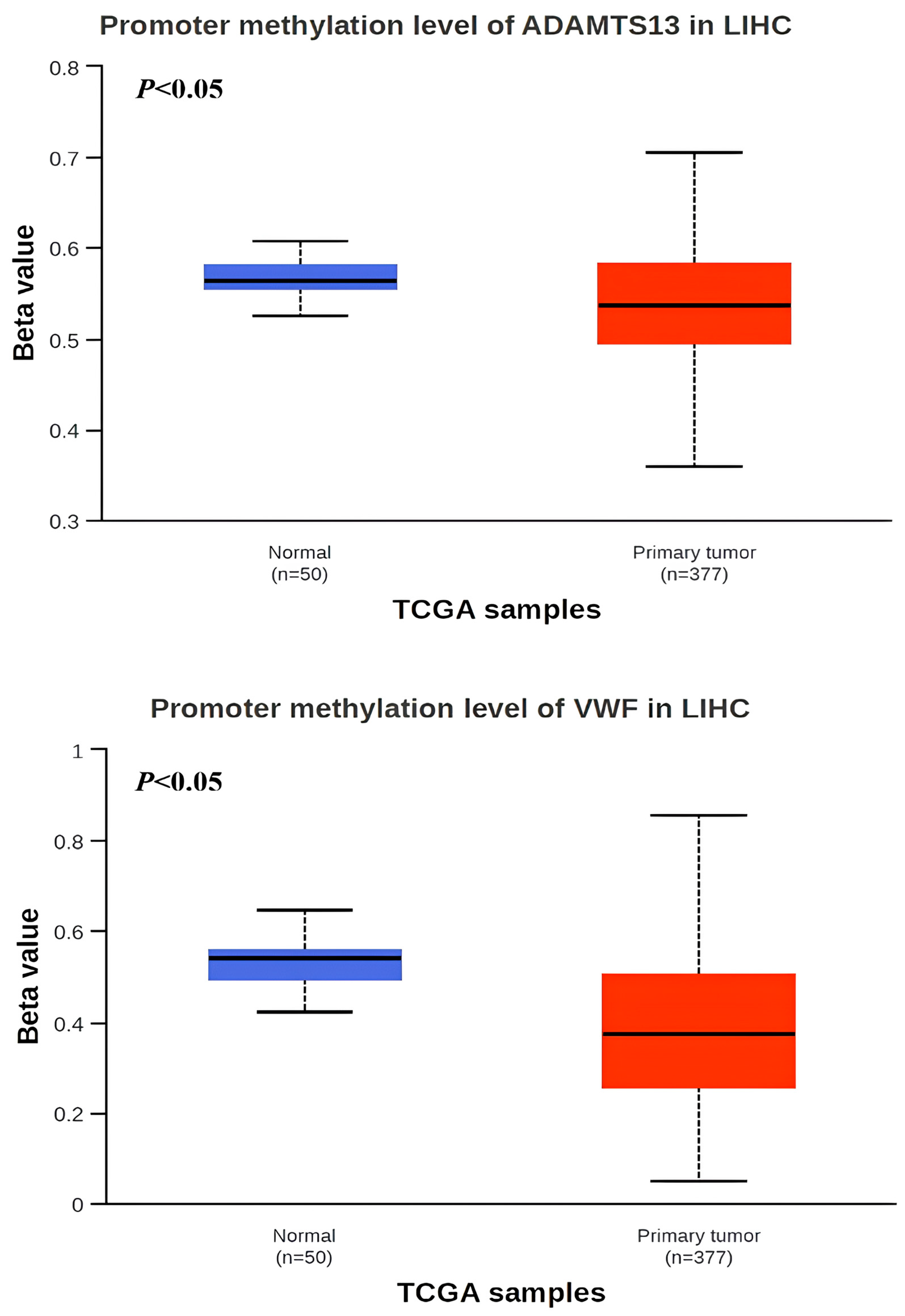
3.2. Promoter Methylation Status of VWF and ADAMTS13
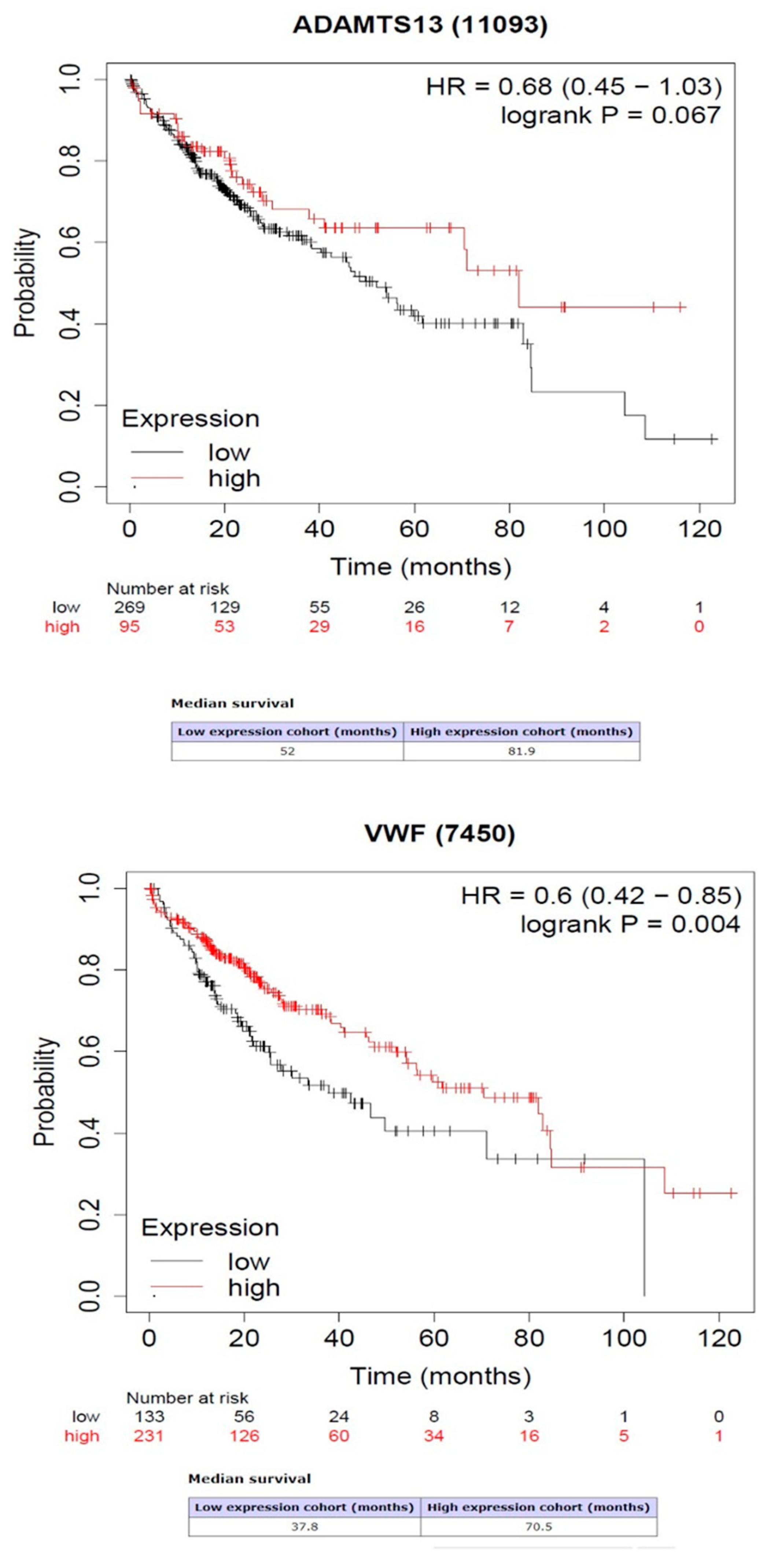
3.3. Survival Analysis Results of VWF and ADAMTS13
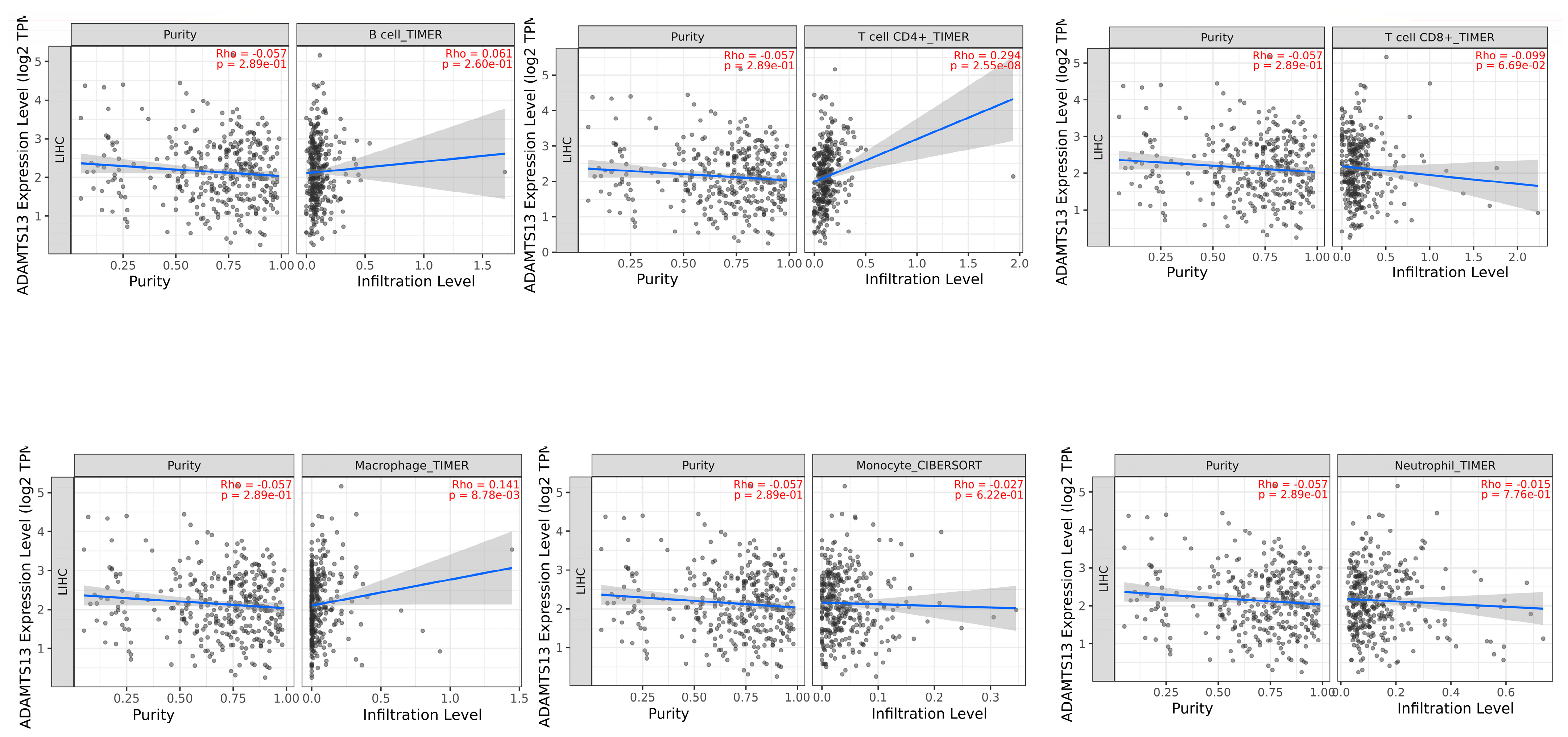
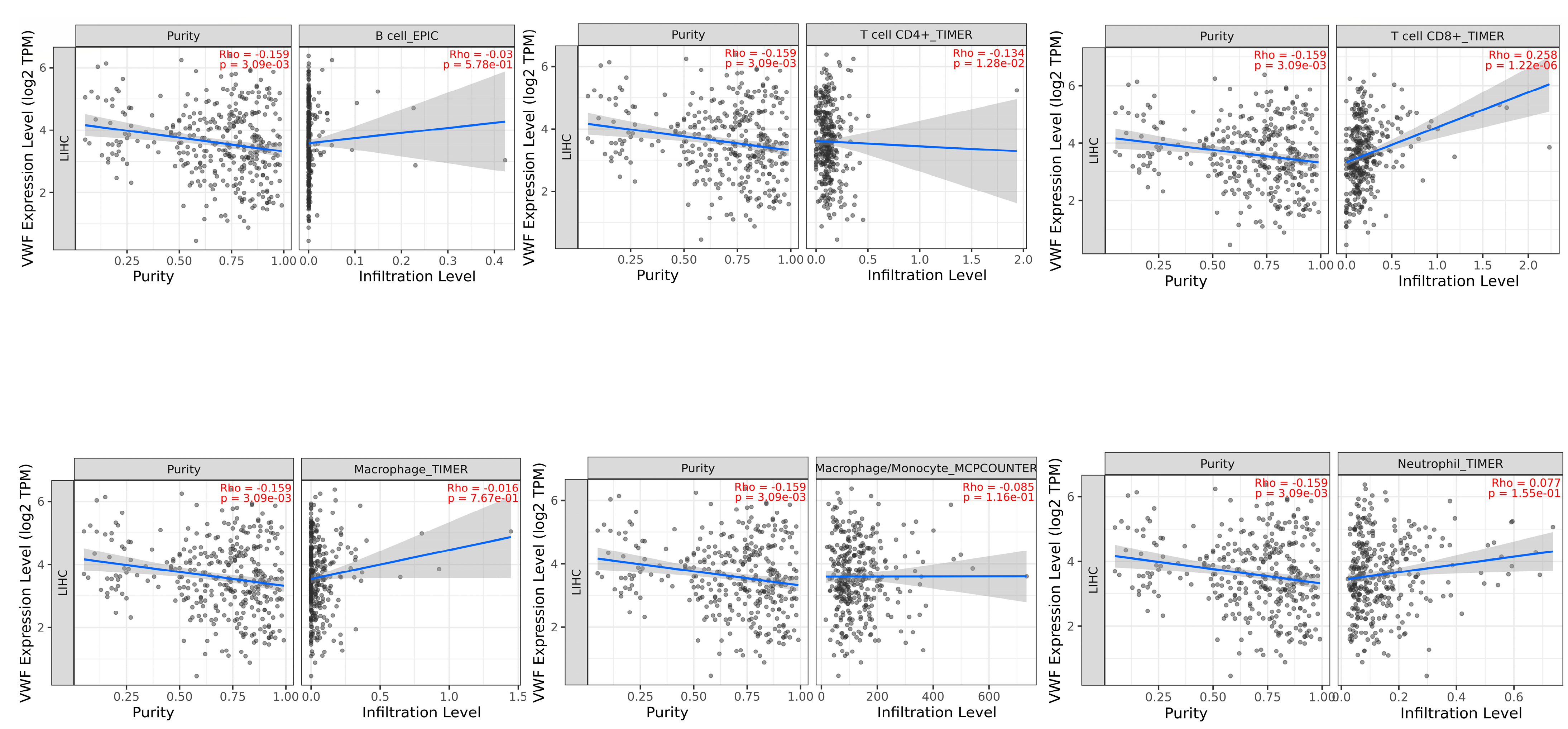
3.4. Immune Infiltrates and Gene Expression of ADAMTS13 and VWF
3.5. TNMplot Analysis of VWF and ADAMTS13
3.6. Gene–Gene Interaction Combined Score Results
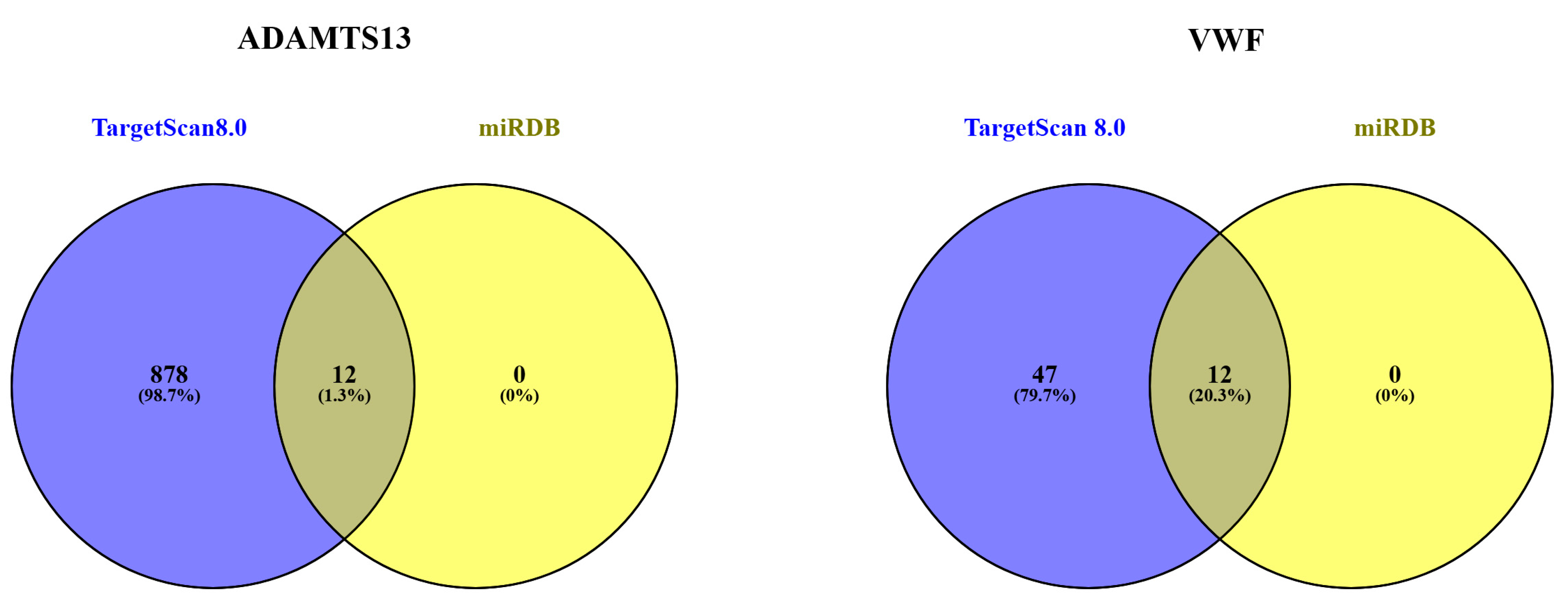
3.7. MicroRNA Target Analysis Result
3.8. Enrichment Analysis of ADAMTS13 and VWF
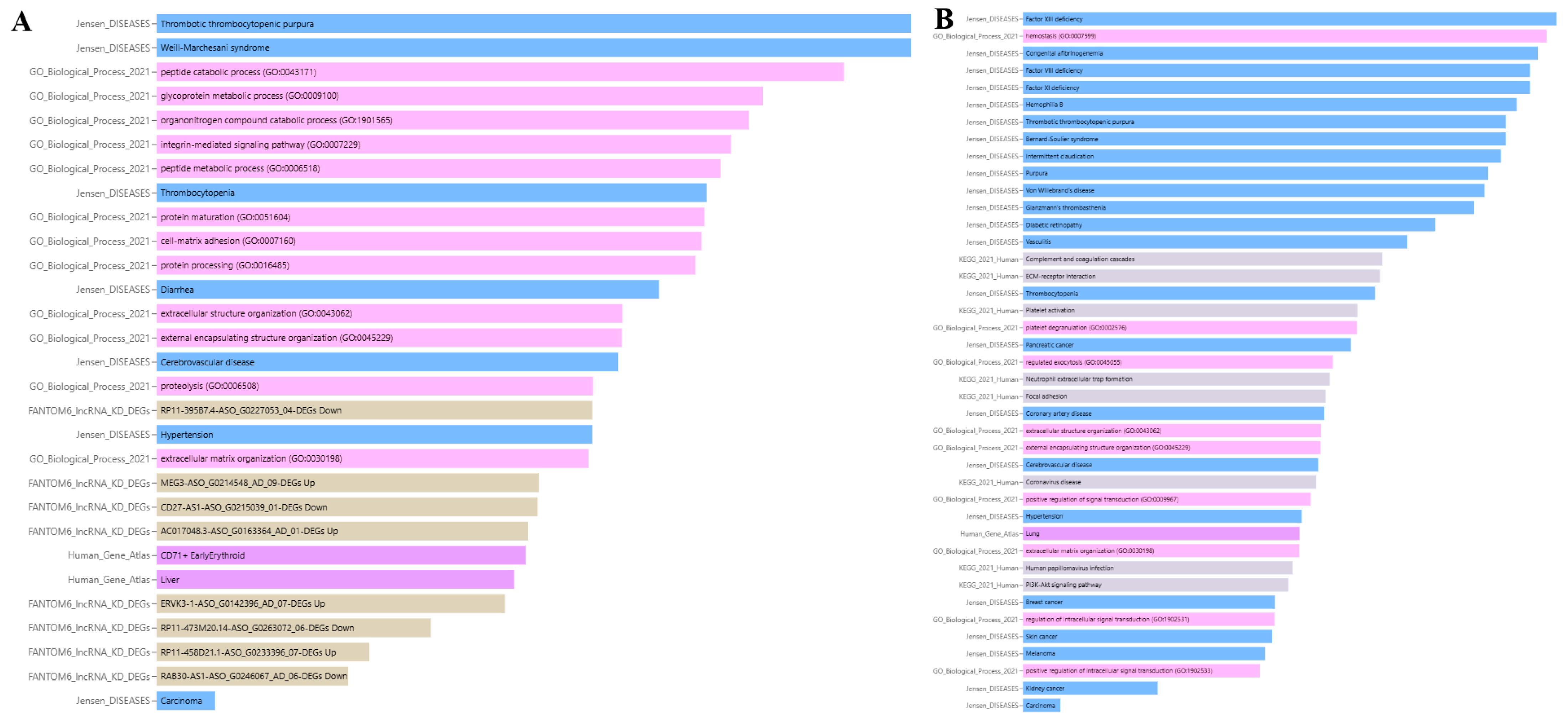
3.8.1. Association of ADAMTS13 with Physiopathological Processes
3.8.2. Association of VWF with Physiopathological Processes
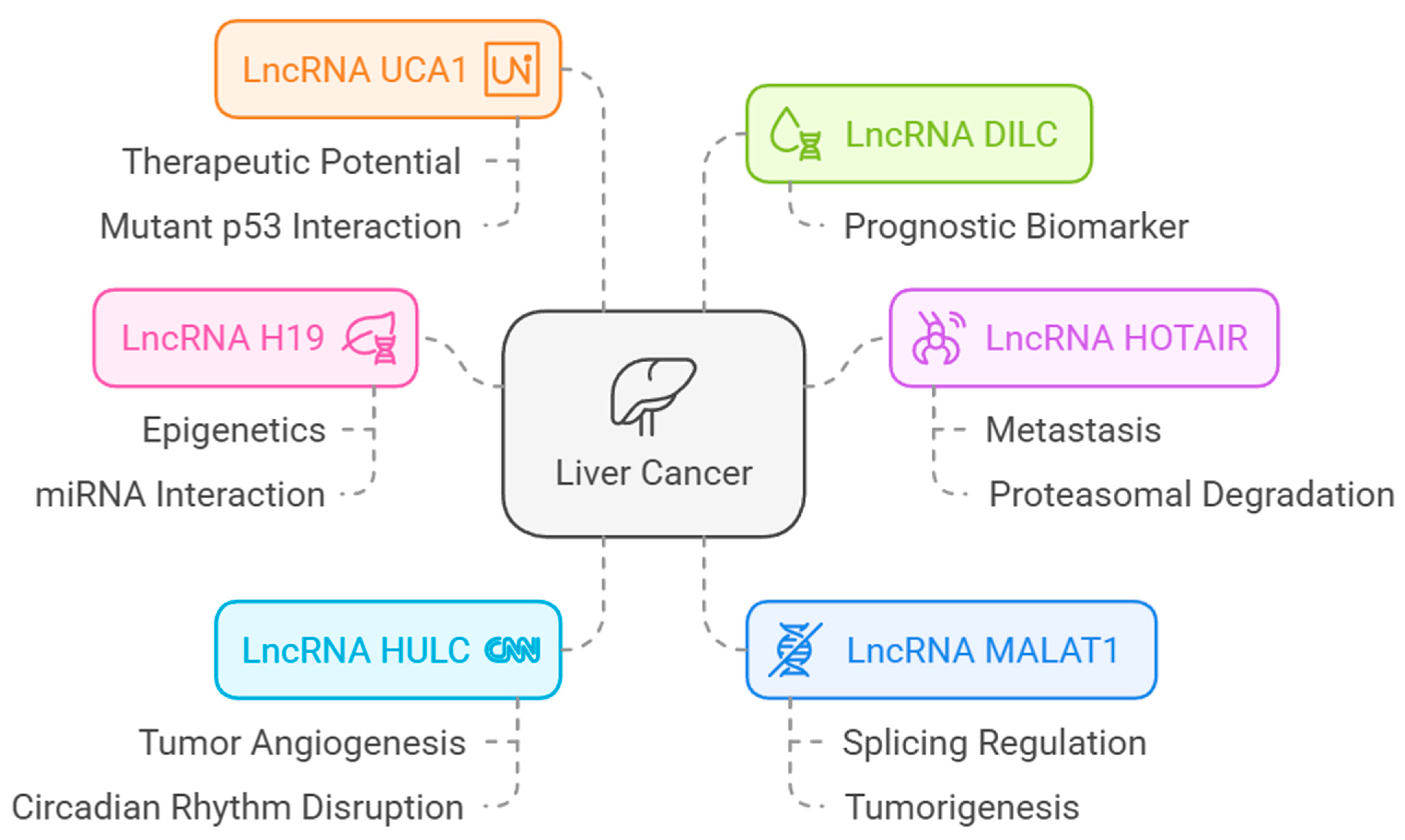
3.8.3. LncRNAs Associated with HCC and Their Association with VWF and ADAMST13
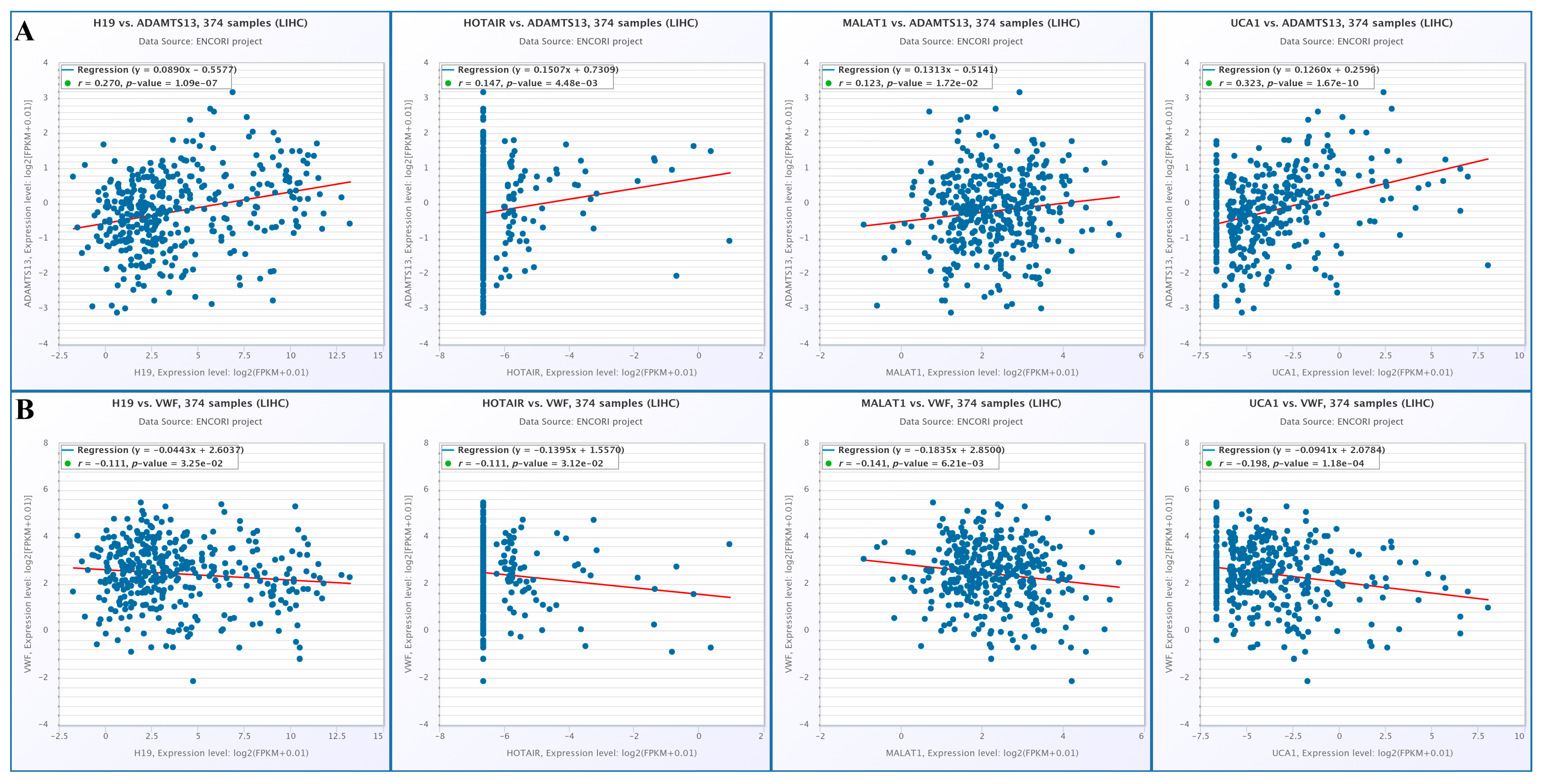
3.8.4. Correlation of lncRNAs with ADAMTS13 and VWF
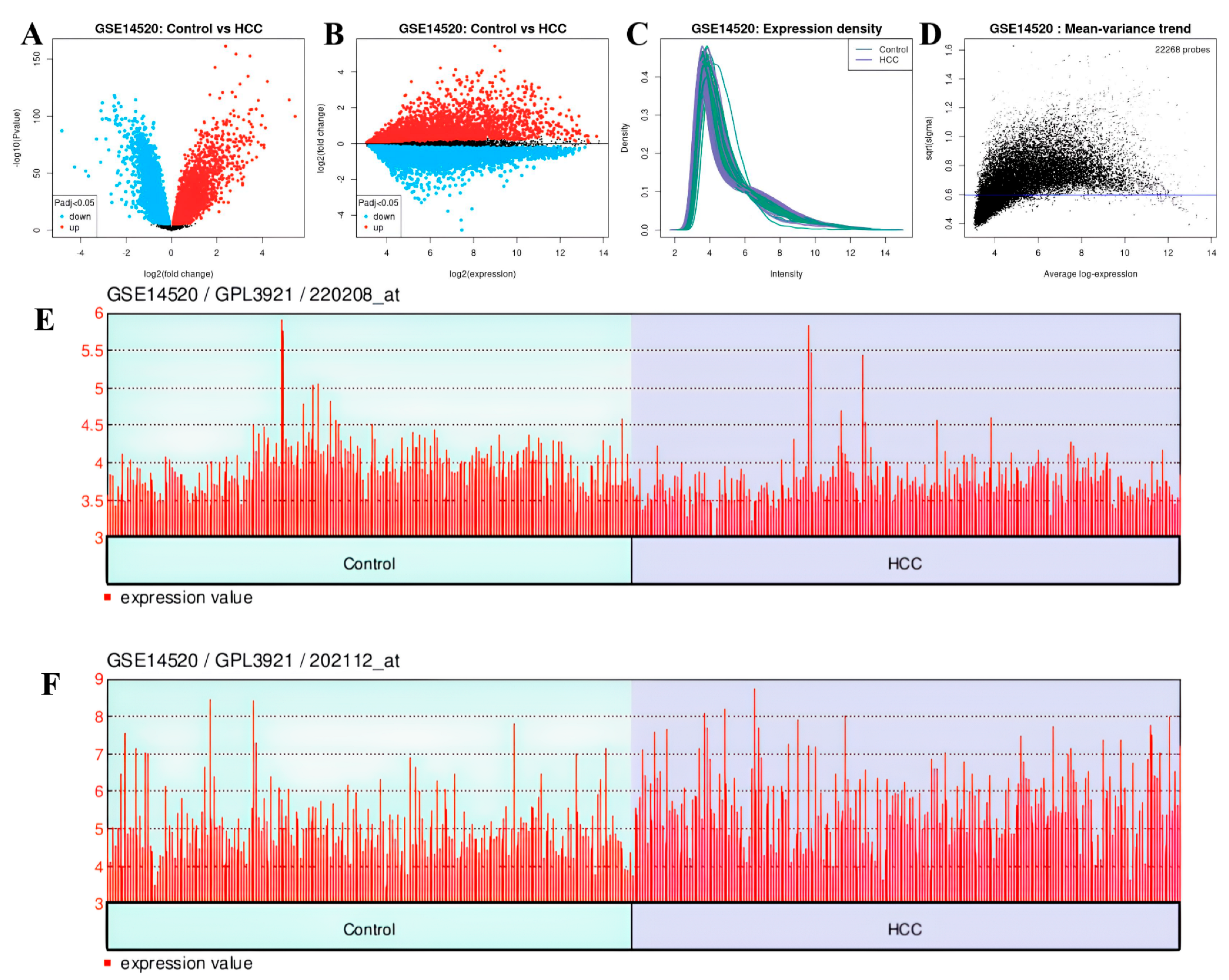
3.9. Analysis Results of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ganesan, P.; Kulik, L.M. Hepatocellular Carcinoma: New Developments. Clin. Liver Dis. 2023, 27, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y. Changing etiology and epidemiology of hepatocellular carcinoma: Asia and worldwide. J. Liver Cancer 2024, 24, 62–70. [Google Scholar] [CrossRef]
- Pan, F.; Lin, X.R.; Hao, L.P.; Chu, X.Y.; Wan, H.J.; Wang, R. The Role of RNA Methyltransferase METTL3 in Hepatocellular Carcinoma: Results and Perspectives. Front. Cell Dev. Biol. 2021, 9, 674919. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Portela, A.; Sayols, S.; Battiston, C.; Hoshida, Y.; Méndez-González, J.; Imbeaud, S.; Letouzé, E.; Hernandez-Gea, V.; Cornella, H.; et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatol. Baltim. Md. 2015, 61, 1945–1956. [Google Scholar] [CrossRef]
- Nagaraju, G.P.; Dariya, B.; Kasa, P.; Peela, S.; El-Rayes, B.F. Epigenetics in hepatocellular carcinoma. Semin. Cancer Biol. 2022, 86 Pt 3, 622–632. [Google Scholar] [CrossRef]
- Wahid, B.; Ali, A.; Rafique, S.; Idrees, M. New Insights into the Epigenetics of Hepatocellular Carcinoma. BioMed Res. Int. 2017, 2017, 1609575. [Google Scholar] [CrossRef]
- Takaya, H.; Namisaki, T.; Moriya, K.; Shimozato, N.; Kaji, K.; Ogawa, H.; Ishida, K.; Tsuji, Y.; Kaya, D.; Takagi, H.; et al. Association between ADAMTS13 activity–VWF antigen imbalance and the therapeutic effect of HAIC in patients with hepatocellular carcinoma. World J. Gastroenterol. 2020, 26, 7232–7241. [Google Scholar] [CrossRef]
- Takaya, H.; Namisaki, T.; Kitade, M.; Kaji, K.; Nakanishi, K.; Tsuji, Y.; Shimozato, N.; Moriya, K.; Seki, K.; Sawada, Y.; et al. VWF/ADAMTS13 ratio as a potential biomarker for early detection of hepatocellular carcinoma. BMC Gastroenterol. 2019, 19, 167. [Google Scholar] [CrossRef]
- Takaya, H.; Namisaki, T.; Shimozato, N.; Kaji, K.; Kitade, M.; Moriya, K.; Sato, S.; Kawaratani, H.; Akahane, T.; Matsumoto, M.; et al. ADAMTS13 and von Willebrand factor are useful biomarkers for sorafenib treatment efficiency in patients with hepatocellular carcinoma. World J. Gastrointest. Oncol. 2019, 11, 424–435. [Google Scholar] [CrossRef]
