Unravelling Convergent Signaling Mechanisms Underlying the Aging-Disease Nexus Using Computational Language Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Large Language Model-Based Data Curation
2.2. Network Function Analysis
2.3. Pathway Enrichment Analysis
2.4. Data Representation and Venn Analyses
2.5. Cell Culture and Treatment
2.6. Immunoblot and Immunoprecipitation
2.7. Statistical Analyses
3. Results
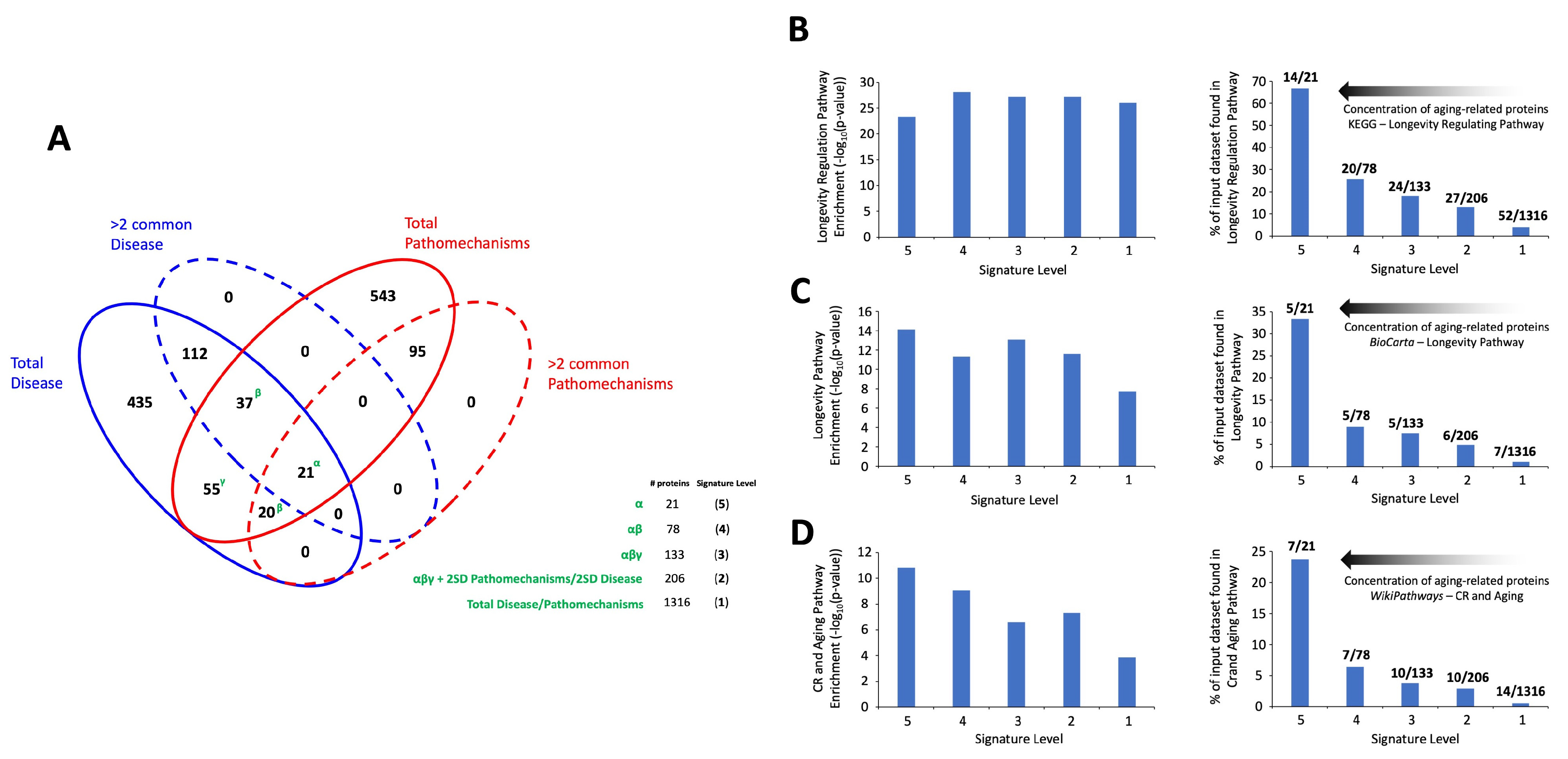
3.1. Signature Generation for Common Diseases and Generic Aging Mechanisms
3.2. Signature Refinement and Analysis for Core Properties
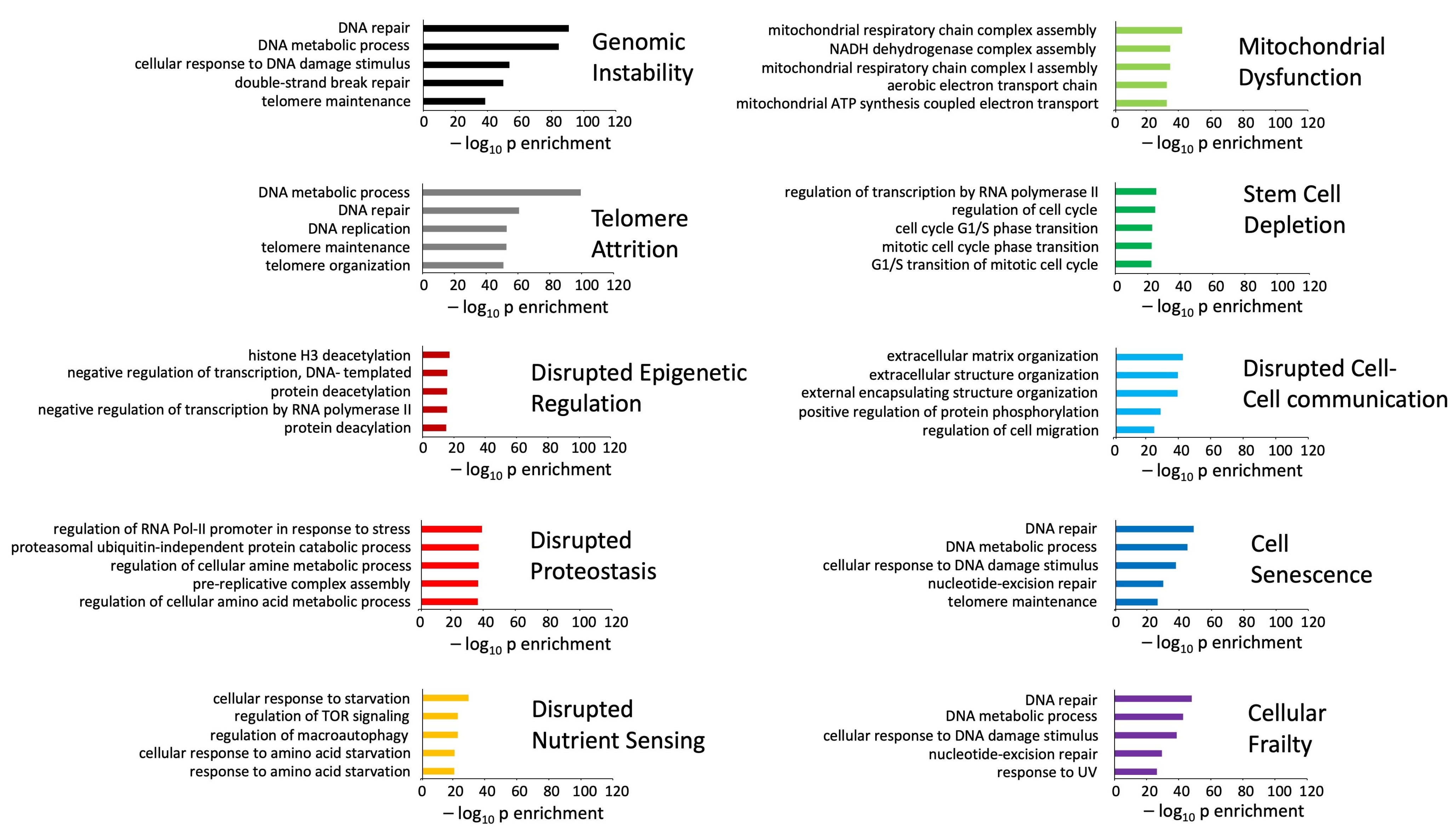
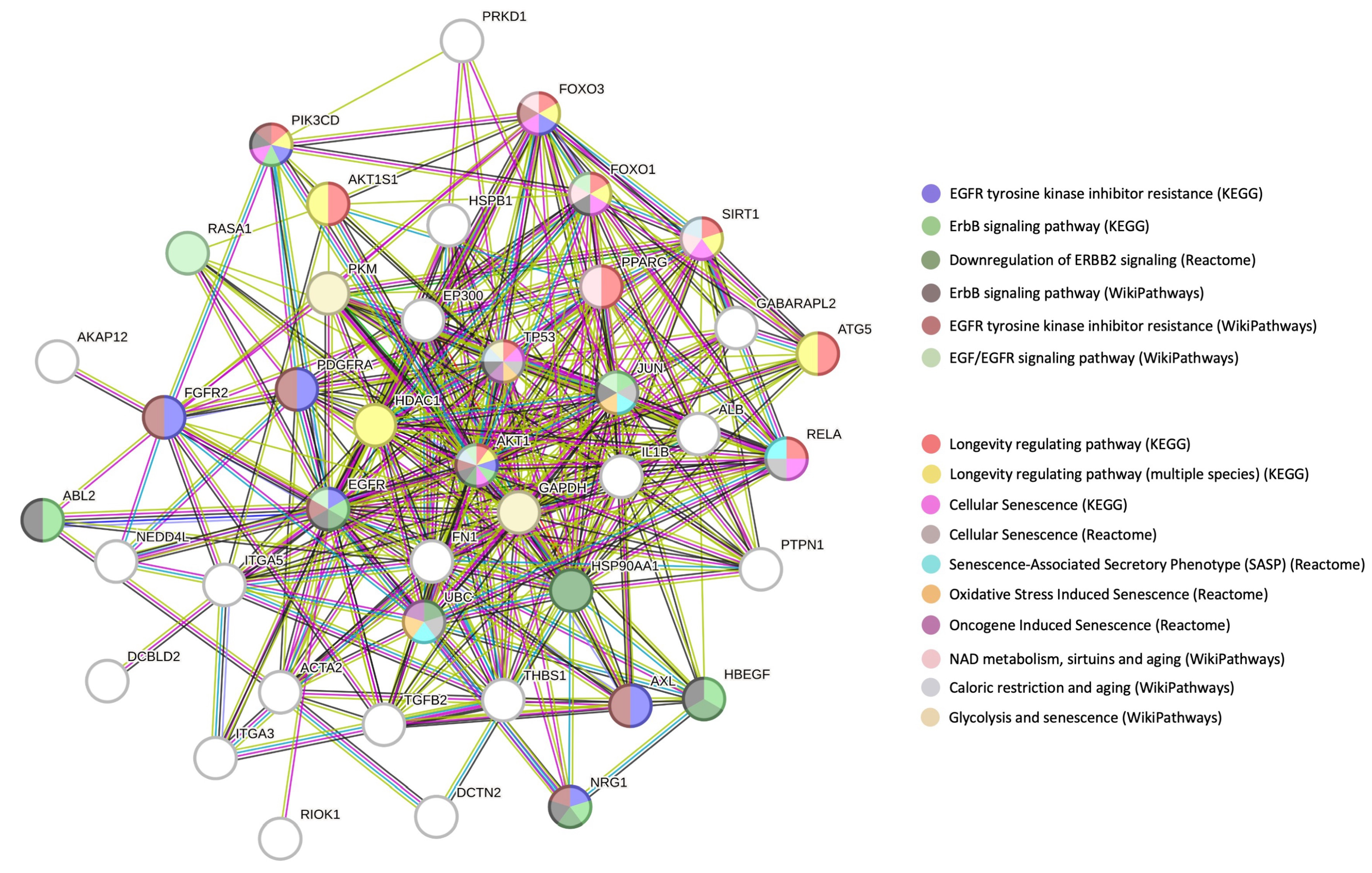
3.3. Multilevel Functional Analyses of Aging Mechanisms and Disease Processes
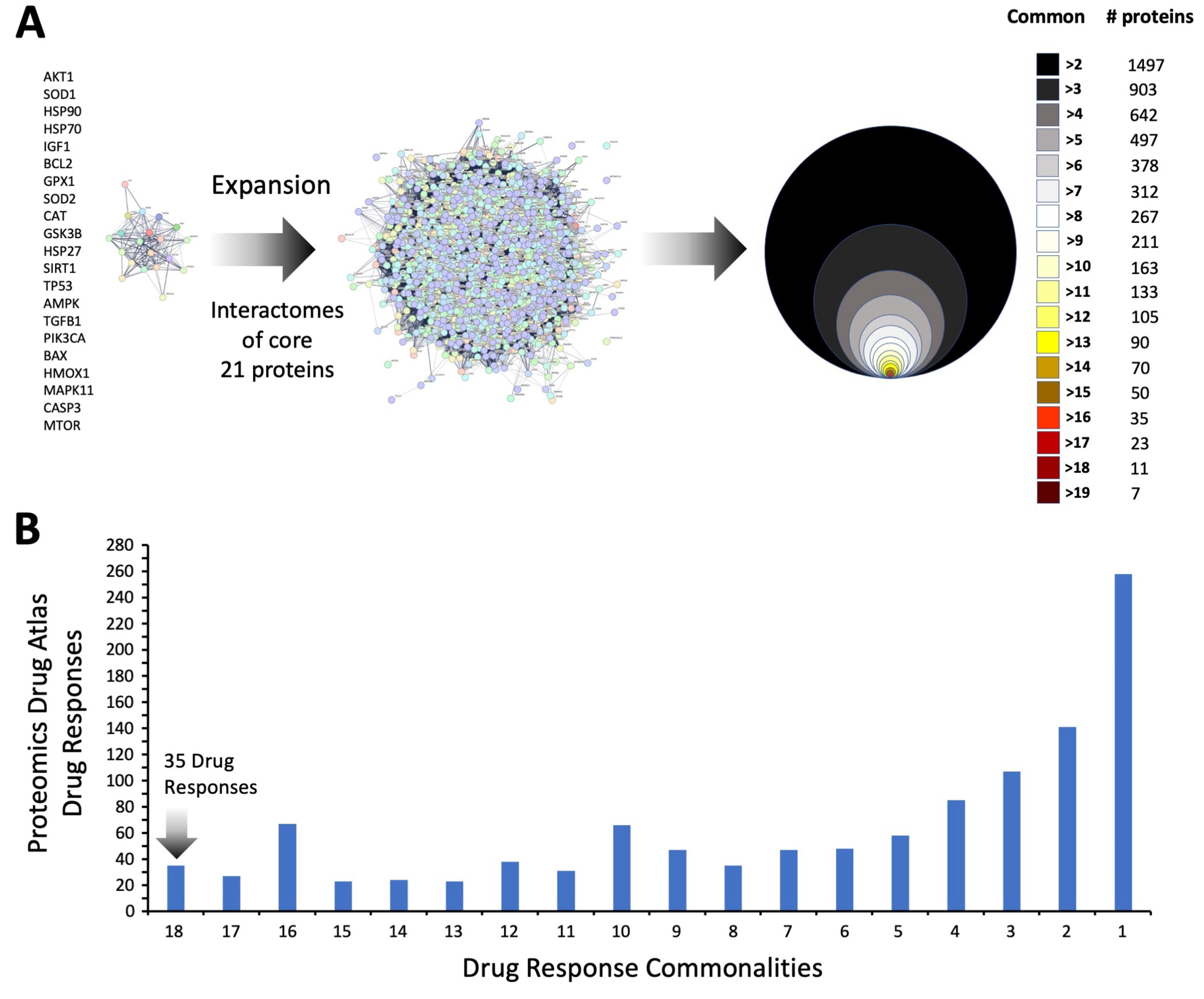
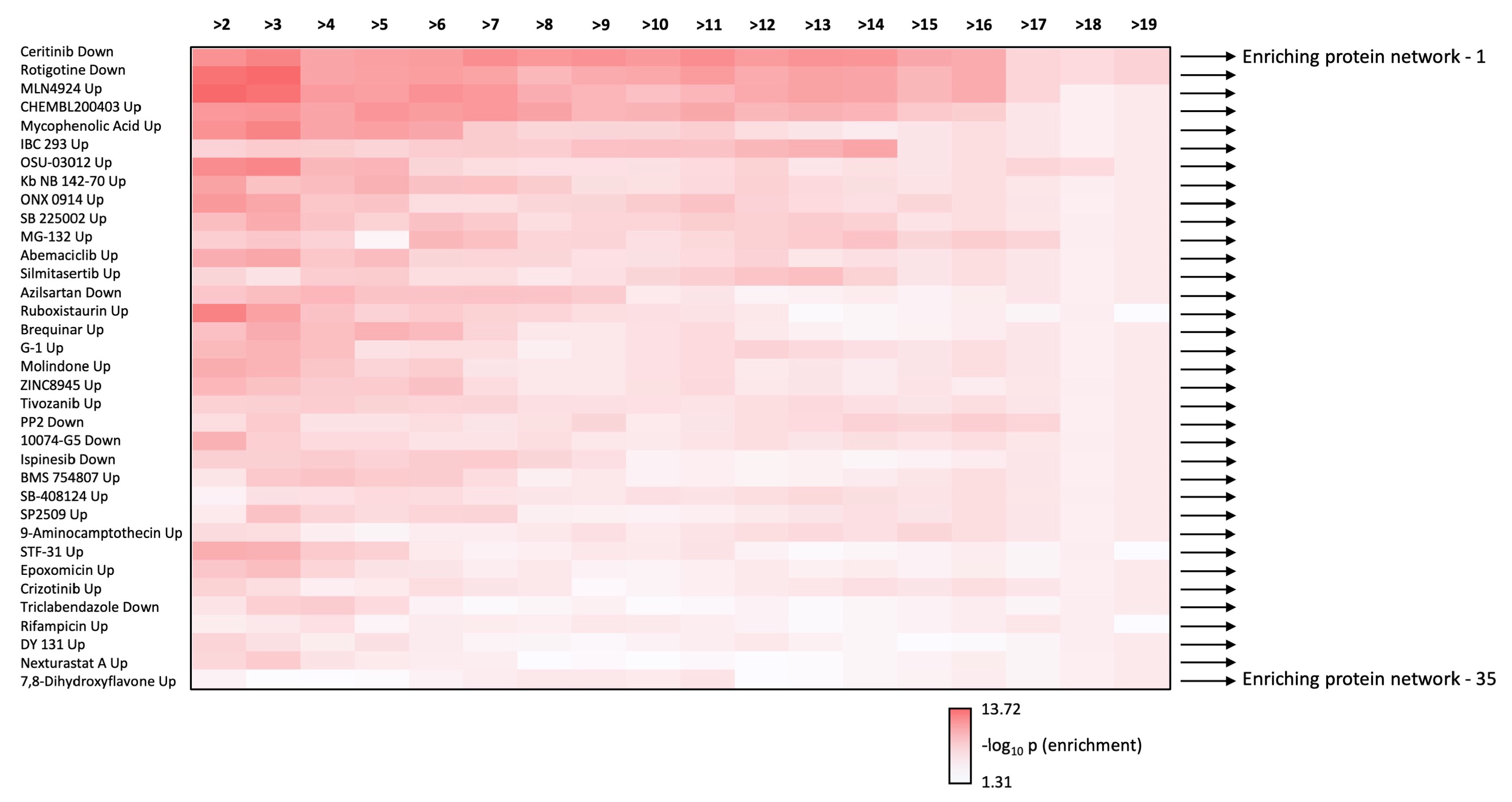
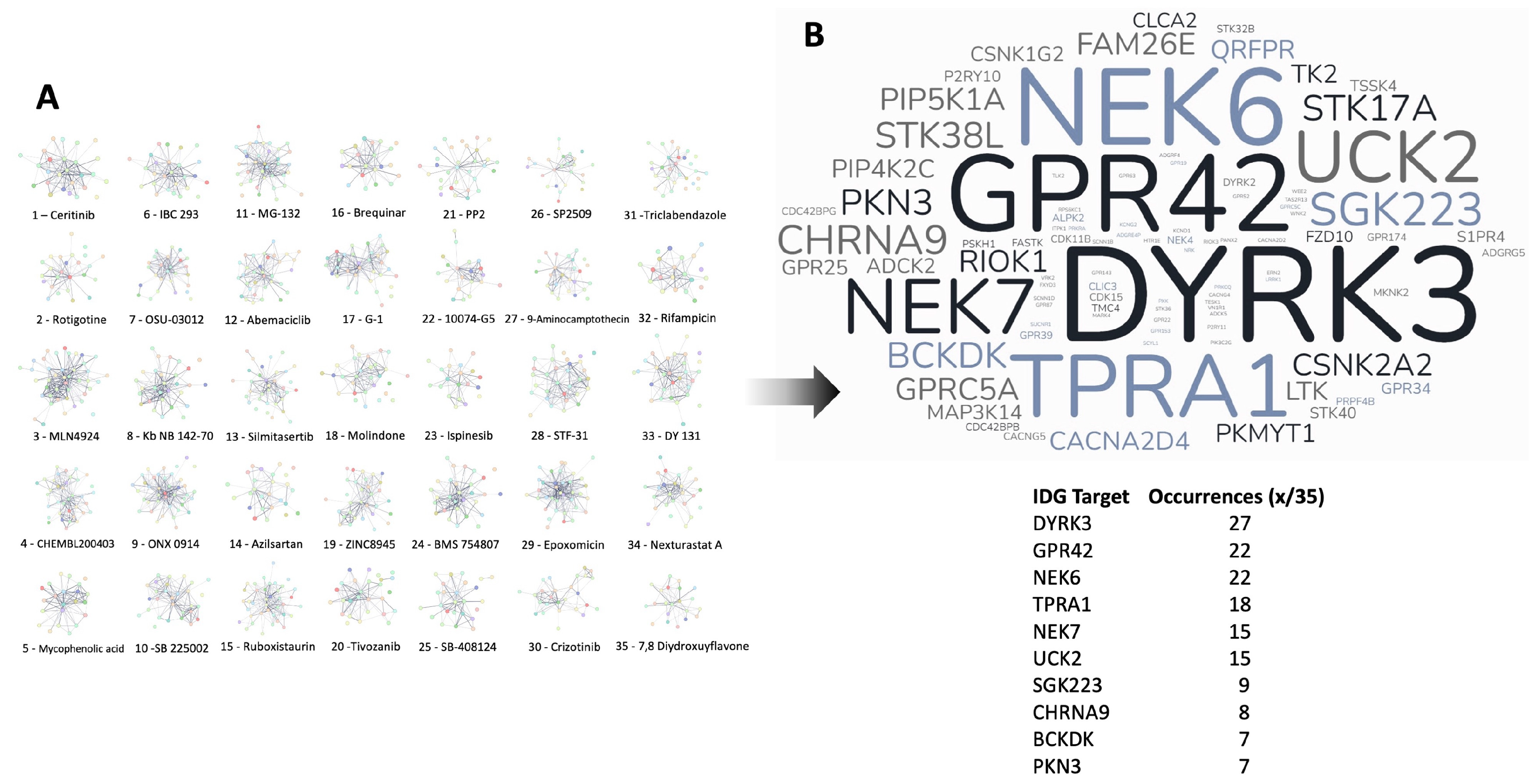
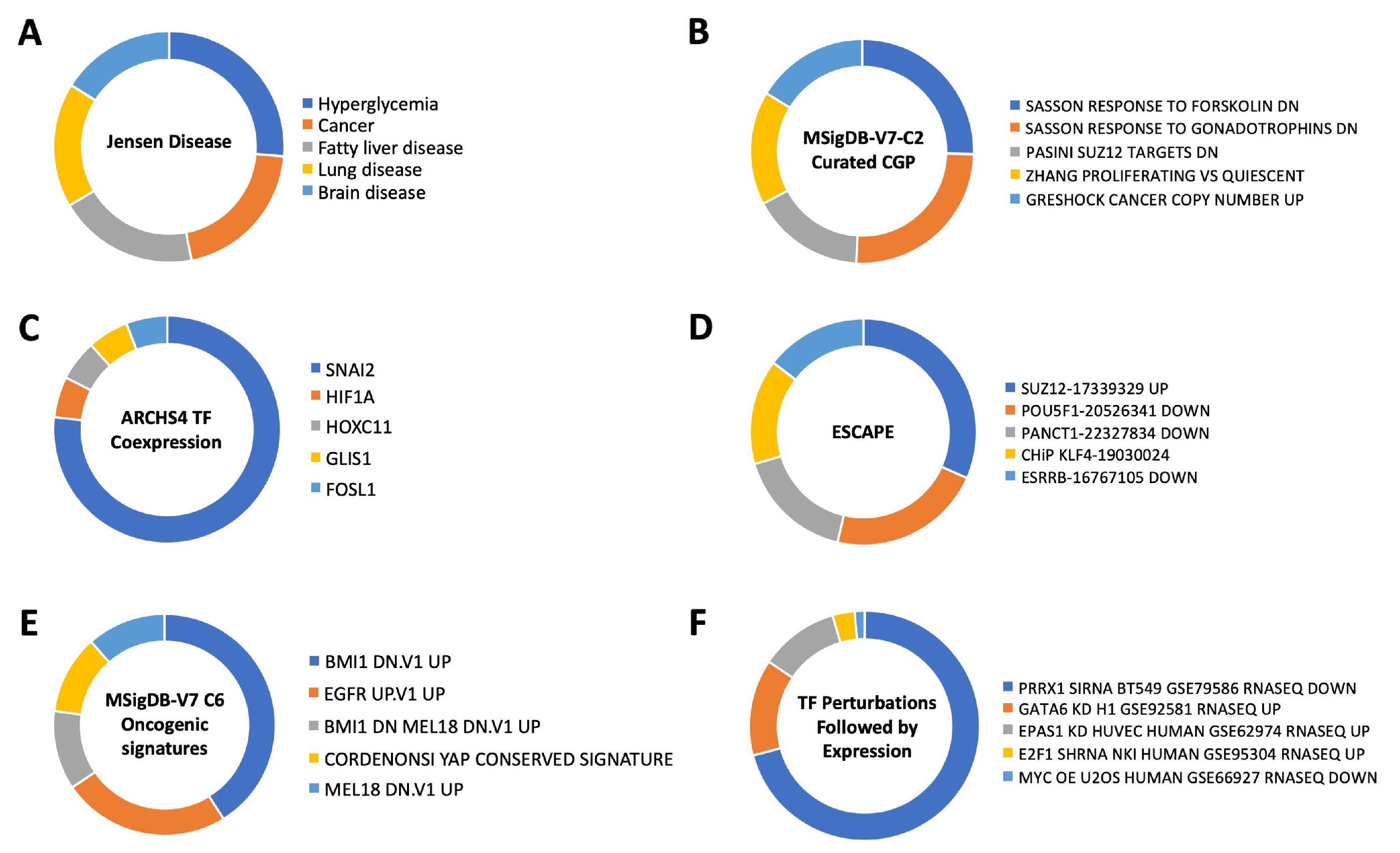
3.4. Therapeutic Signature Analysis of the Aging-Disease Nexus Core Expansion
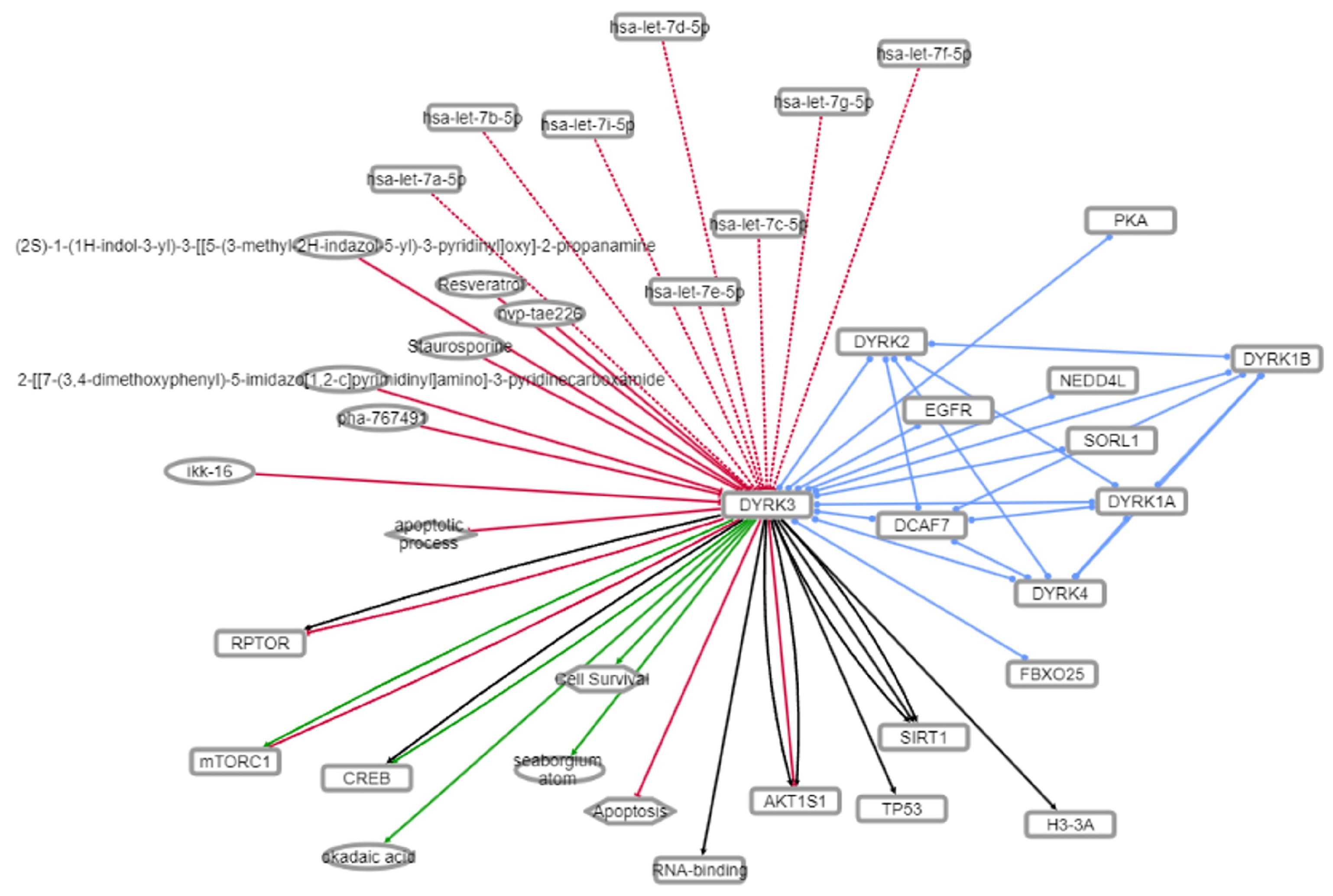
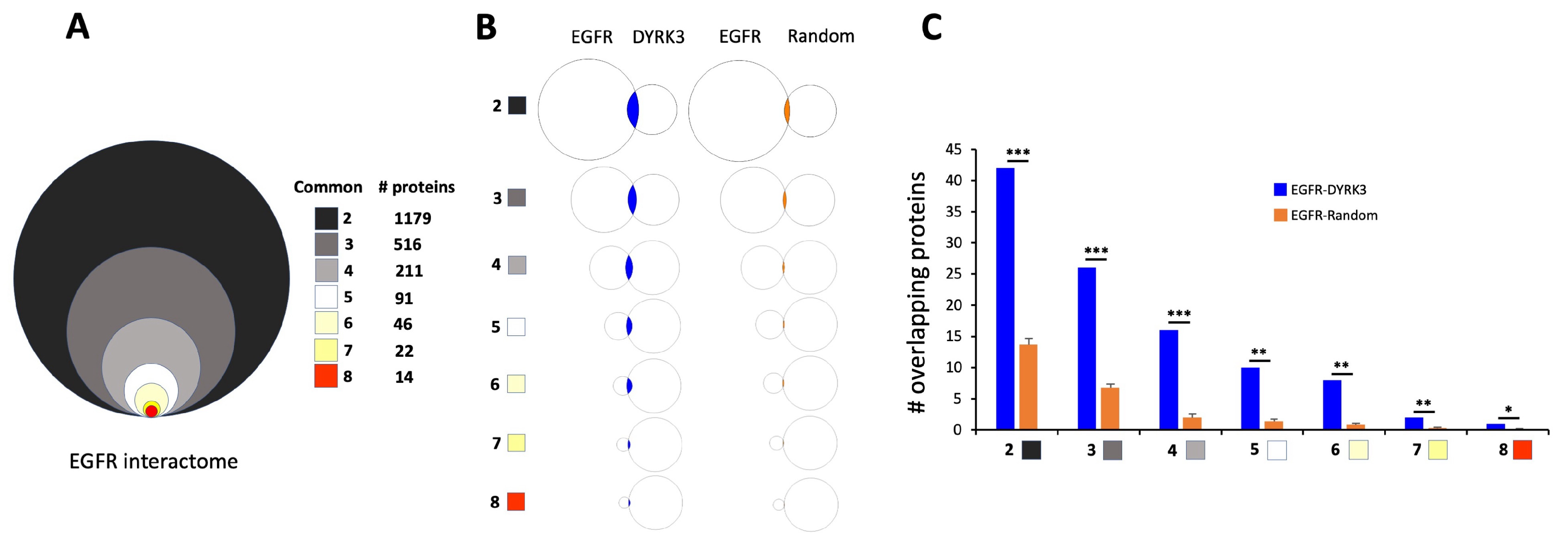
3.5. Mechanistic Investigation of the DYRK3-Aging/Disease Nexus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- van Gastel, J.; Boddaert, J.; Jushaj, A.; Premont, R.T.; Luttrell, L.M.; Janssens, J.; Martin, B.; Maudsley, S. GIT2-A keystone in ageing and age-related disease. Ageing Res. Rev. 2018, 43, 46–63. [Google Scholar] [CrossRef]
- Leysen, H.; van Gastel, J.; Hendrickx, J.O.; Santos-Otte, P.; Martin, B.; Maudsley, S. G Protein-Coupled Receptor Systems as Crucial Regulators of DNA Damage Response Processes. Int. J. Mol. Sci. 2018, 19, 2919. [Google Scholar] [CrossRef]
- McEwen, B.S. Central effects of stress hormones in health and disease: Understanding the protective and damaging effects of stress and stress mediators. Eur. J. Pharmacol. 2008, 583, 174–185. [Google Scholar] [CrossRef]
- Paronetto, M.P.; Passacantilli, I.; Sette, C. Alternative splicing, and cell survival: From tissue homeostasis to disease. Cell Death Differ. 2016, 23, 1919–1929. [Google Scholar] [CrossRef]
- Santos-Otte, P.; Leysen, H.; van Gastel, J.; Hendrickx, J.O.; Martin, B.; Maudsley, S. G Protein-Coupled Receptor Systems and Their Role in Cellular Senescence. Comput. Struct. Biotechnol. J. 2019, 17, 1265–1277. [Google Scholar] [CrossRef]
- Coppens, V.; De Wachter, O.; Goossens, J.; Hendrix, J.; Maudsley, S.; Azmi, A.; van Gastel, J.; Van Saet, A.; Lauwers, T.; Morrens, M. Profiling of the Peripheral Blood Mononuclear Cell Proteome in Schizophrenia and Mood Disorders for the Discovery of Discriminatory Biomarkers: A Proof-of-Concept Study. Neuropsychobiology 2020, 79, 324–334. [Google Scholar] [CrossRef] [PubMed]
- van Gastel, J.; Hendrickx, J.O.; Leysen, H.; Martin, B.; Veenker, L.; Beuning, S.; Coppens, V.; Morrens, M.; Maudsley, S. Enhanced Molecular Appreciation of Psychiatric Disorders Through High-Dimensionality Data Acquisition and Analytics. Methods Mol. Biol. 2019, 2011, 671–723. [Google Scholar] [CrossRef]
- Opdebeeck, B.; Maudsley, S.; Azmi, A.; De Maré, A.; De Leger, W.; Meijers, B.; Verhulst, A.; Evenepoel, P.; D’Haese, P.C.; Neven, E. Indoxyl Sulfate and p-Cresyl Sulfate Promote Vascular Calcification and Associate with Glucose Intolerance. J. Am. Soc. Nephrol. 2019, 30, 751–766. [Google Scholar] [CrossRef]
- Curtis, E.; Litwic, A.; Cooper, C.; Dennison, E. Determinants of Muscle and Bone Aging. J. Cell Physiol. 2015, 230, 2618–2625. [Google Scholar] [CrossRef] [PubMed]
