Identification and Characterization of the RNA Modifying Factors PUS7 and WTAP as Key Components for the Control of Tumor Biological Processes in Renal Cell Carcinomas
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sets
2.2. Data Analysis
3. Results
3.1. Tabular Overview of Analyzed Genes
3.2. Analysis of RCC-Relevant Proliferation and Prognostic Marker Genes
3.3. Validation of Selected Immune Checkpoint Axes in the RCC Data Sets
3.4. Identification of Differentially Expressed Genes Relevant for RNA Pseudouridinylation and RNA Methylation in the RCC Data Sets
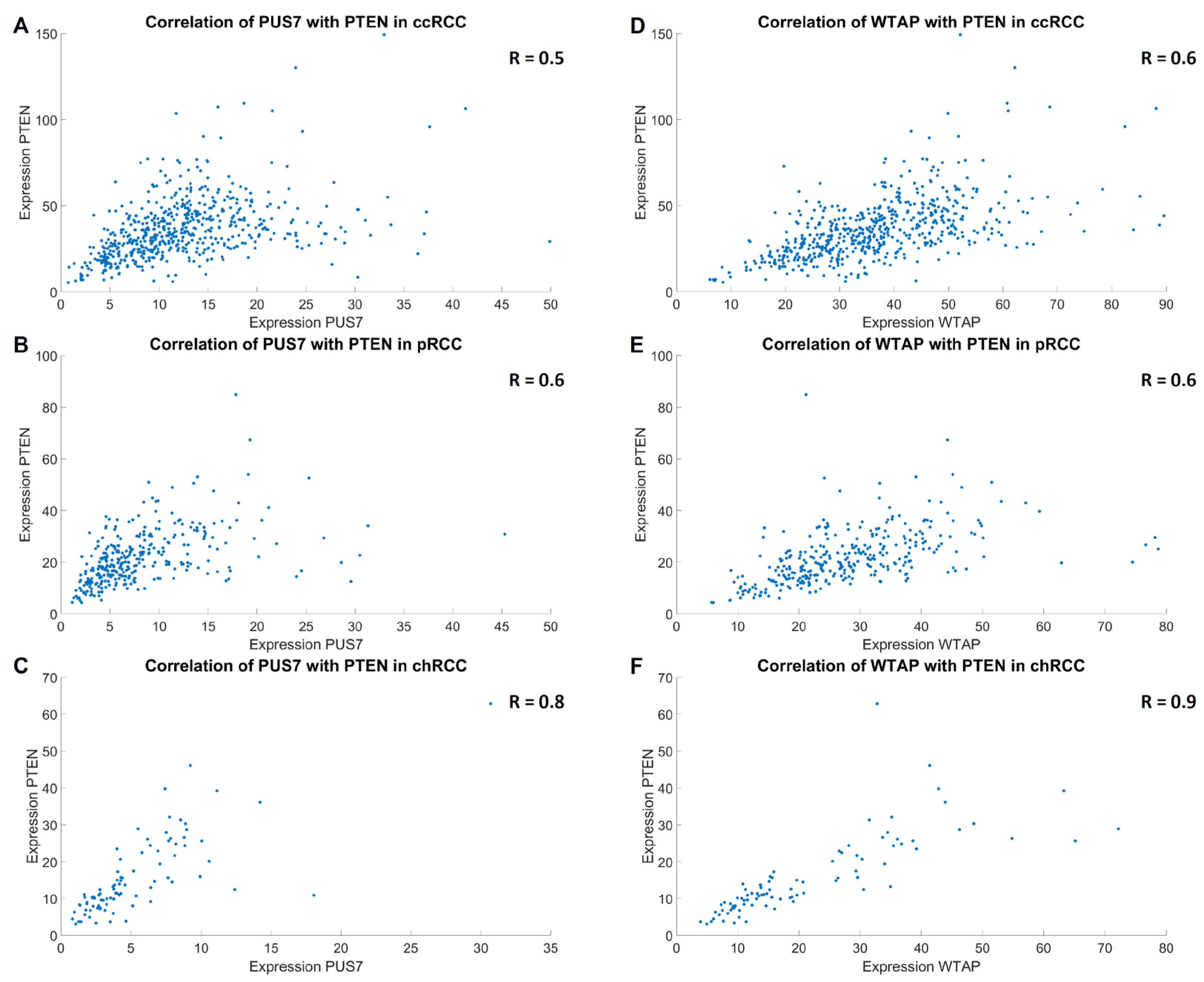
3.5. Examination of Putative Correlations Between the Two RNA-Modifying Factors PUS7 and WTAP as Well as the Differentially Expressed Marker Genes Relevant for Tumor (Immune) Biology
3.6. Identification of Additional PUS7 and WTAP Target Genes in RCCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AS | Alternative splicing |
| DNA | Deoxyribonucleic acid |
| ccRCC | Clear cell RCC |
| CDS | Coding sequence |
| chRCC | Chromophobe RCC |
| HLA | Human leukocyte antigen |
| FDR | False discovery rate |
| ICP | Immune checkpoint |
| lncRNA | Long non-coding RNA |
| mAb | Monoclonal antibody |
| mRNA | Messenger RNA |
| miRNA | Micro RNA |
| PAP | Poly(A) polymerase |
| ORR | Overall response rate |
| pRCC | Papillary RCC |
| RCC | Renal cell carcinoma |
| RNA | Ribonucleic acid |
| RNMTs | RNA methyltransferases |
| rRNA | Ribosomal RNA |
| snRNA | Small nuclear RNA |
| TE | Translational efficiency |
| TPM | Transcripts per million |
| tRNA | Transfer RNA |
| UTR | Untranslated region |
References
- Brown, J.S.; Amend, S.R.; Austin, R.H.; Gatenby, R.A.; Hammarlund, E.U.; Pienta, K.J. Updating the Definition of Cancer. Mol. Cancer Res. 2023, 21, 1142–1147. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Jasinski-Bergner, S.; Blumke, J.; Wickenhauser, C.; Seliger, B. Relevance of 2′-O-Methylation and Pseudouridylation for the Malignant Melanoma. Cancers 2021, 13, 1167. [Google Scholar] [CrossRef]
- Stepinski, D. The Nucleolus, an Ally, and an Enemy of Cancer Cells. Histochem. Cell Biol. 2018, 150, 607–629. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, Q.; Yu, D.; Natchiar, K.; Zhou, C.; Hsu, C.; Hsu, P.-H.; Zhang, X.; Klaholz, B.; Gregory, R.I.; et al. METTL5, an 18S RRNA-Specific m6A Methyltransferase, Modulates Expression of Stress Response Genes. bioRxiv 2020. [Google Scholar] [CrossRef]
- von der Haar, T. Mathematical and Computational Modelling of Ribosomal Movement and Protein Synthesis: An Overview. Comput. Struct. Biotechnol. J. 2012, 1, e201204002. [Google Scholar] [CrossRef]
- Penzo, M.; Montanaro, L.; Trere, D.; Derenzini, M. The Ribosome Biogenesis-Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef]
- Casamassimi, A.; Federico, A.; Rienzo, M.; Esposito, S.; Ciccodicola, A. Transcriptome Profiling in Human Diseases: New Advances and Perspectives. Int. J. Mol. Sci. 2017, 18, 1652. [Google Scholar] [CrossRef]
- Wang, D.; Farhana, A. Biochemistry, RNA Structure. In StatPearls; Treasure Island (FL) ineligible companies. Disclosure: Aisha Farhana declares no relevant financial relationships with ineligible companies; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Eun, J.W.; Jang, S.H.; Kim, J.Y.; Jeong, J.-Y. The Diverse Landscape of RNA Modifications in Cancer Development and Progression. Genes Genom. 2025, 47, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Nabi, S.; Kessler, E.R.; Bernard, B.; Flaig, T.W.; Lam, E.T. Renal Cell Carcinoma: A Review of Biology and Pathophysiology. F1000Research 2018, 7, 307. [Google Scholar] [CrossRef]