- Sonneveld, M.A.H.; de Maat, M.P.M.; Leebeek, F.W.G. Von Willebrand factor and ADAMTS13 in arterial thrombosis: A systematic review and meta-analysis. Blood Rev. 2014, 28, 167–178. [Google Scholar] [PubMed]
- Colonne, C.K.; Favaloro, E.J.; Pasalic, L. The Intriguing Connections between von Willebrand Factor, ADAMTS13 and Cancer. Healthcare 2022, 10, 557. [Google Scholar] [CrossRef] [PubMed]
- Randi, A.M. Angiogenesis and the ADAMTS13-VWF balance. Blood 2017, 130, 1–2. [Google Scholar] [PubMed]
- Théret, N.; Bouezzeddine, F.; Azar, F.; Diab-Assaf, M.; Legagneux, V. ADAM and ADAMTS Proteins, New Players in the Regulation of Hepatocellular Carcinoma Microenvironment. Cancers 2021, 13, 1563. [Google Scholar] [CrossRef]
- Obermeier, H.L.; Riedl, J.; Ay, C.; Koder, S.; Quehenberger, P.; Bartsch, R.; Kaider, A.; Zielinski, C.C.; Pabinger, I. The role of ADAMTS-13 and von Willebrand factor in cancer patients: Results from the Vienna Cancer and Thrombosis Study. Res. Pract. Thromb. Haemost. 2019, 3, 503–514. [Google Scholar]
- Uemura, M.; Fujimura, Y.; Ko, S.; Matsumoto, M.; Nakajima, Y.; Fukui, H. Determination of ADAMTS13 and Its Clinical Significance for ADAMTS13 Supplementation Therapy to Improve the Survival of Patients with Decompensated Liver Cirrhosis. Int. J. Hepatol. 2011, 2011, 759047. [Google Scholar]
- Niu, Z.S.; Niu, X.J.; Wang, W.H. Long non-coding RNAs in hepatocellular carcinoma: Potential roles and clinical implications. World J. Gastroenterol. 2017, 23, 5860–5874. [Google Scholar] [CrossRef]
- Morishita, A.; Oura, K.; Tadokoro, T.; Fujita, K.; Tani, J.; Masaki, T. MicroRNAs in the Pathogenesis of Hepatocellular Carcinoma: A Review. Cancers 2021, 13, 514. [Google Scholar] [CrossRef]
- Koustas, E.; Trifylli, E.M.; Sarantis, P.; Papadopoulos, N.; Papanikolopoulos, K.; Aloizos, G.; Damaskos, C.; Garmpis, N.; Garmpi, A.; Matthaios, D.; et al. An Insight into the Arising Role of MicroRNAs in Hepatocellular Carcinoma: Future Diagnostic and Therapeutic Approaches. Int. J. Mol. Sci. 2023, 24, 7168. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Qiu, L.; Huang, Y.; Li, Z.; Dong, X.; Chen, G.; Xu, H.; Zeng, Y.; Cai, Z.; Liu, X.; Liu, J. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol. Oncol. 2019, 13, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Zhang, X.L.; Jiang, X.; Li, X.; Ding, J.; Zuo, L.J.; Duan, S.S.; Chen, R.; Sun, B.B.; Hu, X.Y.; et al. Long Non-Coding RNA and mRNA Expression Analysis in Liver of Mice With Clonorchis sinensis Infection. Front. Cell. Infect. Microbiol. 2022, 11, 754224. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Fu, Y.; Tan, T.; Hu, J.; Li, F.; Liao, Z.; Peng, J. Screening of lncRNA-miRNA-mRNA Coexpression Regulatory Networks Involved in Acute Traumatic Coagulation Dysfunction Based on CTD, GeneCards, and PharmGKB Databases. Oxid. Med. Cell Longev. 2022, 2022, 7280312. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Nagy, Á.; Lánczky, A.; Menyhárt, O.; Győrffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, J.E.; Xie, Z.; Marino, G.B.; Nguyen, N.; Clarke, D.J.B.; Ma’ayan, A. Enrichr-KG: Bridging enrichment analysis across multiple libraries. Nucleic Acids Res. 2023, 51, W168–W179. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013, 41, D983–D986. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Roessler, S.; Jia, H.L.; Budhu, A.; Forgues, M.; Ye, Q.-H.; Lee, J.-S.; Thorgeirsson, S.S.; Sun, Z.; Tang, Z.-Y.; Qin, L.-X.; et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010, 70, 10202–10212. [Google Scholar] [CrossRef]
