Anthocyanin-Binding Affinity and Non-Covalent Interactions with IIS-Pathway-Related Protein Through Molecular Docking
Abstract
1. Introduction
2. Materials and Methods
2.1. Termite Collection
2.2. RNA Extraction and Illumina Sequencing
2.3. Protein Sequences of Reticulitermes chinensis Castes
2.4. Molecular Docking and Molecular Dynamics (MD) Simulation
2.5. Physicochemical Analysis of Anthocyanin Compounds for Lead-like Properties
2.6. The Top-Binding Ligands and the Inhibitory Ligand Visualization of the Aging-Related Genes
2.7. Quantitative Real-Time PCR (RT-qPCR)
3. Results and Discussion
3.1. Different Anthocyanin Compounds and Known Inhibitory Ligands Are Compared for Their Lead-like Qualities
3.2. Biochemical and Structural Analysis
3.3. Docking of Several Anthocyanin Compounds In Silico to Proteins Associated with Aging
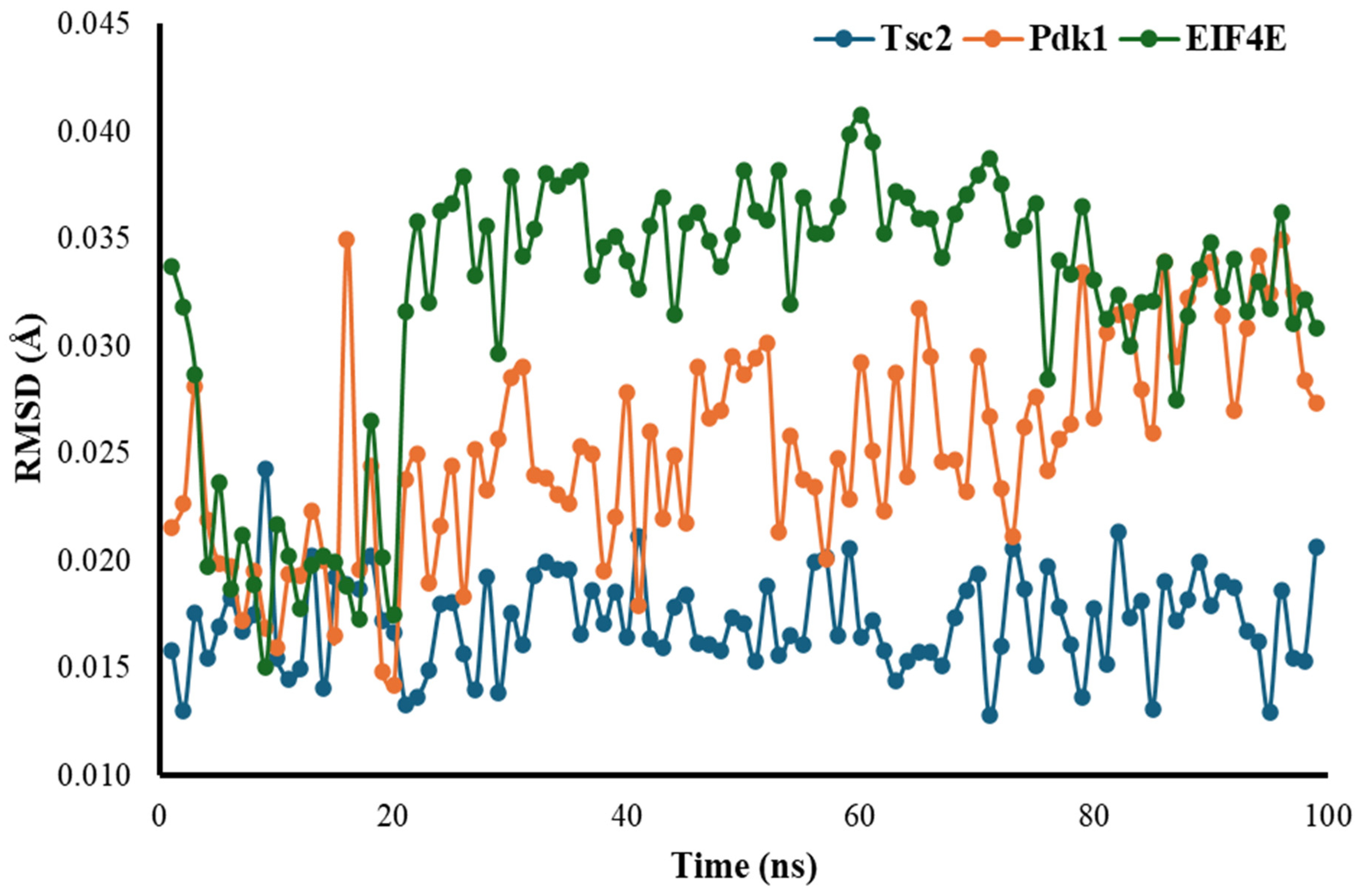
3.4. Root Mean Square Deviation (RMSD)
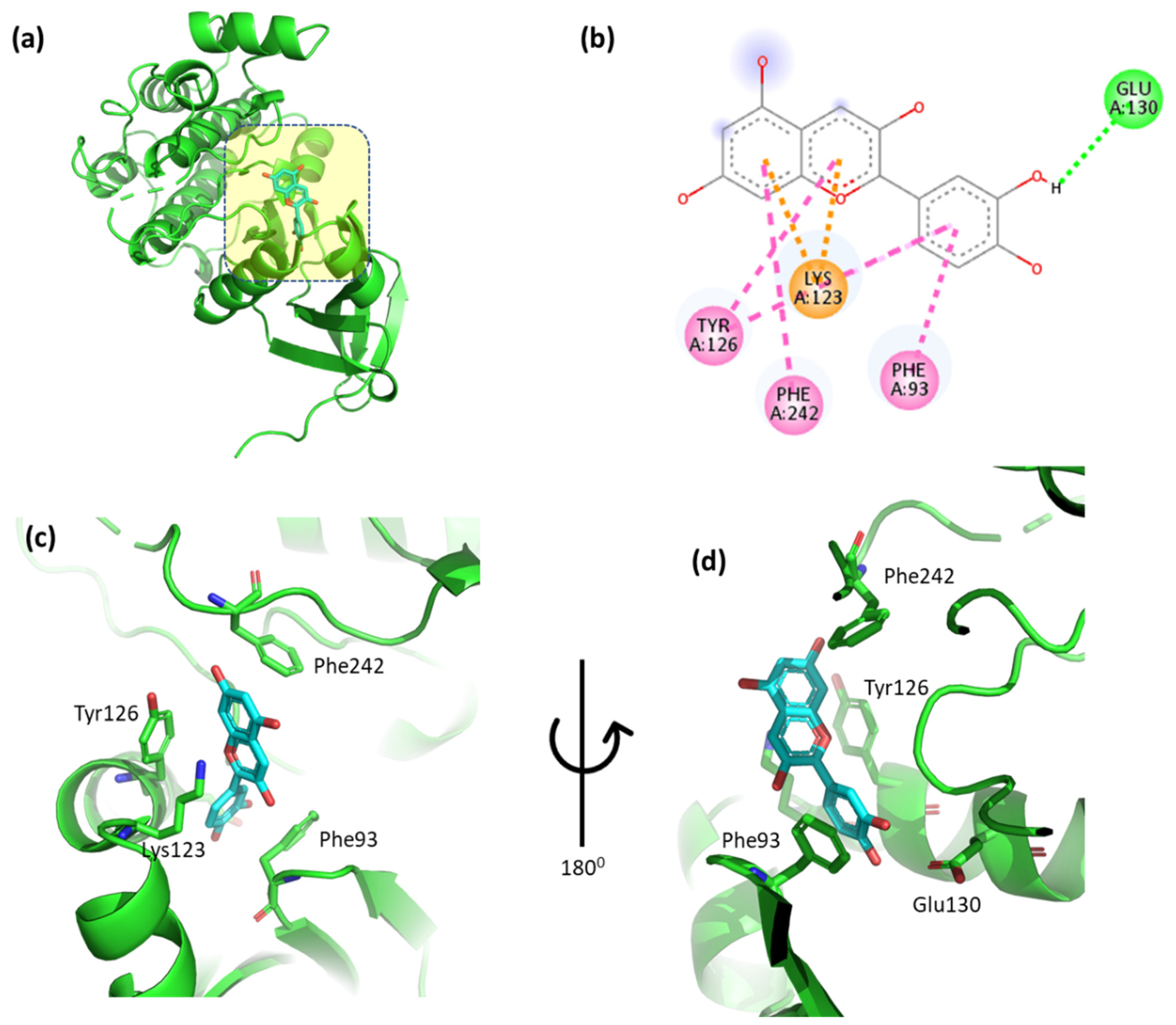
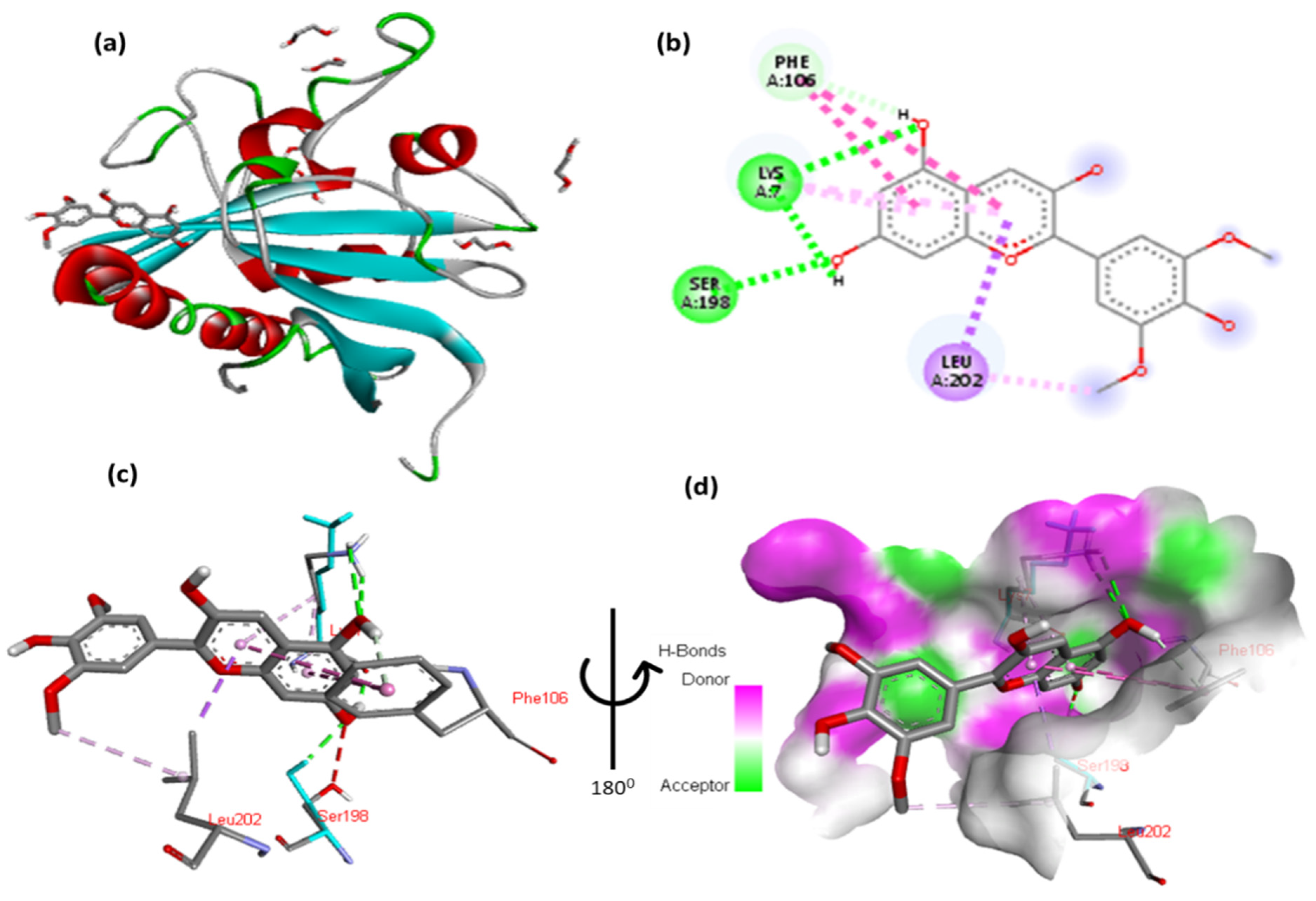
3.5. Evaluation of the Interactions Between the Top-Binding Binders of Anthocyanin
3.6. RT-qPCR Analysis of Gene Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Troen, B.R. The biology of aging. Mt. Sinai J. Med. 2003, 70, 3–22. [Google Scholar] [PubMed]
- Kenyon, C.; Chang, J.; Gensch, E.; Rudner, A.; Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 1993, 366, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef]
- Mattson, M.P.; Arumugam, T.V. Hallmarks of brain aging: Adaptive and pathological modification by metabolic states. Cell Metab. 2018, 27, 1176–1199. [Google Scholar] [CrossRef]
- Licastro, F.; Candore, G.; Lio, D.; Porcellini, E.; Colonna-Romano, G.; Franceschi, C.; Caruso, C. Innate immunity and inflammation in ageing: A key for understanding age-related diseases. Immun. Ageing 2005, 2, 1–14. [Google Scholar] [CrossRef]