- Laursen, K.R.; Hulman, A.; Witte, D.R.; Terkildsen Maindal, H. Social relations, depressive symptoms, and incident type 2 diabetes mellitus: The English Longitudinal Study of Ageing. Diabetes Res. Clin. Pract. 2017, 126, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Neergaard, J.S.; Dragsbæk, K.; Kehlet, S.N.; Hansen, H.B.; Hansen, G.; Byrjalsen, I.; Alexandersen, P.; Lindgren, L.M.; Bihlet, A.R.; Riis, B.J.; et al. Cohort Profile: The Prospective Epidemiological Risk Factor (PERF) study. Int. J. Epidemiol. 2017, 46, 1104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maudsley, S.; Devanarayan, V.; Martin, B.; Geerts, H.; Brain Health Modeling Initiative (BHMI). Intelligent and effective informatic deconvolution of “Big Data” and its future impact on the quantitative nature of neurodegenerative disease therapy. Alzheimer’s Dement. 2018, 14, 961–975. [Google Scholar] [CrossRef]
- Hendrickx, J.O.; van Gastel, J.; Leysen, H.; Martin, B.; Maudsley, S. High-dimensionality Data Analysis of Pharmacological Systems Associated with Complex Diseases. Pharmacol. Rev. 2020, 72, 191–217. [Google Scholar] [CrossRef]
- Pedditzi, E.; Peters, R.; Beckett, N. The risk of overweight/obesity in mid-life and late life for the development of dementia: A systematic review and meta-analysis of longitudinal studies. Age Ageing 2016, 45, 14–21. [Google Scholar] [CrossRef]
- Rogulj, D.; El Aklouk, I.; Konjevoda, P.; Ljubić, S.; Pibernik Okanović, M.; Barbir, A.; Luburić, M.; Radman, M.; Budinski, N.; Vučić Lovrenčić, M. Age-dependent systemic DNA damage in early Type 2 Diabetes mellitus. Acta Biochim. Pol. 2017, 64, 233–238. [Google Scholar] [CrossRef]
- Mendoza-Núñez, V.M.; Mendoza-Soto, A.B. Is Aging a Disease? A Critical Review Within the Framework of Ageism. Cureus 2024, 16, e54834. [Google Scholar] [CrossRef]
- Gladyshev, T.V.; Gladyshev, V.N. A Disease or Not a Disease? Aging As a Pathology. Trends Mol. Med. 2016, 22, 995–996. [Google Scholar] [CrossRef]
- Le Couteur, D.G.; Thillainadesan, J. What Is an Aging-Related Disease? An Epidemiological Perspective. J. Gerontol. A Biol. Sci. Med. Sci. 2022, 77, 2168–2174. [Google Scholar] [CrossRef]
- Franceschi, C.; Garagnani, P.; Morsiani, C.; Conte, M.; Santoro, A.; Grignolio, A.; Monti, D.; Capri, M.; Salvioli, S. The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front. Med. 2018, 5, 61. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Ren, Y.; Wang, Y.; Fang, J.; Yue, H.; Ma, S.; Guan, F. Aging and age-related diseases: From mechanisms to therapeutic strategies. Biogerontology 2021, 22, 165–187. [Google Scholar] [CrossRef]
- Novoselov, V.M. Is aging a disease? Adv. Gerontol. 2017, 30, 836–840. [Google Scholar] [CrossRef] [PubMed]
- Saborido, C.; García-Barranquero, P. Is Aging a Disease? The Theoretical Definition of Aging in the Light of the Philosophy of Medicine. J. Med. Philos. 2022, 47, 770–783. [Google Scholar] [CrossRef]
- Rattan, S.I. Aging is not a disease: Implications for intervention. Aging Dis. 2014, 5, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Bakula, D.; Aliper, A.M.; Mamoshina, P.; Petr, M.A.; Teklu, A.; Baur, J.A.; Campisi, J.; Ewald, C.Y.; Georgievskaya, A.; Gladyshev, V.N.; et al. Aging, and drug discovery. Aging 2018, 10, 3079–3088. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Champ, C.; Day, J.; Aarts, E.; Bahr, B.A.; Bakker, M.; Bánáti, D.; Calabrese, V.; Cederholm, T.; Cryan, J.; et al. Poor cognitive ageing: Vulnerabilities, mechanisms and the impact of nutritional interventions. Ageing Res. Rev. 2018, 42, 40–55. [Google Scholar] [CrossRef]