- Roessler, S.; Long, E.L.; Budhu, A.; Chen, Y.; Zhao, X.; Ji, J.; Walker, R.; Jia, H.; Ye, Q.; Qin, L.; et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 2012, 142, 957–966.e12. [Google Scholar] [CrossRef]
- Zhao, X.; Parpart, S.; Takai, A.; Roessler, S.; Budhu, A.; Yu, Z.; Blank, M.; Zhang, Y.E.; Jia, H.-L.; Ye, Q.-H.; et al. Integrative genomics identifies YY1AP1 as an oncogenic driver in EpCAM(+) AFP(+) hepatocellular carcinoma. Oncogene 2015, 34, 5095–5104. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, B.; Tan, P.Y.; Handoko, Y.A.; Sekar, K.; Deivasigamani, A.; Seshachalam, V.P.; Ouyang, H.; Shi, M.; Xie, C.; et al. Genome-wide CRISPR knockout screens identify NCAPG as an essential oncogene for hepatocellular carcinoma tumor growth. FASEB J. 2019, 33, 8759–8770. [Google Scholar] [CrossRef]
- Sun, Y.; Ji, F.; Kumar, M.R.; Zheng, X.; Xiao, Y.; Liu, N.; Shi, J.; Wong, L.; Forgues, M.; Qin, L.-X.; et al. Transcriptome integration analysis in hepatocellular carcinoma reveals discordant intronic miRNA-host gene pairs in expression. Int. J. Biol. Sci. 2017, 13, 1438–1449. [Google Scholar] [PubMed]
- Lu, Y.; Xu, W.; Ji, J.; Feng, D.; Sourbier, C.; Yang, Y.; Qu, J.; Zeng, Z.; Wang, C.; Chang, X.; et al. Alternative splicing of the cell fate determinant Numb in hepatocellular carcinoma. Hepatology 2015, 62, 1122–1131. [Google Scholar]
- Chen, S.; Fang, H.; Li, J.; Shi, J.; Zhang, J.; Wen, P.; Wang, Z.; Yang, H.; Cao, S.; Zhang, H.; et al. Microarray Analysis For Expression Profiles of lncRNAs and circRNAs in Rat Liver after Brain-Dead Donor Liver Transplantation. BioMed Res. Int. 2019, 2019, 5604843. [Google Scholar] [CrossRef]
- Chen, S.L.; Zhu, Z.X.; Yang, X.; Liu, L.L.; He, Y.-F.; Yang, M.-M.; Guan, X.-Y.; Wang, X.; Yun, J.-P. Cleavage and Polyadenylation Specific Factor 1 Promotes Tumor Progression via Alternative Polyadenylation and Splicing in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2021, 9, 616835. [Google Scholar]
- Wang, C.; Liao, Y.; He, W.; Zhang, H.; Zuo, D.; Liu, W.; Yang, Z.; Qiu, J.; Yuan, Y.; Li, K.; et al. Elafin promotes tumour metastasis and attenuates the anti-metastatic effects of erlotinib via binding to EGFR in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 113. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Kwon, S.M.; Li, D.; Li, L.; Peng, X.; Zhang, J.; Sueyoshi, T.; Raufman, J.-P.; Negishi, M.; Wang, X.W.; et al. Human constitutive androstane receptor represses liver cancer development and hepatoma cell proliferation by inhibiting erythropoietin signaling. J. Biol. Chem. 2022, 298, 101885. [Google Scholar]
- Zhao, N.; Dang, H.; Ma, L.; Martin, S.P.; Forgues, M.; Ylaya, K.; Hewitt, S.M.; Wang, X.W. Intratumoral γδ T-Cell Infiltrates, Chemokine (C-C Motif) Ligand 4/Chemokine (C-C Motif) Ligand 5 Protein Expression and Survival in Patients With Hepatocellular Carcinoma. Hepatology 2021, 73, 1045–1060. [Google Scholar] [CrossRef]