- Muglia, V.F.; Prando, A. Renal Cell Carcinoma: Histological Classification and Correlation with Imaging Findings. Radiol. Bras. 2015, 48, 166–174. [Google Scholar] [CrossRef]
- Alaghehbandan, R.; Perez Montiel, D.; Luis, A.S.; Hes, O. Molecular Genetics of Renal Cell Tumors: A Practical Diagnostic Approach. Cancers 2019, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Akin, O.; Elnajjar, P.; Heller, M.; Jarosz, R.; Erickson, B.J.; Kirk, S.; Lee, Y.; Linehan, M.W.; Gautam, R.; Vikram, R.; et al. The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma Collection (TCGA-KIRC) (Version 3) [Data Set]. In The Cancer Imaging Archive; National Institutes of Health (NIH): Bethesda, MD, USA, 2016. [Google Scholar] [CrossRef]
- Linehan, M.W.; Gautam, R.; Sadow, C.A.; Levine, S. The Cancer Genome Atlas Kidney Chromophobe Collection (TCGA-KICH) (Version 3) [Data Set]. In The Cancer Imaging Archive; National Institutes of Health (NIH): Bethesda, MD, USA, 2016. [Google Scholar] [CrossRef]
- Linehan, M.; Gautam, R.; Kirk, S.; Lee, Y.; Roche, C.; Bonaccio, E.; Filippini, J.; Rieger-Christ, K.; Lemmerman, J.; Jarosz, R. The Cancer Genome Atlas Cervical Kidney Renal Papillary Cell Carcinoma Collection (TCGA-KIRP) (Version 4) [Data Set]. In The Cancer Imaging Archive; National Institutes of Health (NIH): Bethesda, MD, USA, 2016. [Google Scholar] [CrossRef]
- Jones, J.; Otu, H.; Spentzos, D.; Kolia, S.; Inan, M.; Beecken, W.D.; Fellbaum, C.; Gu, X.; Joseph, M.; Pantuck, A.J.; et al. Gene Signatures of Progression and Metastasis in Renal Cell Cancer. Clin Cancer Res 2005, 11, 5730–5739. [Google Scholar] [CrossRef]
- Dalgliesh, G.L.; Furge, K.; Greenman, C.; Chen, L.; Bignell, G.; Butler, A.; Davies, H.; Edkins, S.; Hardy, C.; Latimer, C.; et al. Systematic Sequencing of Renal Carcinoma Reveals Inactivation of Histone Modifying Genes. Nature 2010, 463, 360–363. [Google Scholar] [CrossRef]
- Ding, Y.; Huang, D.; Zhang, Z.; Smith, J.; Petillo, D.; Looyenga, B.D.; Feenstra, K.; Mackeigan, J.P.; Furge, K.A.; Teh, B.T. Combined Gene Expression Profiling and RNAi Screening in Clear Cell Renal Cell Carcinoma Identify PLK1 and Other Therapeutic Kinase Targets. Cancer Res. 2011, 71, 5225–5234. [Google Scholar] [CrossRef]
- Li, L.; Zhu, C.; Xu, S.; Xu, Q.; Xu, D.; Gan, S.; Cui, X.; Tang, C. PUS1 Is a Novel Biomarker for Evaluating Malignancy of Human Renal Cell Carcinoma. Aging 2023, 15, 5215–5227. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Hohmann, T.; Hohmann, U.; Dehghani, F.; Grisk, O.; Jasinski-Bergner, S. Analyzing the Impact of the Highest Expressed Epstein-Barr Virus-Encoded MicroRNAs on the Host Cell Transcriptome. Int. J. Mol. Sci. 2024, 25, 7838. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-J.; Yoon, B.-H.; Kim, S.-K.; Kim, S.-Y. GENT2: An Updated Gene Expression Database for Normal and Tumor Tissues. BMC Med. Genom. 2019, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Creed, J.H.; Gerke, T.A.; Berglund, A.E. MatSurv: Survival Analysis and Visualization in MATLAB. J. Open Source Softw. 2020, 5, 1830. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Kobayashi, G.; Hayashi, T.; Sentani, K.; Uraoka, N.; Fukui, T.; Kido, A.; Katsuya, N.; Ishikawa, A.; Babasaki, T.; Sekino, Y.; et al. MCM4 Expression Is Associated with High-Grade Histology, Tumor Progression and Poor Prognosis in Urothelial Carcinoma. Diagn. Pathol. 2023, 18, 106. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Z.; Liu, Z.; Chen, Z.; Jiang, J.; Ji, Y.; Zhang, Y.; Zhu, H.; Zheng, B. CENPF Promotes the Proliferation of Renal Cell Carcinoma in Vitro. Transl. Androl. Urol. 2023, 12, 320–329. [Google Scholar] [CrossRef]