- Bland, J.S. Age as a Modifiable Risk Factor for Chronic Disease. Integr. Med. 2018, 17, 16–19. [Google Scholar]
- MacNee, W.; Rabinovich, R.A.; Choudhury, G. Ageing and the border between health and disease. Eur. Respir. J. 2014, 44, 1332–1352. [Google Scholar] [CrossRef]
- Kourtis, N.; Tavernarakis, N. Cellular Stress response pathways and ageing: Intricate molecular relationships. EMBO J. 2011, 30, 2520–2531. [Google Scholar] [CrossRef]
- Tenchov, R.; Sasso, J.M.; Wang, X.; Zhou, Q.A. Aging Hallmarks and Progression and Age-Related Diseases: A Landscape View of Research Advancement. ACS Chem. Neurosci. 2024, 15, 1–30. [Google Scholar] [CrossRef]
- Gaspar-Silva, F.; Trigo, D.; Magalhaes, J. Ageing in the brain: Mechanisms and rejuvenating strategies. Cell Mol. Life Sci. 2023, 80, 190. [Google Scholar] [CrossRef] [PubMed]
- Ho, E.; Qualls, C.; Villareal, D.T. Effect of Diet, Exercise, or Both on Biological Age and Healthy Aging in Older Adults with Obesity: Secondary Analysis of a Randomized Controlled Trial. J. Nutr. Health Aging 2022, 26, 552–557. [Google Scholar] [CrossRef]
- Tong, J.; Hei, T.K. Aging and age-related health effects of ionizing radiation. Radiat. Med. Prot. 2020, 1, 15–23. [Google Scholar] [CrossRef]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757–772. [Google Scholar] [CrossRef]
- Schumacher, B.; Pothof, J.; Vijg, J.; Hoeijmakers, J.H.J. The central role of DNA damage in the ageing process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, X.; Luo, J.; Bao, T.; Wang, S.; Wu, X. Molecular mechanisms of aging and anti-aging strategies. Cell Commun. Signal 2024, 22, 285. [Google Scholar] [CrossRef] [PubMed]
- Yousefzadeh, M.; Henpita, C.; Vyas, R.; Soto-Palma, C.; Robbins, P.; Niedernhofer, L. DNA damage-how and why we age? Elife 2021, 10, e62852. [Google Scholar] [CrossRef]
- van Gastel, J.; Leysen, H.; Boddaert, J.; Vangenechten, L.; Luttrell, L.M.; Martin, B.; Maudsley, S. Aging-related modifications to G protein-coupled receptor signaling diversity. Pharmacol. Ther. 2021, 223, 107793. [Google Scholar] [CrossRef]
- Guo, J.; Huang, X.; Dou, L.; Yan, M.; Shen, T.; Tang, W.; Li, J. Aging, and aging-related diseases: From molecular mechanisms to interventions and treatments. Signal Transduct. Target. Ther. 2022, 7, 391. [Google Scholar] [CrossRef]
- Ukraintseva, S.; Arbeev, K.; Duan, M.; Akushevich, I.; Kulminski, A.; Stallard, E.; Yashin, A. Decline in biological resilience as key manifestation of aging: Potential mechanisms and role in health and longevity. Mech. Ageing Dev. 2021, 194, 111418. [Google Scholar] [CrossRef] [PubMed]
- Lau, E.; Venkatraman, V.; Thomas, C.T.; Wu, J.C.; Van Eyk, J.E.; Lam, M.P.Y. Identifying High-Priority Proteins Across the Human Diseasome Using Semantic Similarity. J. Proteome Res. 2018, 17, 4267–4278. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Pillich, R.T.; Chen, J.; Churas, C.; Fong, D.; Gyori, B.M.; Ideker, T.; Karis, K.; Liu, S.N.; Ono, K.; Pico, A.; et al. NDEx IQuery: A multi-method network gene set analysis leveraging the Network Data Exchange. Bioinformatics 2023, 39, btad118. [Google Scholar] [CrossRef]