- Wu, B.; Liu, D.A.; Guan, L.; Myint, P.K.; Chin, L.; Dang, H.; Xu, Y.; Ren, J.; Li, T.; Yu, Z.; et al. Stiff matrix induces exosome secretion to promote tumour growth. Nat. Cell Biol. 2023, 25, 415–424. [Google Scholar]
- Long, Y.; Wang, W.; Liu, S.; Wang, X.; Tao, Y. The survival prediction analysis and preliminary study of the biological function of YEATS2 in hepatocellular carcinoma. Cell. Oncol. 2024, 47, 2297–2316. [Google Scholar]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar]
- Avtaeva, Y.N.; Melnikov, I.S.; Vasiliev, S.A.; Gabbasov, Z.A. The role of von Willebrand factor in hemostasis pathology. Атерoтрoмбoз 2023, 12, 79–102. [Google Scholar] [CrossRef]
- Tao, Q.; Qi, Y.; Gu, J.; Yu, D.; Lu, Y.; Liu, J.; Liang, X. Breast cancer cells-derived Von Willebrand Factor promotes VEGF-A-related angiogenesis through PI3K/Akt-miR-205-5p signaling pathway. Toxicol. Appl. Pharmacol. 2022, 440, 115927. [Google Scholar] [CrossRef]
- Nishida, N.; Kudo, M.; Nishimura, T.; Arizumi, T.; Takita, M.; Kitai, S.; Yada, N.; Hagiwara, S.; Inoue, T.; Minami, Y.; et al. Unique association between global DNA hypomethylation and chromosomal alterations in human hepatocellular carcinoma. PLoS ONE 2013, 8, e72312. [Google Scholar] [CrossRef]
- Matsui, D.; Nagai, H.; Mukozu, T.; Ogino, Y.U.; Sumino, Y. VEGF in patients with advanced hepatocellular carcinoma receiving intra-arterial chemotherapy. Anticancer. Res. 2015, 35, 2205–2210. [Google Scholar]
- Li, Z.; Zhang, Z.; Fang, L.; Zhao, J.; Niu, Z.; Chen, H.; Cao, G. Tumor Microenvironment Composition and Related Therapy in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2023, 10, 2083–2099. [Google Scholar] [CrossRef]
- Bian, J.; Lin, J.; Long, J.; Yang, X.; Yang, X.; Lu, X.; Sang, X.; Zhao, H. T lymphocytes in hepatocellular carcinoma immune microenvironment: Insights into human immunology and immunotherapy. Am. J. Cancer Res. 2020, 10, 4585–4606. [Google Scholar] [PubMed]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Zheng, X.; Jin, W.; Wang, S.; Ding, H. Progression on the Roles and Mechanisms of Tumor-Infiltrating T Lymphocytes in Patients With Hepatocellular Carcinoma. Front. Immunol. 2021, 12, 729705. [Google Scholar] [CrossRef]
- Fu, J.; Xu, D.; Liu, Z.; Shi, M.; Zhao, P.; Fu, B.; Zhang, Z.; Yang, H.; Zhang, H.; Zhou, C.; et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 2007, 132, 2328–2339. [Google Scholar] [CrossRef]
- Fisicaro, P.; Barili, V.; Rossi, M.; Montali, I.; Vecchi, A.; Acerbi, G.; Laccabue, D.; Zecca, A.; Penna, A.; Missale, G.; et al. Pathogenetic Mechanisms of T Cell Dysfunction in Chronic HBV Infection and Related Therapeutic Approaches. Front. Immunol. 2020, 11, 849. [Google Scholar] [CrossRef] [PubMed]
- Ramzan, M.; Sturm, N.; Decaens, T.; Bioulac-Sage, P.; Bancel, B.; Merle, P.; Tran Van Nhieu, J.; Slama, R.; Letoublon, C.; Zarski, J.P.; et al. Liver-infiltrating CD8(+) lymphocytes as prognostic factor for tumour recurrence in hepatitis C virus-related hepatocellular carcinoma. Liver Int. Off. J. Int. Assoc. Study Liver 2016, 36, 434–444. [Google Scholar]
- Fu, J.; Zhang, Z.; Zhou, L.; Qi, Z.; Xing, S.; Lv, J.; Shi, J.; Fu, B.; Liu, Z.; Zhang, J.-Y.; et al. Impairment of CD4+ cytotoxic T cells predicts poor survival and high recurrence rates in patients with hepatocellular carcinoma. Hepatol. Baltim. Md. 2013, 58, 139–149. [Google Scholar]