- Mehdi, M.Z.; Nagi, A.H.; Naseem, N. MCM—2 and Ki—67 as Proliferation Markers in Renal Cell Carcinoma: A Quantitative and Semi—Quantitative Analysis. Int. Braz. J. Urol. 2016, 42, 1121–1128. [Google Scholar] [CrossRef][Green Version]
- de la Taille, A.; Buttyan, R.; Katz, A.E.; McKiernan, J.; Burchardt, M.; Burchardt, T.; Chopin, D.K.; Sawczuk, I.S. Biomarkers of Renal Cell Carcinoma. Past and Future Considerations. Urol. Oncol. 2000, 5, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Farber, N.J.; Kim, C.J.; Modi, P.K.; Hon, J.D.; Sadimin, E.T.; Singer, E.A. Renal Cell Carcinoma: The Search for a Reliable Biomarker. Transl. Cancer Res. 2017, 6, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Guglas, K.; Kolodziejczak, I.; Kolenda, T.; Kopczynska, M.; Teresiak, A.; Sobocinska, J.; Blizniak, R.; Lamperska, K. YRNAs and YRNA-Derived Fragments as New Players in Cancer Research and Their Potential Role in Diagnostics. Int. J. Mol. Sci. 2020, 21, 5682. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, T.; Mori, K.; Matsukawa, A.; Kawada, T.; Katayama, S.; Bekku, K.; Laukhtina, E.; Rajwa, P.; Quhal, F.; Pradere, B.; et al. Updated Systematic Review and Network Meta-Analysis of First-Line Treatments for Metastatic Renal Cell Carcinoma with Extended Follow-up Data. Cancer Immunol. Immunother. 2024, 73, 38. [Google Scholar] [CrossRef]
- Xiong, W.D.; Zhao, Y.C.; Wei, Z.L.; Li, C.F.; Zhao, R.Z.; Ge, J.B.; Shi, B. N1-Methyladenosine Formation, Gene Regulation, Biological Functions, and Clinical Relevance. Mol. Ther. 2023, 31, 308–330. [Google Scholar] [CrossRef]
- Borchardt, E.K.; Martinez, N.M.; Gilbert, W. V Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020, 54, 309–336. [Google Scholar] [CrossRef]
- Ding, H.; Liu, N.; Wang, Y.; Adam, S.A.; Jin, J.; Feng, W.; Sun, J. Implications of RNA Pseudouridylation for Cancer Biology and Therapeutics: A Narrative Review. J. Transl. Med. 2024, 22, 906. [Google Scholar] [CrossRef]
- Mongan, N.P.; Emes, R.D.; Archer, N. Detection and Analysis of RNA Methylation. F1000Research 2019, 8, 559. [Google Scholar] [CrossRef]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Peng, Y.; Yu, G.; Tian, S.; Li, H. Co-Expression and Co-Purification of Archaeal and Eukaryal Box C/D RNPs. PLoS ONE 2014, 9, e103096. [Google Scholar] [CrossRef]
- Wu, Y.; Pu, X.; Wu, S.; Zhang, Y.; Fu, S.; Tang, H.; Wang, X.; Xu, M. PCIF1, the Only Methyltransferase of N6,2-O-Dimethyladenosine. Cancer Cell Int. 2023, 23, 226. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-Methyladenosine and Its Role in Cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef]
- Ruszkowska, A. METTL16, Methyltransferase-Like Protein 16: Current Insights into Structure and Function. Int. J. Mol. Sci. 2021, 22, 2176. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Hao, F.; Song, S.; Zhang, J.; Zhou, H.; Zhang, J.; Li, Y. METTL Family in Healthy and Disease. Mol. Biomed. 2024, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.D. RNA Binding by the M6A Methyltransferases METTL16 and METTL3. Biology 2024, 13, 391. [Google Scholar] [CrossRef]