- Khansari, N.; Shakiba, Y.; Mahmoudi, M. Chronic inflammation and oxidative stress as a major cause of age-related diseases and cancer. Recent Pat. Inflamm. Allergy Drug Discov. 2009, 3, 73–80. [Google Scholar] [CrossRef]
- Leszek, J.; Barreto, G.E.; Gasiorowski, K.; Koutsouraki, E.; Aliev, G. Inflammatory mechanisms and oxidative stress as key factors responsible for progression of neurodegeneration: Role of brain innate immune system. CNS Neurol. Disord.-Drug Targets (Former. Curr. Drug Targets-CNS Neurol. Disord.) 2016, 15, 329–336. [Google Scholar] [CrossRef]
- Rajawat, Y.S.; Hilioti, Z.; Bossis, I. Aging: Central role for autophagy and the lysosomal degradative system. Ageing Res. Rev. 2009, 8, 199–213. [Google Scholar] [CrossRef]
- Frankowska, N.; Lisowska, K.; Witkowski, J.M. Proteolysis dysfunction in the process of aging and age-related diseases. Front. Aging 2022, 3, 927630. [Google Scholar] [CrossRef]
- Paaby, A.B.; Schmidt, P.S. Dissecting the genetics of longevity in Drosophila melanogaster. Fly 2009, 3, 29–38. [Google Scholar] [CrossRef]
- Ogienko, A.A.; Omelina, E.S.; Bylino, O.V.; Batin, M.A.; Georgiev, P.G.; Pindyurin, A.V. Drosophila as a model organism to study basic mechanisms of longevity. Int. J. Mol. Sci. 2022, 23, 11244. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Smagghe, G. Roles of the insulin signaling pathway in insect development and organ growth. Peptides 2019, 122, 169923. [Google Scholar] [CrossRef]