- Pal, I.; Mandal, M. PI3K and Akt as molecular targets for cancer therapy: Current clinical outcomes. Acta Pharmacol. Sin. 2012, 33, 1441–1458. [Google Scholar] [CrossRef]
- Mallela, V.R.; Rajtmajerová, M.; Trailin, A.; Liška, V.; Hemminki, K.; Ambrozkiewicz, F. miRNA and lncRNA as potential tissue biomarkers in hepatocellular carcinoma. Non-Coding RNA Res. 2024, 9, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Abbastabar, M.; Sarfi, M.; Golestani, A.; Khalili, E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018, 17, 900–913. [Google Scholar]
- Hou, Z.; Xu, X.; Zhou, L.; Fu, X.; Tao, S.; Zhou, J.; Tan, D.; Liu, S. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. 2017, 39, 1010428317718135. [Google Scholar]
- Peng, N.; He, J.; Li, J.; Huang, H.; Huang, W.; Liao, Y.; Zhu, S. Long noncoding RNA MALAT1 inhibits the apoptosis and autophagy of hepatocellular carcinoma cell by targeting the microRNA-146a/PI3K/Akt/mTOR axis. Cancer Cell Int. 2020, 20, 165. [Google Scholar]
- Cantile, M.; Belli, V.; Scognamiglio, G.; Martorana, A.; De Pietro, G.; Tracey, M.; Budillon, A. The role of HOTAIR in the modulation of resistance to anticancer therapy. Front. Mol. Biosci. 2024, 11, 1414651. [Google Scholar]
- Wang, F.; Ying, H.Q.; He, B.S.; Pan, Y.Q.; Deng, Q.W.; Sun, H.L.; Chen, J.; Liu, X.; Wang, S.K. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget 2015, 6, 7899–7917. [Google Scholar] [CrossRef] [PubMed]
- Tietze, L.; Kessler, S.M. The Good, the Bad, the Question-H19 in Hepatocellular Carcinoma. Cancers 2020, 12, 126. [Google Scholar] [CrossRef]
- Wu, B.; Zhang, Y.; Yu, Y.; Zhong, C.; Lang, Q.; Liang, Z.; Lv, C.; Xu, F.; Tian, Y. Long Noncoding RNA H19: A Novel Therapeutic Target Emerging in Oncology Via Regulating Oncogenic Signaling Pathways. Front. Cell Dev. Biol. 2021, 9, 796740. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, Y.; Li, Z.; Li, H.; Li, X.; Yan, L.; Mao, J.; Shen, J.; Chen, W.; Xue, F. Long non-coding RNA H19 is involved in sorafenib resistance in hepatocellular carcinoma by upregulating miR-675. Oncol. Rep. 2020, 44, 165–173. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef] [PubMed]
- Belghiti, J.; Kianmanesh, R. Surgical treatment of hepatocellular carcinoma. HPB 2005, 7, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J.S. Current perspectives on radiotherapy in hepatocellular carcinoma management: A comprehensive review. J. Liver Cancer 2024, 24, 33–46. [Google Scholar] [CrossRef]
- Periferakis, A.; Tsigas, G.; Periferakis, A.T.; Badarau, I.A.; Scheau, A.E.; Tampa, M.; Georgescu, S.R.; Didilescu, A.C.; Scheau, C.; Caruntu, C. Antitumoral and Anti-inflammatory Roles of Somatostatin and Its Analogs in Hepatocellular Carcinoma. Anal. Cell Pathol. 2021, 2021, 1840069. [Google Scholar] [CrossRef]
- Mandlik, D.S.; Mandlik, S.K.; Choudhary, H.B. Immunotherapy for hepatocellular carcinoma: Current status and future perspectives. World J. Gastroenterol. 2023, 29, 1054–1075. [Google Scholar] [CrossRef]
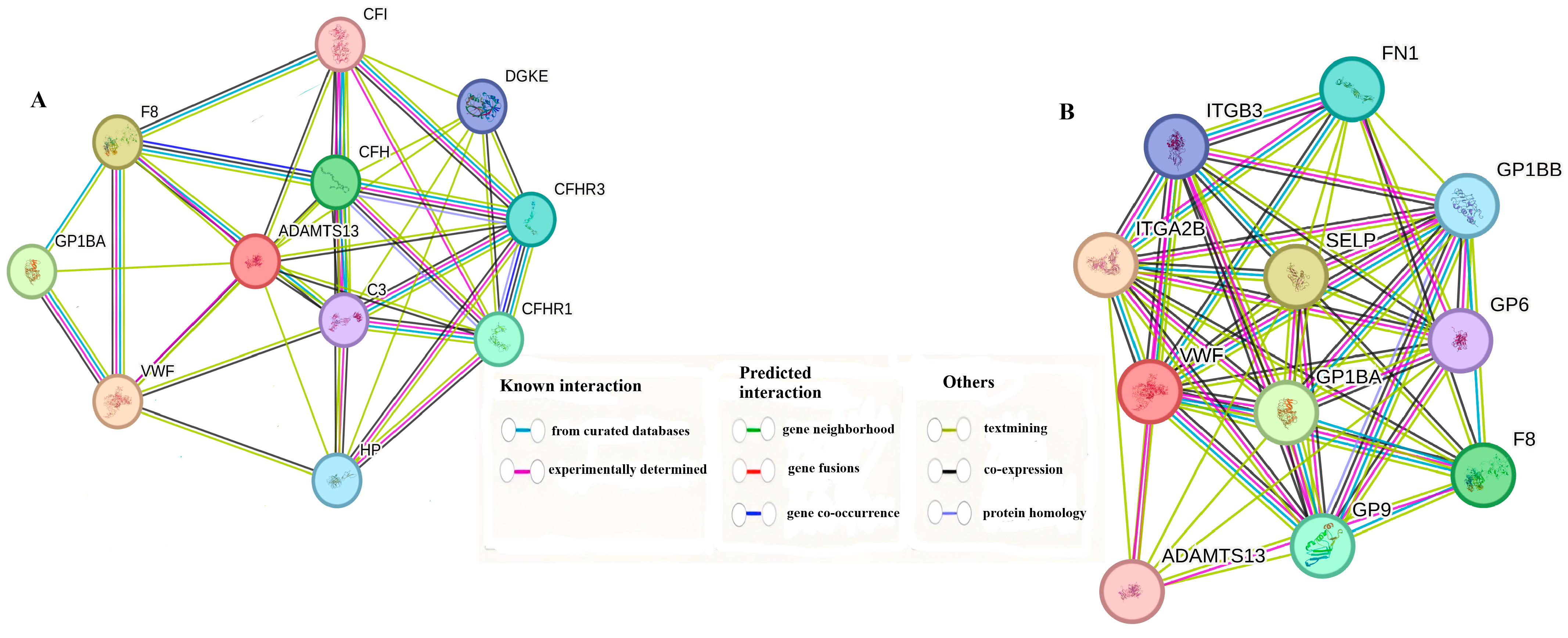
| Gene-1 | Gene-2 | Protein Annotation | Combined Score |
|---|---|---|---|
| VWF | ADAMTS13 | Von Willebrand antigen 2 | 0.997 |