- Pinto, R.; Vagbo, C.B.; Jakobsson, M.E.; Kim, Y.; Baltissen, M.P.; O’Donohue, M.F.; Guzman, U.H.; Malecki, J.M.; Wu, J.; Kirpekar, F.; et al. The Human Methyltransferase ZCCHC4 Catalyses N6-Methyladenosine Modification of 28S Ribosomal RNA. Nucleic Acids Res. 2020, 48, 830–846. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, S.; Gao, X.; Ru, Y.; Gu, X.; Hu, X. Research Progress of N1-Methyladenosine RNA Modification in Cancer. Cell Commun. Signal. 2024, 22, 79. [Google Scholar] [CrossRef]
- Song, H.; Zhang, J.; Liu, B.; Xu, J.; Cai, B.; Yang, H.; Straube, J.; Yu, X.; Ma, T. Biological Roles of RNA m(5)C Modification and Its Implications in Cancer Immunotherapy. Biomark. Res. 2022, 10, 15. [Google Scholar] [CrossRef]
- Li, M.; Tao, Z.; Zhao, Y.; Li, L.; Zheng, J.; Li, Z.; Chen, X. 5-Methylcytosine RNA Methyltransferases and Their Potential Roles in Cancer. J. Transl. Med. 2022, 20, 214. [Google Scholar] [CrossRef]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of Ribosomal RNA by NSUN5 Is a Conserved Mechanism Modulating Organismal Lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef]
- Wu, D.; Li, X.; Khan, F.A.; Yuan, C.; Pandupuspitasari, N.S.; Huang, C.; Sun, F.; Guan, K. TRNA Modifications and TRNA-Derived Small RNAs: New Insights of TRNA in Human Disease. Cell Biol. Toxicol. 2024, 40, 76. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Yao, Y.; Wu, P.; Zi, X.; Sun, N.; He, J. The Potential Role of N(7)-Methylguanosine (M7G) in Cancer. J. Hematol. Oncol. 2022, 15, 63. [Google Scholar] [CrossRef] [PubMed]
- Alhammadi, M.A.; Bajbouj, K.; Talaat, I.M.; Hamoudi, R. The Role of RNA-Modifying Proteins in Renal Cell Carcinoma. Cell Death Dis. 2024, 15, 227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Luo, X.; Qiao, S. METTL14-Mediated N6-Methyladenosine Modification of Pten MRNA Inhibits Tumour Progression in Clear-Cell Renal Cell Carcinoma. Br. J. Cancer 2022, 127, 30–42. [Google Scholar] [CrossRef]
- Guegueniat, J.; Halabelian, L.; Zeng, H.; Dong, A.; Li, Y.; Wu, H.; Arrowsmith, C.H.; Kothe, U. The Human Pseudouridine Synthase PUS7 Recognizes RNA with an Extended Multi-Domain Binding Surface. Nucleic Acids Res. 2021, 49, 11810–11822. [Google Scholar] [CrossRef]
- Dong, B.; Wang, B.; Fan, M.; Zhang, J.; Zhao, Z. Comprehensive Analysis to Identify PUS7 as a Prognostic Biomarker from Pan-Cancer Analysis to Osteosarcoma Validation. Aging 2024, 16, 9188–9203. [Google Scholar] [CrossRef]
- Zhang, G.; Cheng, C.; Wang, X.; Wang, S. N6-Methyladenosine Methylation Modification in Breast Cancer: Current Insights. J. Transl. Med. 2024, 22, 971. [Google Scholar] [CrossRef]
- Liu, M.; Yu, B.; Tian, Y.; Li, F. Regulatory Function and Mechanism Research for M6A Modification WTAP via SUCLG2-AS1- MiR-17-5p-JAK1 Axis in AML. BMC Cancer 2024, 24, 98. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Sun, H.; Gao, Z.; Zhu, Z.; Yuan, K. Role of WTAP in Cancer: From Mechanisms to the Therapeutic Potential. Biomolecules 2022, 12, 1224. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.; Wang, W.; Wang, Z.; Zhang, Y.; Pan, X.; Wen, X.; Leng, H.; Guo, J.; Ma, X.X. WTAP/IGF2BP3 Mediated M6A Modification of the EGR1/PTEN Axis Regulates the Malignant Phenotypes of Endometrial Cancer Stem Cells. J. Exp. Clin. Cancer Res. 2024, 43, 204. [Google Scholar] [CrossRef]