- Berginski, M.E.; Moret, N.; Liu, C.; Goldfarb, D.; Sorger, P.K.; Gomez, S.M. The Dark Kinase Knowledgebase: An online compendium of knowledge and experimental results of understudied kinases. Nucleic Acids Res. 2021, 49, D529–D535. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Gerstner, N.; Kehl, T.; Lenhof, K.; Müller, A.; Mayer, C.; Eckhart, L.; Grammes, N.L.; Diener, C.; Hart, M.; Hahn, O.; et al. Walter J, Wyss-Coray T, Meese E, Keller A, Lenhof HP. GeneTrail 3: Advanced high-throughput enrichment analysis. Nucleic Acids Res. 2020, 48, W515–W520. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Maudsley, S.; Pierce, K.L.; Zamah, A.M.; Miller, W.E.; Ahn, S.; Daaka, Y.; Lefkowitz, R.J.; Luttrell, L.M. The beta(2)-adrenergic receptor mediates extracellular signal-regulated kinase activation via assembly of a multi-receptor complex with the epidermal growth factor receptor. J. Biol. Chem. 2000, 275, 9572–9580. [Google Scholar] [CrossRef] [PubMed]
- van Gastel, J.; Leysen, H.; Santos-Otte, P.; Hendrickx, J.O.; Azmi, A.; Martin, B.; Maudsley, S. The RXFP3 receptor is functionally associated with cellular responses to oxidative stress and DNA damage. Aging 2019, 11, 11268–11313. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012, 40, D857–D861. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, A.; Schilder, B.M.; Wojciechowicz, M.L.; Torre, D.; Kuleshov, M.V.; Keenan, A.B.; Ma’ayan, A. Geneshot: Search engine for ranking genes from arbitrary text queries. Nucleic Acids Res. 2019, 47, W571–W577. [Google Scholar] [CrossRef]
- Panneerselvam, K.; Porras, P.; Del-Toro, N.; Perfetto, L.; Shrivastava, A.; Ragueneau, E.; Reyes, J.J.M.; Orchard, S.; Hermjakob, H. IMEx Consortium Curators. IntAct Database for Accessing IMEx’s Contextual Metadata of Molecular Interactions. Curr. Protoc. 2024, 4, e70018. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.S.; Krishnan, A.; Wong, A.K.; Ricciotti, E.; Zelaya, R.A.; Himmelstein, D.S.; Zhang, R.; Hartmann, B.M.; Zaslavsky, E.; Sealfon, S.C.; et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015, 47, 569–576. [Google Scholar] [CrossRef]
- Mitchell, D.C.; Kuljanin, M.; Li, J.; Van Vranken, J.G.; Bulloch, N.; Schweppe, D.K.; Huttlin, E.L.; Gygi, S.P. A proteome-wide atlas of drug mechanism of action. Nat. Biotechnol. 2023, 41, 845–857. [Google Scholar] [CrossRef]
- Yoshida, S.; Yoshida, K. New insights into the roles for DYRK family in mammalian development and congenital diseases. Genes. Dis. 2022, 10, 758–770. [Google Scholar] [CrossRef]
- Boni, J.; Rubio-Perez, C.; López-Bigas, N.; Fillat, C.; de la Luna, S. The DYRK Family of Kinases in Cancer: Molecular Functions and Therapeutic Opportunities. Cancers 2020, 12, 2106. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Williams, J.G.; Schug, T.T.; Li, X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J. Biol. Chem. 2010, 285, 13223–13232. [Google Scholar] [CrossRef]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Santos-Durán, G.N.; Barreiro-Iglesias, A. Roles of dual specificity tyrosine-phosphorylation-regulated kinase 2 in nervous system development and disease. Front. Neurosci. 2022, 16, 994256. [Google Scholar] [CrossRef] [PubMed]
- Belal, A.; Abdel Gawad, N.M.; Mehany, A.B.M.; Abourehab, M.A.S.; Elkady, H.; Al-Karmalawy, A.A.; Ismael, A.S. Design, synthesis, and molecular docking of new fused 1H-pyrroles, pyrrolo [3,2-d]pyrimidines and pyrrolo[3,2-e][1, 4]diazepine derivatives as potent EGFR/CDK2 inhibitors. J. Enzyme Inhib. Med. Chem. 2022, 37, 1884–1902. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.; Sharma, S.D.; Brichkina, A.; Pfefferle, P.; Keber, U.; Pagenstecher, A.; Lauth, M. DYRK3 contributes to differentiation and hypoxic control in neuroblastoma. Biochem. Biophys. Res. Commun. 2021, 567, 215–221. [Google Scholar] [CrossRef]