- Biglou, S.G.; Bendena, W.G.; Chin-Sang, I. An overview of the insulin signaling pathway in model organisms Drosophila melanogaster and Caenorhabditis elegans. Peptides 2021, 145, 170640. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Chao, L.; Bergstrom, C.T.; Doebeli, M. On the evolutionary origin of aging. Aging Cell 2007, 6, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Flatt, T.; Schmidt, P.S. Integrating evolutionary and molecular genetics of aging. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2009, 1790, 951–962. [Google Scholar] [CrossRef]
- Lee, H.Y.; Lee, S.H.; Min, K.J. Insects as a model system for aging studies. Entomol. Res. 2015, 45, 1–8. [Google Scholar] [CrossRef]
- Gershon, H.; Gershon, D. The budding yeast, Saccharomyces cerevisiae, as a model for aging research: A critical review. Mech. Ageing Dev. 2000, 120, 1–22. [Google Scholar] [CrossRef]
- Partridge, L.; Gems, D. Benchmarks for ageing studies. Nature 2007, 450, 165–167. [Google Scholar] [CrossRef]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span—From yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
- Cohen, A.A. Aging across the tree of life: The importance of a comparative perspective for the use of animal models in aging. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2018, 1864, 2680–2689. [Google Scholar] [CrossRef]
- Bitto, A.; Wang, A.M.; Bennett, C.F.; Kaeberlein, M. Biochemical genetic pathways that modulate aging in multiple species. Cold Spring Harb. Perspect. Med. 2015, 5, a025114. [Google Scholar] [CrossRef] [PubMed]
- Colman, R.J. Non-human primates as a model for aging. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2018, 1864, 2733–2741. [Google Scholar] [CrossRef] [PubMed]