- Zhao, Z.; Wan, J.; Guo, M.; Wang, Y.; Yang, Z.; Zhou, F.; Li, Z.; Ming, L. Expression and Prognostic Significance of M6A-Related Genes in TP53-Mutant Non-Small-Cell Lung Cancer. J. Clin. Lab. Anal. 2022, 36, e24118. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.Y.; Wang, T.; Su, X.; Guo, S. Identification of the m(6)A RNA Methylation Regulators WTAP as a Novel Prognostic Biomarker and Genomic Alterations in Cutaneous Melanoma. Front. Mol. Biosci. 2021, 8, 665222. [Google Scholar] [CrossRef] [PubMed]
- Jasinski-Bergner, S.; Eckstein, M.; Taubert, H.; Wach, S.; Fiebig, C.; Strick, R.; Hartmann, A.; Seliger, B. The Human Leukocyte Antigen G as an Immune Escape Mechanism and Novel Therapeutic Target in Urological Tumors. Front. Immunol. 2022, 13, 811200. [Google Scholar] [CrossRef] [PubMed]
- Jasinski-Bergner, S.; Reches, A.; Stoehr, C.; Massa, C.; Gonschorek, E.; Huettelmaier, S.; Braun, J.; Wach, S.; Wullich, B.; Spath, V.; et al. Identification of Novel MicroRNAs Regulating HLA-G Expression and Investigating Their Clinical Relevance in Renal Cell Carcinoma. Oncotarget 2016, 7, 26866–26878. [Google Scholar] [CrossRef]
- Jasinski-Bergner, S.; Stoehr, C.; Bukur, J.; Massa, C.; Braun, J.; Hüttelmaier, S.; Spath, V.; Wartenberg, R.; Legal, W.; Taubert, H.; et al. Clinical Relevance of MiR-Mediated HLA-G Regulation and the Associated Immune Cell Infiltration in Renal Cell Carcinoma. Oncoimmunology 2015, 4, e1008805. [Google Scholar] [CrossRef]
- Bukur, J.; Jasinski, S.; Seliger, B. The Role of Classical and Non-Classical HLA Class I Antigens in Human Tumors. Semin. Cancer Biol. 2012, 22, 350–358. [Google Scholar] [CrossRef]
- Bauer, M.; Schobel, C.M.; Wickenhauser, C.; Seliger, B.; Jasinski-Bergner, S. Deciphering the Role of Alternative Splicing in Neoplastic Diseases for Immune-Oncological Therapies. Front. Immunol. 2024, 15, 1386993. [Google Scholar] [CrossRef]
- Andrzejczak, A.; Tupikowski, K.; Tomkiewicz, A.; Malkiewicz, B.; Ptaszkowski, K.; Domin, A.; Szydelko, T.; Karabon, L. The Variations’ in Genes Encoding TIM-3 and Its Ligand, Galectin-9, Influence on CcRCC Risk and Prognosis. Int. J. Mol. Sci. 2023, 24, 2042. [Google Scholar] [CrossRef]
- Yildirim, C. Galectin-9, a pro-Survival Factor Inducing Immunosuppression, Leukemic Cell Transformation and Expansion. Mol. Biol. Rep. 2024, 51, 571. [Google Scholar] [CrossRef]
- London, M.; Gallo, E. Critical Role of EphA3 in Cancer and Current State of EphA3 Drug Therapeutics. Mol. Biol. Rep. 2020, 47, 5523–5533. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, X.; Wu, H.; Du, S.; Wang, Z.; Xie, S.; Zhang, R.; Chen, G.; Chen, H. Identification of CREB5 as a Prognostic and Immunotherapeutic Biomarker in Glioma through Multi-Omics Pan-Cancer Analysis. Comput. Biol. Med. 2024, 173, 108307. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Li, W.; Zhang, L.; Xue, B.; Chen, Y.; Zhang, Z.; Wang, D.; Wu, B. CDH13 Is a Prognostic Biomarker and a Potential Therapeutic Target for Patients with Clear Cell Renal Cell Carcinoma. Am. J. Cancer Res. 2022, 12, 4520–4544. [Google Scholar] [PubMed]
- Tang, Q.; Li, W.; Zheng, X.; Ren, L.; Liu, J.; Li, S.; Wang, J.; Du, G. MELK Is an Oncogenic Kinase Essential for Metastasis, Mitotic Progression, and Programmed Death in Lung Carcinoma. Signal Transduct. Target. Ther. 2020, 5, 279. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; van Ommen, G.-J.B. Antisense-Mediated Exon Skipping: A Versatile Tool with Therapeutic and Research Applications. RNA 2007, 13, 1609–1624. [Google Scholar] [CrossRef]