| F8 | ADAMTS13 | Factor VIIIa heavy chain | 0.950 |
| GP1BA | ADAMTS13 | Platelet glycoprotein Ib alpha chain | 0.931 |
| CFH | ADAMTS13 | Complement factor H | 0.853 |
| CFHR1 | ADAMTS13 | Complement factor H-related protein 1 | 0.833 |
| CFHR3 | ADAMTS13 | Complement factor H-related protein 3 | 0.824 |
| HP | ADAMTS13 | Haptoglobin alpha chain | 0.773 |
| DGKE | ADAMTS13 | Diacylglycerol kinase epsilon | 0.767 |
| C3 | ADAMTS13 | Complement C3c alpha’ chain fragment 1 | 0.735 |
| CFI | ADAMTS13 | Complement factor I heavy chain | 0.721 |
| ITGA2B | VWF | Integrin alpha-IIb light chain | 0.999 |
| SELP | VWF | Selectin P | 0.999 |
| GP1BA | VWF | Platelet glycoprotein Ib alpha chain | 0.999 |
| F8 | VWF | Factor VIIIa heavy chain | 0.999 |
| GP9 | VWF | Platelet glycoprotein IX | 0.998 |
| FN1 | VWF | Fibronectin | 0.998 |
| GP1BB | VWF | Platelet glycoprotein Ib beta chain | 0.998 |
| ITGB3 | VWF | Integrin beta-3; Integrin alpha-V/beta-3 | 0.998 |
| GP6 | VWF | Platelet glycoprotein VI | 0.997 |
| ADAMTS13 | VWF | A disintegrin and metalloproteinase with thrombospondin motifs 13 | 0.997 |
| Predicted miRNAs for ADAMTS13 | |
|---|---|
| miRDB databases TargetScanHuman8.0 | hsa-miR-7850-5p, hsa-miR-4520-2-3p, hsa-miR-4516, hsa-miR-4434, hsa-miR-1263, hsa-miR-4462, hsa-miR-5703, hsa-miR-596, hsa-miR-3978, hsa-miR-6848-5p, hsa-miR-6846-5p, hsa-miR-3148 |
| Predicted miRNAs for VWF | |
| miRDB databases TargetScanHuman8.0 | hsa-miR-4296, hsa-miR-4322, hsa-miR-4265, hsa-miR-6759-5p, hsa-miR-6796-5p, hsa-miR-1972, hsa-miR-4437, hsa-miR-4468, hsa-miR-2278, hsa-miR-450b-5p, hsa-miR-2467-3p, hsa-miR-3190-5p |
| Term | Library | p-Value | q-Value | z-Score | Combined Score |
|---|---|---|---|---|---|
| thrombotic thrombocytopenic purpura | Jensen_DISEASES | 0.00065 | 0.002275 | 19,987 | 146,700 |
| Weill–Marchesani syndrome | Jensen_DISEASES | 0.00065 | 0.002275 | 19,987 | 146,700 |
| peptide catabolic process (GO:0043171) | GO_Biological_Process_2021 | 0.00125 | 0.00795 | 19,975 | 133,500 |
| glycoprotein metabolic process (GO:0009100) | GO_Biological_Process_2021 | 0.00275 | 0.00795 | 19,945 | 117,600 |
| organonitrogen compound catabolic process (GO:1901565) | GO_Biological_Process_2021 | 0.00315 | 0.00795 | 19,937 | 114,800 |
| integrin-mediated signaling pathway (GO:0007229) | GO_Biological_Process_2021 | 0.00375 | 0.00795 | 19,925 | 111,300 |
| peptide metabolic process (GO:0006518) | GO_Biological_Process_2021 | 0.00415 | 0.00795 | 19,917 | 109,200 |
| thrombocytopenia | Jensen_DISEASES | 0.00475 | 0.01108 | 19,905 | 106,500 |
| protein maturation (GO:0051604) | GO_Biological_Process_2021 | 0.00485 | 0.00795 | 19,903 | 106,100 |
| cell–matrix adhesion (GO:0007160) | GO_Biological_Process_2021 | 0.005 | 0.00795 | 19,900 | 105,400 |
| protein processing (GO:0016485) | GO_Biological_Process_2021 | 0.0053 | 0.00795 | 19,894 | 104,200 |
| diarrhea | Jensen_DISEASES | 0.00755 | 0.01321 | 19,849 | 96,990 |
| extracellular structure organization (GO:0043062) | GO_Biological_Process_2021 | 0.0108 | 0.01302 | 19,784 | 89,590 |
| external encapsulating structure organization (GO:0045229) | GO_Biological_Process_2021 | 0.01085 | 0.01302 | 19,783 | 89,490 |