- Guo, Y.; Cui, Y.; Li, Y.; Jin, X.; Wang, D.; Lei, M.; Chen, F.; Liu, Y.; Xu, J.; Yao, G.; et al. Cytoplasmic YAP1-mediated ESCRT-III assembly promotes autophagic cell death and is ubiquitinated by NEDD4L in breast cancer. Cancer Commun. 2023, 43, 582–612. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, J.; Fu, J.; Du, L.; Jeong, H.; West, T.; Xiang, L.; Peng, Q.; Hou, Z.; Cai, H.; et al. Neuroprotective role of Sirt1 in mammalian models of Huntington’s disease through activation of multiple Sirt1 targets. Nat. Med. 2011, 18, 153–158. [Google Scholar] [CrossRef]
- Mota-Martorell, N.; Jové, M.; Pamplona, R. mTOR Complex 1 Content and Regulation Is Adapted to Animal Longevity. Int. J. Mol. Sci. 2022, 23, 8747. [Google Scholar] [CrossRef]
- Ganner, A.; Gerber, J.; Ziegler, A.K.; Li, Y.; Kandzia, J.; Matulenski, T.; Kreis, S.; Breves, G.; Klein, M.; Walz, G.; et al. CBP-1/p300 acetyltransferase regulates SKN-1/Nrf cellular levels, nuclear localization, and activity in C. elegans. Exp. Gerontol. 2019, 126, 110690. [Google Scholar] [CrossRef]
- Ciummo, S.L.; D’Antonio, L.; Sorrentino, C.; Fieni, C.; Lanuti, P.; Stassi, G.; Todaro, M.; Di Carlo, E. The C-X-C Motif Chemokine Ligand 1 Sustains Breast Cancer Stem Cell Self-Renewal and Promotes Tumor Progression and Immune Escape Programs. Front. Cell Dev. Biol. 2021, 9, 689286. [Google Scholar] [CrossRef] [PubMed]
- Doshida, Y.; Sano, H.; Iwabuchi, S.; Aigaki, T.; Yoshida, M.; Hashimoto, S.; Ishigami, A. Age-associated changes in the transcriptomes of non-cultured adipose-derived stem cells from young and old mice assessed via single-cell transcriptome analysis. PLoS ONE 2020, 15, e0242171. [Google Scholar] [CrossRef] [PubMed]
- Gross, K.M.; Zhou, W.; Breindel, J.L.; Ouyang, J.; Jin, D.X.; Sokol, E.S.; Gupta, P.B.; Huber, K.; Zou, L.; Kuperwasser, C. Loss of Slug Compromises DNA Damage Repair and Accelerates Stem Cell Aging in Mammary Epithelium. Cell Rep. 2019, 28, 394–407.e6. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, S.; Fang, M.; Ni, B.; Lu, D.; Martin, B.; Maudsley, S. Central role of the EGF receptor in neurometabolic aging. Int. J. Endocrinol. 2012, 2012, 739428. [Google Scholar] [CrossRef]
- Vermeulen, Z.; Hervent, A.S.; Dugaucquier, L.; Vandekerckhove, L.; Rombouts, M.; Beyens, M.; Schrijvers, D.M.; De Meyer, G.R.Y.; Maudsley, S.; De Keulenaer, G.W.; et al. Inhibitory actions of the NRG-1/ErbB4 pathway in macrophages during tissue fibrosis in the heart, skin, and lung. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H934–H945. [Google Scholar] [CrossRef]
- Akhoon, B.A.; Rathor, L.; Pandey, R. Withanolide A extends the lifespan in human EGFR-driven cancerous Caenorhabditis elegans. Exp. Gerontol. 2018, 104, 113–117. [Google Scholar] [CrossRef]
- Won, H.Y.; Lee, J.Y.; Shin, D.H.; Park, J.H.; Nam, J.S.; Kim, H.C.; Kong, G. Loss of Mel-18 enhances breast cancer stem cell activity and tumorigenicity through activating Notch signaling mediated by the Wnt/TCF pathway. FASEB J. 2012, 26, 5002–5013. [Google Scholar] [CrossRef]
- Yoon, M.H.; Kang, S.M.; Lee, S.J.; Woo, T.G.; Oh, A.Y.; Park, S.; Ha, N.C.; Park, B.J. p53 induces senescence through Lamin A/C stabilization-mediated nuclear deformation. Cell Death Dis. 2019, 10, 107. [Google Scholar] [CrossRef]
- Miki, J.; Fujimura, Y.; Koseki, H.; Kamijo, T. Polycomb complexes regulate cellular senescence by repression of ARF in cooperation with E2F3. Genes Cells 2007, 12, 1371–1382. [Google Scholar] [CrossRef]
- Kennerdell, J.R.; Liu, N.; Bonini, N.M. MiR-34 inhibits polycomb repressive complex 2 to modulate chaperone expression and promote healthy brain aging. Nat. Commun. 2018, 9, 4188. [Google Scholar] [CrossRef]