- Brusson, M.; Miccio, A. A CRISPR/Cas Approach to β-Haemoglobinopathies. Med. Sci. 2025, 41, 33–39. [Google Scholar] [CrossRef]
- Minskaia, E.; Galieva, A.; Egorov, A.D.; Ivanov, R.; Karabelsky, A. Viral Vectors in Gene Replacement Therapy. Biochemistry 2023, 88, 2157–2178. [Google Scholar] [CrossRef]
- Padda, I.S.; Mahtani, A.U.; Patel, P.; Parmar, M. Small Interfering RNA (SiRNA) Therapy; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]


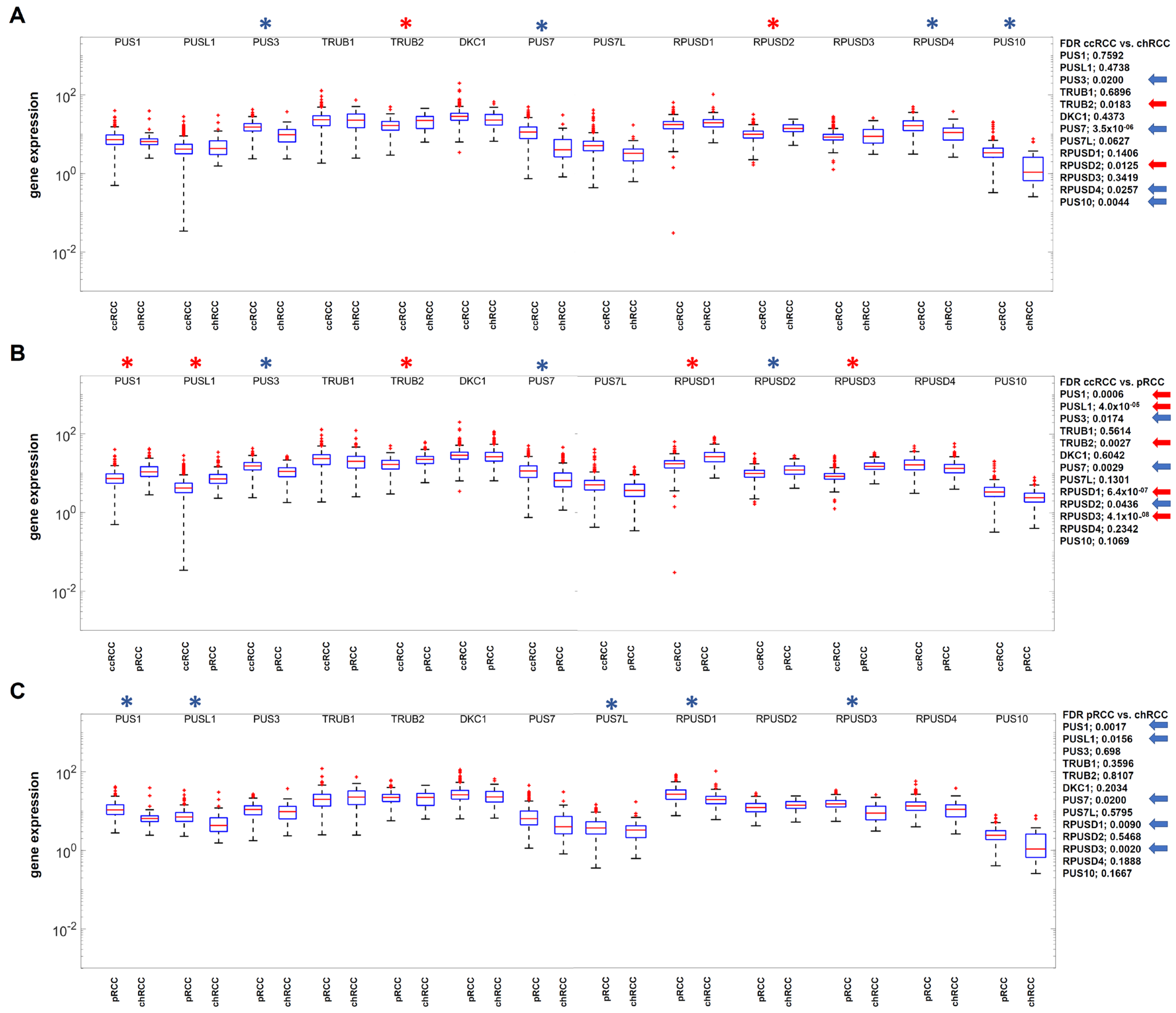



| Proliferation Marker | Prognosis Marker | Tumor Immune Checkpoints | RNA Pseudouridinylation | RNA Ortho-Methylation | RNA N-Methylation |
|---|---|---|---|---|---|
| MKI67 | TP53 | CD47 | PUS1 | FBL | PCIF1 |
| PCNA | BCL2 | CD24 | PUSL1 | NOP56 | METTL3 |
| MCM2 | BIRC5 | STC1 | PUS3 | NOP58 | METTL14 |
| MCM4 | PTEN | CD274 | TRUB1 | SNU13 | WTAP |
| CENPF | NRAS | HLA-G | TRUB2 | VIRMA | |
| CXCR4 | TSC1 | HLA-E | DKC1 | RBM15 | |
| TSC2 | LGALS3 | PUS7 | ZC3H13 | ||
| CDKN2A | FGL1 | PUS7L | METTL16 | ||
| LGALS9 | RPUSD1 | METTL5 | |||
| HMGB1 | RPUSD2 | NSUN2 | |||
| PVR | RPUSD3 | TRMT10C | |||
| RPUSD4 | ZCCHC4 | ||||
| PUS10 | TRMT6 | ||||
| TRMT61A | |||||
| TRMT61B | |||||
| RRP8 | |||||
| DNMT1 | |||||
| TRDMT1 | |||||
| NSUN4 | |||||
| NSUN5 | |||||
| RNMT | |||||
| CMTR1 | |||||
| WDR4 | |||||
| METTL1 |
| ccRCC | pRCC | chRCC | ΣRCC | Non-Tumorous Kidney | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PUS7 | WTAP | PUS7 | WTAP | PUS7 | WTAP | PUS7 | WTAP | PUS7 | WTAP | |
| CXCR4 | R = 0.4 | R = 0.4 | R = 0.1 | R = 0.4 | R = 0.4 | R = 0.5 | R = 0.5 | R = 0.5 | R = 0.31 | R = 0.42 |
| [0.32; 0.47] | [0.29; 0.44] | [0.02; 0.23] | [0.28; 0.47] | [0.25; 0.60] | [0.26; 0.61] | [0.48; 0.57] | [0.47; 0.57] | [0.12; 0.49] | [0.23; 0.58] | |
| p < 1.0 × 10−10 | p < 1.0 × 10−10 | p = 0.0273 | p < 1.0 × 10−10 | p = 1.4 × 10−5 | p = 8.6 × 10−6 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p = 0.0013 | p = 1.2 × 10−5 | |
| TP53 | R = 0.5 | R = 0.5 | R = 0.6 | R = 0.5 | R = 0.7 | R = 0.8 | R = 0.5 | R = 0.5 | R = 0.61 | R = 0.50 |
| [0.37; 0.52] | [0.47; 0.60] | [0.51; 0.66] | [0.41; 0.58] | [0.53; 0.80] | [0.67; 0.87] | [0.58; 0.61] | [0.59; 0.61] | [0.45; 0.74] | [0.31; 0.66] | |
| p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p = 1.6 × 10−6 | |
| PTEN | R = 0.5 | R = 0.6 | R = 0.6 | R = 0.6 | R = 0.8 | R = 0.9 | R = 0.7 | R = 0.7 | R = 0.74 | R = 0.80 |
| [0.45; 0.58] | [0.55; 0.66] | [0.50; 0.67] | [0.56; 0.71] | [0.71; 0.87] | [0.83; 0.92] | [0.63; 0.71] | [0.67; 0.74] | [0.64; 0.82] | [0.70; 0.87] | |
| p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | |
| NRAS | R = 0.7 | R = 0.7 | R = 0.6 | R = 0.6 | R = 0.8 | R = 0.9 | R = 0.73 | R = 0.7 | R = 0.62 | R = 0.66 |
| [0.61; 0.71] | [0.62; 0.71] | [0.50; 0.66] | [0.49; 0.65] | [0.65; 0.83] | [0.87; 0.95] | [0.70; 0.76] | [0.70; 0.76] | [0.46; 0.73] | [0.51; 0.78] | |
| p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | |
| HLA-G | R = 0.2 | R = 0.2 | R = 0 | R = 0.2 | R = 0.3 | R = 0.3 | R = 0.3 | R = 0.4 | R = 0.38 | R = 0.41 |
| [0.09; 0.25] | [0.16; 0.32] | [−0.14; 0.08] | [0.08; 0.30] | [0.05; 0.44] | [0.14; 0.52] | [0.22; 0.34] | [0.30; 0.41] | [0.19; 0.55] | [0.22; 0.58] | |