| cerebrovascular disease | Jensen_DISEASES | 0.01125 | 0.01575 | 19,775 | 88,740 |
| proteolysis (GO:0006508) | GO_Biological_Process_2021 | 0.01435 | 0.015 | 19,713 | 83,660 |
| RP11-395B7.4-ASO_G0227053_04-DEGs down | FANTOM6_lncRNA_KD_DEGs | 0.01445 | 0.0539 | 19,711 | 83,520 |
| hypertension | Jensen_DISEASES | 0.01445 | 0.01686 | 19,711 | 83,520 |
| extracellular matrix organization (GO:0030198) | GO_Biological_Process_2021 | 0.015 | 0.015 | 19,700 | 82,730 |
| MEG3-ASO_G0214548_AD_09-DEGs Up | FANTOM6_lncRNA_KD_DEGs | 0.0243 | 0.0539 | 19,514 | 72,540 |
| CD27-AS1-ASO_G0215039_01-DEGs Down | FANTOM6_lncRNA_KD_DEGs | 0.0246 | 0.0539 | 19,508 | 72,280 |
| AC017048.3-ASO_G0163364_AD_01-DEGs Up | FANTOM6_lncRNA_KD_DEGs | 0.02695 | 0.0539 | 19,461 | 70,330 |
| CD71+ early erythroid | Human_Gene_Atlas | 0.02765 | 0.0309 | 19,447 | 69,780 |
| liver | Human_Gene_Atlas | 0.0309 | 0.0309 | 19,382 | 67,390 |
| ERVK3-1-ASO_G0142396_AD_07-DEGs Up | FANTOM6_lncRNA_KD_DEGs | 0.0338 | 0.05408 | 19,324 | 65,460 |
| RP11-473M20.14-ASO_G0263072_06-DEGs down | FANTOM6_lncRNA_KD_DEGs | 0.0695 | 0.09267 | 18,610 | 49,620 |
| RP11-458D21.1-ASO_G0233396_07-DEGs Up | FANTOM6_lncRNA_KD_DEGs | 0.1262 | 0.1443 | 17,475 | 36,160 |
| RAB30-AS1-ASO_G0246067_AD_06-DEGs down | FANTOM6_lncRNA_KD_DEGs | 0.1552 | 0.1552 | 16,895 | 31,470 |
| carcinoma | Jensen_DISEASES | 0.5659 | 0.5659 | 8682 | 4943 |
| Term | Library | p-Value | q-Value | z-Score | Combined Score |
|---|---|---|---|---|---|
| Factor XIII deficiency | Jensen_DISEASES | 0.0003 | 0.002012 | 19,994 | 162,200 |
| Hemostasis (GO:0007599) | GO_Biological_Process_2021 | 0.00035 | 0.00315 | 19,993 | 159,100 |
| Congenital afibrinogenemia | Jensen_DISEASES | 0.0004 | 0.002012 | 19,992 | 156,400 |
| Factor VIII deficiency | Jensen_DISEASES | 0.00045 | 0.002012 | 19,991 | 154,100 |
| Factor XI deficiency | Jensen_DISEASES | 0.00045 | 0.002012 | 19,991 | 154,100 |
| Hemophilia B | Jensen_DISEASES | 0.00055 | 0.002012 | 19,989 | 150,000 |
| Thrombotic thrombocytopenic purpura | Jensen_DISEASES | 0.00065 | 0.002012 | 19,987 | 146,700 |
| Bernard–Soulier syndrome | Jensen_DISEASES | 0.00065 | 0.002012 | 19,987 | 146,700 |
| Intermittent claudication | Jensen_DISEASES | 0.0007 | 0.002012 | 19,986 | 145,200 |
| Purpura | Jensen_DISEASES | 0.00085 | 0.00207 | 19,983 | 141,300 |
| Von Willebrand’s disease | Jensen_DISEASES | 0.0009 | 0.00207 | 19,982 | 140,100 |
| Glanzmann’s thrombasthenia | Jensen_DISEASES | 0.00105 | 0.002195 | 19,979 | 137,000 |
| Diabetic retinopathy | Jensen_DISEASES | 0.0019 | 0.003642 | 19,962 | 125,100 |
| Vasculitis | Jensen_DISEASES | 0.0029 | 0.005131 | 19,942 | 116,500 |
| Complement and coagulation cascades | KEGG_2021_Human | 0.00425 | 0.01547 | 19,915 | 108,800 |
| ECM-receptor interaction | KEGG_2021_Human | 0.0044 | 0.01547 | 19,912 | 108,000 |
| Thrombocytopenia | Jensen_DISEASES | 0.00475 | 0.007803 | 19,905 | 106,500 |
| Platelet activation | KEGG_2021_Human | 0.0062 | 0.01547 | 19,876 | 101,000 |
| Platelet degranulation (GO:0002576) | GO_Biological_Process_2021 | 0.00625 | 0.0189 | 19,875 | 100,900 |
| Pancreatic cancer | Jensen_DISEASES | 0.00685 | 0.0105 | 19,863 | 98,990 |
| Regulated exocytosis (GO:0045055) | GO_Biological_Process_2021 | 0.009 | 0.0189 | 19,820 | 93,360 |