- Ito, T.; Teo, Y.V.; Evans, S.A.; Neretti, N.; Sedivy, J.M. Regulation of Cellular Senescence by Polycomb Chromatin Modifiers through Distinct DNA Damage- and Histone Methylation-Dependent Pathways. Cell Rep. 2018, 22, 3480–3492. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Mukherjee, A.K.; Roy, S.S.; Bagri, S.; Lier, S.; Verma, M.; Sengupta, A.; Kumar, M.; Nesse, G.; Pandey, D.P.; et al. Human telomerase is directly regulated by non-telomeric TRF2-G-quadruplex interaction. Cell Rep. 2021, 35, 109154. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ang, Y.S.; Sevilla, A.; Lemischka, I.R.; Ma’ayan, A. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput. Biol. 2014, 10, e1003777. [Google Scholar] [CrossRef]
- Xu, H.; Baroukh, C.; Dannenfelser, R.; Chen, E.Y.; Tan, C.M.; Kou, Y.; Kim, Y.E.; Lemischka, I.R.; Ma’ayan, A. ESCAPE: Database for integrating high-content published data collected from human and mouse embryonic stem cells. Database 2013, 2013, bat045. [Google Scholar] [CrossRef]
- Shi, L.; Tang, X.; Qian, M.; Liu, Z.; Meng, F.; Fu, L.; Wang, Z.; Zhu, W.G.; Huang, J.D.; Zhou, Z.; et al. A SIRT1-centered circuitry regulates breast cancer stemness and metastasis. Oncogene 2018, 37, 6299–6315. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, H.; Zheng, Y.; Zhou, J.; Yuan, J.; Yu, Y.; Wang, J. Resveratrol Exerts Antioxidant Effects by Activating SIRT2 to Deacetylate Prx1. Biochemistry 2017, 56, 6325–6328. [Google Scholar] [CrossRef]
- Mo, C.; Guo, J.; Qin, J.; Zhang, X.; Sun, Y.; Wei, H.; Cao, D.; Zhang, Y.; Zhao, C.; Xiong, Y.; et al. Single-cell transcriptomics of LepR-positive skeletal cells reveals heterogeneous stress-dependent stem and progenitor pools. EMBO J. 2022, 41, e108415. [Google Scholar] [CrossRef]
- Sun, L.; Han, T.; Zhang, X.; Liu, X.; Li, P.; Shao, M.; Dong, S.; Li, W. PRRX1 isoform PRRX1A regulates the stemness phenotype and epithelial-mesenchymal transition (EMT) of cancer stem-like cells (CSCs) derived from non-small cell lung cancer (NSCLC). Transl. Lung Cancer Res. 2020, 9, 731–744. [Google Scholar] [CrossRef]
- Liu, T.; Li, K.; Wang, Y.; Li, H.; Zhao, H. Evaluating the Utilities of Foundation Models in Single-cell Data Analysis. bioRxiv 2024, preprint. [Google Scholar] [CrossRef]
- Agathokleous, E.; Saitanis, C.J.; Fang, C.; Yu, Z. Use of ChatGPT: What does it mean for biology and environmental science? Sci. Total Environ. 2023, 888, 164154. [Google Scholar] [CrossRef]
- Li, T.; Shetty, S.; Kamath, A.; Jaiswal, A.; Jiang, X.; Ding, Y.; Kim, Y. CancerGPT: Few-shot Drug Pair Synergy Prediction using Large Pre-trained Language Models. arXiv 2023, arXiv:2304.10946v1. [Google Scholar] [CrossRef]
- Fawaz, A.; Ferraresi, A.; Isidoro, C. Systems Biology in Cancer Diagnosis Integrating Omics Technologies, and Artificial Intelligence to Support Physician Decision Making. J. Pers. Med. 2023, 13, 1590. [Google Scholar] [CrossRef]
- Wu, X.; Li, W.; Tu, H. Big data, and artificial intelligence in cancer research. Trends Cancer. 2023, 10, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Vandenberk, B.; Chew, D.S.; Prasana, D.; Gupta, S.; Exner, D.V. Successes and challenges of artificial intelligence in cardiology. Front. Digit. Health 2023, 5, 1201392. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, Y.; Hua, Z.; Lin, F.; Zheng, T.; Zhou, H.; Zhang, S.; Gao, J.; Wang, Z.; Shao, H.; Li, W.; et al. Detection, and classification of breast lesions using multiple information on contrast-enhanced mammography by a multiprocess deep-learning system: A multicenter study. Chin. J. Cancer Res. 2023, 35, 408–423. [Google Scholar] [CrossRef] [PubMed]