| p = 3.9 × 10−5 | p = 6.2 × 10−9 | p = 0.6350 | p = 0.0004 | p = 0.0168 | p = 0.0010 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p = 8.2 × 10−5 | p = 1.6 × 10−5 | |
| LGALS9 | R = 0.2 | R = 0.3 | R = 0.1 | R = 0.2 | R = 0.4 | R = 0.4 | R = 0.4 | R = 0.4 | R = 0.49 | R = 0.43 |
| [0.14; 0.31] | [0.17; 0.33] | [−0.04; 0.18] | [0.13; 0.34] | [0.20; 0.56] | [0.26; 0.59] | [0.29; 0.41] | [0.32; 0.44] | [0.32; 0.62] | [0.26; 0.58] | |
| p = 5.5 × 10−8 | p = 3.1 × 10−10 | p = 0.2035 | p = 1.1 × 10−5 | p = 0.0001 | p = 1.9 × 10−5 | p < 1.0 × 10−10 | p < 1.0 × 10−10 | p = 3.0 × 10−7 | p = 6.5 × 10−6 | |
| PUS7 | WTAP | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Expression | Correlation Coefficient | p-Value | Gene | Expression | Correlation Coefficient | p-Value |
| ACLY | 190.0238 | 0.7465 | 4.68 × 10−178 | KLF10 | 687.2348 | 0.5013 | 1.73 × 10−64 |
| NUF2 | 179.1750 | −0.3738 | 2.20 × 10−34 | FRZB | 119.7849 | 0.6163 | 2.97 × 10−105 |
| SST | 140.5689 | 0.6030 | 1.20 × 10−99 | CPA3 | 104.5162 | 0.6406 | 3.96 × 10−116 |
| NPTX2 | 111.1566 | 0.5450 | 3.81 × 10−78 | CLEC4E | 103.6079 | 0.5289 | 7.08 × 10−73 |
| MELK | 103.6079 | 0.4901 | 2.56 × 10−61 | RAPGEF5 | 75.9460 | 0.5316 | 9.49 × 10−74 |
| HAVCR1 | 78.9817 | 0.4859 | 3.91 × 10−60 | POSTN | 75.4483 | 0.3515 | 2.45 × 10−30 |
| REL | 78.8732 | 0.5333 | 2.88 × 10−74 | EPHA3 | 73.3262 | 0.4336 | 6.45 × 10−47 |
| TNFAIP6 | 77.3074 | 0.5625 | 3.41 × 10−84 | ARHGAP29 | 53.2551 | 0.6308 | 1.29 × 10−111 |
| IL1RAP | 75.8770 | 0.5011 | 1.90 × 10−64 | IFI44L | 47.7831 | 0.4633 | 3.90 × 10−54 |
| VCAN | 73.3262 | 0.4964 | 4.26 × 10−63 | ADGRG6 | 44.4220 | 0.7105 | 7.15 × 10−154 |
| EDN1 | 66.8430 | 0.6020 | 3.08 × 10−99 | SFRP2 | 29.5562 | 0.6880 | 1.39 × 10−140 |
| SCGN | 62.5494 | 0.4680 | 2.39 × 10−55 | ITGA4 | 24.6503 | 0.4976 | 1.99 × 10−63 |
| CREB5 | 60.7807 | 0.7228 | 1.04 × 10−161 | SCARA3 | 22.4505 | 0.4419 | 7.25 × 10−49 |
| PBK | 55.8713 | 0.5313 | 1.21 × 10−73 | CDH13 | 17.8840 | 0.6109 | 6.22 × 10−103 |
| ARHGAP11A | 50.1871 | 0.4210 | 4.64 × 10−44 | IRAK3 | 14.5509 | 0.5764 | 2.98 × 10−89 |
| FRZB | 40.2235 | 0.6295 | 4.95 × 10−111 | RGS5 | 11.9904 | 0.6551 | 3.70 × 10−123 |
| LOXL2 | 39.4410 | 0.6048 | 2.12 × 10−100 | ECEL1 | 10.0061 | 0.6368 | 2.32 × 10−114 |
| QRFPR | 28.1993 | 0.5818 | 2.69 × 10−91 | VNN2 | 9.5169 | 0.4689 | 1.36 × 10−55 |
| COL5A1 | 25.9432 | 0.5086 | 1.20 × 10−66 | GJA1 | 9.2066 | 0.5597 | 3.32 × 10−83 |
| SLC5A1 | 24.9524 | 0.3168 | 1.20 × 10−24 | ITGB6 | 8.5897 | 0.5524 | 1.23 × 10−80 |
| ADGRG6 | 24.6503 | 0.5560 | 6.72 × 10−82 | INHA | 7.9855 | 0.6423 | 5.94 × 10−117 |
| SLC7A2 | 20.7983 | 0.4573 | 1.23 × 10−52 | NID2 | 6.4530 | 0.5376 | 1.15 × 10−75 |
| RECQL | 20.2517 | 0.6875 | 2.65 × 10−140 | APOLD1 | 5.9691 | 0.5043 | 2.35 × 10−65 |
| RRM2 | 19.5924 | −0.4178 | 2.35 × 10−43 | TGFB2 | 3.8323 | 0.6762 | 4.63 × 10−134 |
| RAPGEF5 | 17.8840 | 0.6231 | 3.42 × 10−108 | OLFM4 | 3.6288 | 0.5691 | 1.44 × 10−86 |
| ZC3HAV1L | 15.7110 | 0.4402 | 1.83 × 10−48 | AC003092.1 | 2.0515 | 0.4798 | 1.76 × 10−58 |
| NID2 | 14.5509 | 0.5850 | 1.68 × 10−92 | ELK3 | 1.8317 | 0.4500 | 8.06 × 10−51 |
| FAM111B | 14.5212 | 0.5169 | 4.01 × 10−69 | ||||
| MKI67 | 14.0369 | −0.4819 | 4.80 × 10−59 | ||||
| SSPN | 13.6805 | 0.7057 | 6.72 × 10−151 | ||||
| GAS2L3 | 13.2700 | 0.6528 | 5.13 × 10−122 | ||||
| P4HA3 | 11.7873 | 0.7959 | 7.82 × 10−219 | ||||
| LRRK2 | 11.1293 | 0.6397 | 9.65 × 10−116 | ||||
| TPX2 | 11.0676 | 0.5789 | 3.60 × 10−90 | ||||