| Neutrophil extracellular trap formation | KEGG_2021_Human | 0.00945 | 0.01547 | 19,811 | 92,350 |
| Focal adhesion | KEGG_2021_Human | 0.01005 | 0.01547 | 19,799 | 91,080 |
| Coronary artery disease | Jensen_DISEASES | 0.01025 | 0.01473 | 19,795 | 90,670 |
| Extracellular structure organization (GO:0043062) | GO_Biological_Process_2021 | 0.0108 | 0.0189 | 19,784 | 89,590 |
| External encapsulating structure organization (GO:0045229) | GO_Biological_Process_2021 | 0.01085 | 0.0189 | 19,783 | 89,490 |
| Cerebrovascular disease | Jensen_DISEASES | 0.01125 | 0.01522 | 19,775 | 88,740 |
| Coronavirus disease | KEGG_2021_Human | 0.0116 | 0.01547 | 19,768 | 88,100 |
| Positive regulation of signal transduction (GO:0009967) | GO_Biological_Process_2021 | 0.0126 | 0.0189 | 19,748 | 86,380 |
| Hypertension | Jensen_DISEASES | 0.01445 | 0.01846 | 19,711 | 83,520 |
| Lung | Human_Gene_Atlas | 0.01495 | 0.01495 | 19,701 | 82,800 |
| Extracellular matrix organization (GO:0030198) | GO_Biological_Process_2021 | 0.015 | 0.01929 | 19,700 | 82,730 |
| Human papillomavirus infection | KEGG_2021_Human | 0.01655 | 0.0177 | 19,669 | 80,670 |
| PI3K-Akt signaling pathway | KEGG_2021_Human | 0.0177 | 0.0177 | 19,646 | 79,260 |
| Breast cancer | Jensen_DISEASES | 0.0217 | 0.0261 | 19,566 | 74,950 |
| Regulation of intracellular signal transduction (GO:1902531) | GO_Biological_Process_2021 | 0.02185 | 0.02458 | 19,563 | 74,800 |
| Skin cancer | Jensen_DISEASES | 0.0227 | 0.0261 | 19,546 | 73,990 |
| Melanoma | Jensen_DISEASES | 0.02525 | 0.02765 | 19,495 | 71,720 |
| Positive regulation of intracellular signal transduction (GO:1902533) | GO_Biological_Process_2021 | 0.0273 | 0.0273 | 19,454 | 70,050 |
| Kidney cancer | Jensen_DISEASES | 0.1292 | 0.1351 | 17,416 | 35,640 |
| Carcinoma | Jensen_DISEASES | 0.5659 | 0.5659 | 8682 | 4943 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayan, D.; Bozkurt Polat, Ş.B.; Bayram, E.; Özmen, E.; Aydın, F.E.; Ersan, S. A Comparative Analysis of the Roles of von Willebrand Factor and ADAMTS13 in Hepatocellular Carcinoma: A Bioinformatics and Microarray-Based Study. Curr. Issues Mol. Biol. 2025, 47, 270. https://doi.org/10.3390/cimb47040270
Ayan D, Bozkurt Polat ŞB, Bayram E, Özmen E, Aydın FE, Ersan S. A Comparative Analysis of the Roles of von Willebrand Factor and ADAMTS13 in Hepatocellular Carcinoma: A Bioinformatics and Microarray-Based Study. Current Issues in Molecular Biology. 2025; 47(4):270. https://doi.org/10.3390/cimb47040270
Chicago/Turabian StyleAyan, Durmuş, Şerife Buket Bozkurt Polat, Ergül Bayram, Esma Özmen, Fatma Esin Aydın, and Serpil Ersan. 2025. "A Comparative Analysis of the Roles of von Willebrand Factor and ADAMTS13 in Hepatocellular Carcinoma: A Bioinformatics and Microarray-Based Study" Current Issues in Molecular Biology 47, no. 4: 270. https://doi.org/10.3390/cimb47040270
APA StyleAyan, D., Bozkurt Polat, Ş. B., Bayram, E., Özmen, E., Aydın, F. E., & Ersan, S. (2025). A Comparative Analysis of the Roles of von Willebrand Factor and ADAMTS13 in Hepatocellular Carcinoma: A Bioinformatics and Microarray-Based Study. Current Issues in Molecular Biology, 47(4), 270. https://doi.org/10.3390/cimb47040270