- Van Meenen, J.; Leysen, H.; Chen, H.; Baccarne, R.; Walter, D.; Martin, B.; Maudsley, S. Making Biomedical Sciences publications more accessible for machines. Med. Health Care Philos. 2022, 25, 179–190. [Google Scholar] [CrossRef]
- Gerussi, A.; Verda, D.; Cappadona, C.; Cristoferi, L.; Bernasconi, D.P.; Bottaro, S.; Carbone, M.; Muselli, M.; Invernizzi, P.; Asselta, R.; et al. LLM-PBC: Logic Learning Machine-Based Explainable Rules Accurately Stratify the Genetic Risk of Primary Biliary Cholangitis. J. Pers. Med. 2022, 12, 1587. [Google Scholar] [CrossRef] [PubMed]
- Benary, M.; Wang, X.D.; Schmidt, M.; Soll, D.; Hilfenhaus, G.; Nassir, M.; Sigler, C.; Knödler, M.; Keller, U.; Beule, D.; et al. Leveraging Large Language Models for Decision Support in Personalized Oncology. JAMA Netw. Open. 2023, 6, e2343689. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, H.; Liu, H.; Jiang, Y.; Ran, T.; Jin, X.; Xiao, X.; Lin, Z.; Chen, H.; Niu, Z. An extensive benchmark study on biomedical text generation and mining with ChatGPT. Bioinformatics 2023, 39, btad557. [Google Scholar] [CrossRef]
- Joachimiak, M.P.; Caufield, J.H.; Harris, N.L.; Kim, H.; Mungall, C.J. Gene Set Summarization using Large Language Models. arXiv 2023, arXiv:2305.13338v2. [Google Scholar]
- Xxxx, S.; Ahmad, M.H.; Rani, L.; Mondal, A.C. Convergent Molecular Pathways in Type 2 Diabetes Mellitus and Parkinson’s Disease: Insights into Mechanisms and Pathological Consequences. Mol. Neurobiol. 2022, 59, 4466–4487. [Google Scholar] [CrossRef] [PubMed]
- Argueti-Ostrovsky, S.; Alfahel, L.; Kahn, J.; Israelson, A. All Roads Lead to Rome: Different Molecular Players Converge to Common Toxic Pathways in Neurodegeneration. Cells 2021, 10, 2438. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.A.; De Smet, M.; Grant, G.D.; Khoo, T.K.; Pountney, D.L. Investigating the Convergent Mechanisms between Major Depressive Disorder and Parkinson’s Disease. Complex. Psychiatry 2021, 6, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, Y.; Shi, Y.; Zhang, Z.; Huang, C.; He, W.; Wang, C.; Shen, H.M. Autophagy in health and disease: From molecular mechanisms to therapeutic target. MedComm 2022, 3, e150. [Google Scholar] [CrossRef]
- You, Y.; Liang, W. SIRT1 and SIRT6: The role in aging-related diseases. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166815. [Google Scholar] [CrossRef]
- Li, K.; Zhao, S.; Karur, V.; Wojchowski, D.M. DYRK3 activation, engagement of protein kinase A/cAMP response element-binding protein, and modulation of progenitor cell survival. J. Biol. Chem. 2002, 277, 47052–47060. [Google Scholar] [CrossRef]
- Gallo, R.; Rai, A.K.; McIntyre, A.B.R.; Meyer, K.; Pelkmans, L. DYRK3 enables secretory trafficking by maintaining the liquid-like state of ER exit sites. Dev. Cell. 2023, 58, 1880–1897.e11. [Google Scholar] [CrossRef]
- Zhang, C.; Rabouille, C. Membrane-Bound Meet Membraneless in Health, and Disease. Cells 2019, 8, 1000. [Google Scholar] [CrossRef]
- Mediani, L.; Antoniani, F.; Galli, V.; Vinet, J.; Carrà, A.D.; Bigi, I.; Tripathy, V.; Tiago, T.; Cimino, M.; Leo, G.; et al. Hsp90-mediated regulation of DYRK3 couples stress granule disassembly and growth via mTORC1 signaling. EMBO Rep. 2021, 22, e51740. [Google Scholar] [CrossRef]
- Roudabush, F.L.; Pierce, K.L.; Maudsley, S.; Khan, K.D.; Luttrell, L.M. Transactivation of the EGF receptor mediates IGF-1-stimulated shc phosphorylation and ERK1/2 activation in COS-7 cells. J. Biol. Chem. 2000, 275, 22583–22589. [Google Scholar] [CrossRef]