| TOP2A | 10.5442 | 0.6520 | 1.33 × 10−121 | ||||
| PREX2 | 10.0566 | 0.5676 | 5.03 × 10−86 | ||||
| KCNK3 | 8.0663 | 0.6998 | 2.28 × 10−147 | ||||
| EPHA3 | 6.4530 | 0.5480 | 3.85 × 10−79 | ||||
| IL18R1 | 5.7964 | 0.6681 | 9.00 × 10−130 | ||||
| FRMD6 | 5.6745 | 0.6742 | 5.56 × 10−133 | ||||
| ENPP3 | 5.4524 | 0.6710 | 2.93 × 10−131 | ||||
| OSMR | 5.0688 | 0.6223 | 7.87 × 10−108 | ||||
| ANLN | 5.0446 | 0.6949 | 1.54 × 10−144 | ||||
| FOXM1 | 4.9233 | 0.5749 | 1.13 × 10−88 | ||||
| ARL4C | 4.7428 | 0.5523 | 1.31 × 10−80 | ||||
| CDON | 4.6661 | 0.6809 | 1.28 × 10−136 | ||||
| SACS | 4.4960 | 0.6107 | 7.38 × 10−103 | ||||
| CEP55 | 4.3923 | 0.6164 | 2.75 × 10−105 | ||||
| TNNT1 | 4.3796 | 0.3098 | 1.34 × 10−23 | ||||
| CDCA7 | 4.2515 | 0.5939 | 5.30 × 10−96 | ||||
| PGM2L1 | 4.0652 | −0.3533 | 1.20 × 10−30 | ||||
| SEMA3D | 4.0290 | 0.5363 | 2.90 × 10−75 | ||||
| AGMO | 4.0251 | 0.6211 | 2.55 × 10−107 | ||||
| KIF20B | 3.8307 | 0.7649 | 4.79 × 10−192 | ||||
| NTM | 3.6732 | 0.5229 | 5.31 × 10−71 | ||||
| BUB1 | 3.5337 | 0.5834 | 6.58 × 10−92 | ||||
| BUB1B | 3.5335 | 0.3494 | 5.62 × 10−30 | ||||
| MALL | 3.4257 | 0.6727 | 3.52 × 10−132 | ||||
| CENPF | 3.2824 | 0.6335 | 7.55 × 10−113 | ||||
| USP37 | 2.9702 | 0.6771 | 1.44 × 10−134 | ||||
| PRR11 | 2.9327 | 0.6409 | 2.59 × 10−116 | ||||
| KIF4A | 2.9095 | 0.6239 | 1.45 × 10−108 | ||||
| CCNA2 | 2.8841 | 0.5970 | 3.13 × 10−97 | ||||
| CDCA2 | 2.6303 | 0.5656 | 2.82 × 10−85 | ||||
| HMMR | 2.5460 | 0.5881 | 1.02 × 10−93 | ||||
| BRCA1 | 2.4850 | 0.7127 | 3.06 × 10−155 | ||||
| DLGAP5 | 2.4775 | 0.6151 | 9.80 × 10−105 | ||||
| NEK2 | 2.2389 | 0.6021 | 2.72 × 10−99 | ||||
| XIST | 2.0515 | 0.4606 | 1.91 × 10−53 | ||||
| FZD1 | 2.0050 | 0.6155 | 6.59 × 10−105 | ||||
| NCAPG | 1.9583 | 0.6394 | 1.41 × 10−115 | ||||
| EDIL3 | 1.8830 | 0.5784 | 5.38 × 10−90 | ||||
| DTL | 1.8648 | 0.6236 | 2.12 × 10−108 | ||||
| DKK1 | 1.8317 | 0.4001 | 1.38 × 10−39 | ||||
| CENPK | 1.7248 | 0.6565 | 7.56 × 10−124 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hohmann, T.; Hohmann, U.; Dehghani, F.; Grisk, O.; Jasinski-Bergner, S. Identification and Characterization of the RNA Modifying Factors PUS7 and WTAP as Key Components for the Control of Tumor Biological Processes in Renal Cell Carcinomas. Curr. Issues Mol. Biol. 2025, 47, 266. https://doi.org/10.3390/cimb47040266
Hohmann T, Hohmann U, Dehghani F, Grisk O, Jasinski-Bergner S. Identification and Characterization of the RNA Modifying Factors PUS7 and WTAP as Key Components for the Control of Tumor Biological Processes in Renal Cell Carcinomas. Current Issues in Molecular Biology. 2025; 47(4):266. https://doi.org/10.3390/cimb47040266
Chicago/Turabian StyleHohmann, Tim, Urszula Hohmann, Faramarz Dehghani, Olaf Grisk, and Simon Jasinski-Bergner. 2025. "Identification and Characterization of the RNA Modifying Factors PUS7 and WTAP as Key Components for the Control of Tumor Biological Processes in Renal Cell Carcinomas" Current Issues in Molecular Biology 47, no. 4: 266. https://doi.org/10.3390/cimb47040266
APA StyleHohmann, T., Hohmann, U., Dehghani, F., Grisk, O., & Jasinski-Bergner, S. (2025). Identification and Characterization of the RNA Modifying Factors PUS7 and WTAP as Key Components for the Control of Tumor Biological Processes in Renal Cell Carcinomas. Current Issues in Molecular Biology, 47(4), 266. https://doi.org/10.3390/cimb47040266