- Chadwick, W.; Keselman, A.; Park, S.S.; Zhou, Y.; Wang, L.; Brenneman, R.; Martin, B.; Maudsley, S. Repetitive peroxide exposure reveals pleiotropic mitogen-activated protein kinase signaling mechanisms. J. Signal Transduct. 2011, 2011, 636951. [Google Scholar] [CrossRef] [PubMed]
- Donlon, T.A.; Morris, B.J.; He, Q.; Chen, R.; Masaki, K.H.; Allsopp, R.C.; Willcox, D.C.; Tranah, G.J.; Parimi, N.; Evans, D.S.; et al. Association of Polymorphisms in Connective Tissue Growth Factor and Epidermal Growth Factor Receptor Genes With Human Longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Detienne, G.; De Haes, W.; Ernst, U.R.; Schoofs, L.; Temmerman, L. Royalactin extends lifespan of Caenorhabditis elegans through epidermal growth factor signaling. Exp. Gerontol. 2014, 60, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Siebold, A.P.; Banerjee, R.; Tie, F.; Kiss, D.L.; Moskowitz, J.; Harte, P.J. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc. Natl. Acad. Sci. USA 2010, 107, 169–174. [Google Scholar] [CrossRef]
- Chu, L.; Qu, Y.; An, Y.; Hou, L.; Li, J.; Li, W.; Fan, G.; Song, B.L.; Li, E.; Zhang, L.; et al. Induction of senescence-associated secretory phenotype underlies the therapeutic efficacy of PRC2 inhibition in cancer. Cell Death Dis. 2022, 13, 155. [Google Scholar] [CrossRef]
- Adibfar, S.; Elveny, M.; Kashikova, H.S.; Mikhailova, M.V.; Farhangnia, P.; Vakili-Samiani, S.; Tarokhian, H.; Jadidi-Niaragh, F. The molecular mechanisms and therapeutic potential of EZH2 in breast cancer. Life Sci. 2021, 286, 120047. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L.; Fan, Z. The role and mechanisms of polycomb repressive complex 2 on the regulation of osteogenic and neurogenic differentiation of stem cells. Cell Prolif. 2021, 54, e13032. [Google Scholar] [CrossRef]
- Wang, W.; Qin, X.; Wang, R.; Xu, J.; Wu, H.; Khalid, A.; Jiang, H.; Liu, D.; Pan, F. EZH2 is involved in vulnerability to neuroinflammation and depression-like behaviors induced by chronic stress in different aged mice. J. Affect. Disord. 2020, 272, 452–464. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Junyent, M.; Noori, H.; De Schepper, R.; Frajdenberg, S.; Elsaigh, R.K.A.H.; McDonald, P.H.; Duckett, D.; Maudsley, S. Unravelling Convergent Signaling Mechanisms Underlying the Aging-Disease Nexus Using Computational Language Analysis. Curr. Issues Mol. Biol. 2025, 47, 189. https://doi.org/10.3390/cimb47030189
Junyent M, Noori H, De Schepper R, Frajdenberg S, Elsaigh RKAH, McDonald PH, Duckett D, Maudsley S. Unravelling Convergent Signaling Mechanisms Underlying the Aging-Disease Nexus Using Computational Language Analysis. Current Issues in Molecular Biology. 2025; 47(3):189. https://doi.org/10.3390/cimb47030189
Chicago/Turabian StyleJunyent, Marina, Haki Noori, Robin De Schepper, Shanna Frajdenberg, Razan Khalid Abdullah Hussen Elsaigh, Patricia H. McDonald, Derek Duckett, and Stuart Maudsley. 2025. "Unravelling Convergent Signaling Mechanisms Underlying the Aging-Disease Nexus Using Computational Language Analysis" Current Issues in Molecular Biology 47, no. 3: 189. https://doi.org/10.3390/cimb47030189
APA StyleJunyent, M., Noori, H., De Schepper, R., Frajdenberg, S., Elsaigh, R. K. A. H., McDonald, P. H., Duckett, D., & Maudsley, S. (2025). Unravelling Convergent Signaling Mechanisms Underlying the Aging-Disease Nexus Using Computational Language Analysis. Current Issues in Molecular Biology, 47(3), 189. https://doi.org/10.3390/cimb47030189