- Taormina, G.; Ferrante, F.; Vieni, S.; Grassi, N.; Russo, A.; Mirisola, M.G. Longevity: Lesson from model organisms. Genes 2019, 10, 518. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Ma, L.; Zhang, Y.; Feng, Y.; Zhang, J.; Wang, S. Current animal model systems for ovarian aging research. Aging Dis. 2022, 13, 1183. [Google Scholar] [CrossRef]
- Guo, S.; Wang, X.; Kang, L. Special significance of non-Drosophila insects in aging. Front. Cell Dev. Biol. 2020, 8, 576571. [Google Scholar]
- Rincon, M.; Muzumdar, R.; Atzmon, G.; Barzilai, N. The paradox of the insulin/IGF-1 signaling pathway in longevity. Mech. Ageing Dev. 2004, 125, 397–403. [Google Scholar]
- Borrás, C.; Gambini, J.; Gómez-Cabrera, M.C.; Sastre, J.; Pallardó, F.V.; Mann, G.E.; Viña, J. 17β-oestradiol up-regulates longevity-related, antioxidant enzyme expression via the ERK1 and ERK2 [MAPK]/NFκB cascade. Aging Cell 2005, 4, 113–118. [Google Scholar] [CrossRef]
- Greer, E.L.; Brunet, A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 2009, 8, 113–127. [Google Scholar] [CrossRef]
- Papaconstantinou, J.; Hsieh, C.C. Activation of Senescence and Aging Characteristics by Mitochondrially Generated ROS: How Are They Linked? Cell Cycle 2010, 9, 3831–3833. [Google Scholar] [CrossRef]
- Davis, T.; Rokicki, M.J.; Bagley, M.C.; Kipling, D. The effect of small-molecule inhibition of MAPKAPK2 on cell ageing phenotypes of fibroblasts from human Werner syndrome. Chem. Cent. J. 2013, 7, 1–4. [Google Scholar] [CrossRef]
- Palacios, O.M.; Carmona, J.J.; Michan, S.; Chen, K.Y.; Manabe, Y.; Ward, J.L., III; Goodyear, L.J.; Tong, Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1α in skeletal muscle. Aging 2009, 1, 771. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Brown, K.; Hirschey, M.D.; Verdin, E.; Chen, D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010, 12, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, Z.; ElAlieh, H.Z.; Nakamura, E.; Nguyen, M.T.; Mackem, S.; Clemens, T.L.; Bikle, D.D.; Chang, W. IGF-1R signaling in chondrocytes modulates growth plate development by interacting with the PTHrP/Ihh pathway. J. Bone Miner. Res. 2011, 26, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Cheung, H.P.; Tong, Y.; Lu, J.; Ng, T.B.; Zhang, Y.B.; Zhang, Z.-J.; Lee, K.F.; Lam, J.K.W.; Sze, S.C.W. Steroidogenic effect of Erxian decoction for relieving menopause via the p-Akt/PKB pathway in vitro and in vivo. J. Ethnopharmacol. 2017, 195, 188–195. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.M.; Lei, L.; Liu, Y.; Wang, X.; Ma, K.Y.; Chen, Z.-Y. Cranberry anthocyanin extract prolongs lifespan of fruit flies. Exp. Gerontol. 2015, 69, 189–195. [Google Scholar] [CrossRef]
- Wu, T.; Yin, J.; Zhang, G.; Long, H.; Zheng, X. Mulberry and cherry anthocyanin consumption prevents oxidative stress and inflammation in diet-induced obese mice. Mol. Nutr. Food Res. 2016, 60, 687–694. [Google Scholar] [CrossRef]
- Yan, F.; Chen, Y.; Azat, R.; Zheng, X. Mulberry anthocyanin extract ameliorates oxidative damage in HepG2 cells and prolongs the lifespan of Caenorhabditis elegans through MAPK and Nrf2 pathways. Oxidative Med. Cell. Longev. 2017, 2017, 7956158. [Google Scholar] [CrossRef]
- Meiers, S.; Kemény, M.; Weyand, U.; Gastpar, R.; von Angerer, E.; Marko, D. The anthocyanidins cyanidin and delphinidin are potent inhibitors of the epidermal growth-factor receptor. J. Agric. Food Chem. 2001, 49, 958–962. [Google Scholar] [CrossRef]
- Chen, P.N.; Chu, S.C.; Chiou, H.L.; Chiang, C.L.; Yang, S.F.; Hsieh, Y.S. Cyanidin 3-glucoside and peonidin 3-glucoside inhibit tumor cell growth and induce apoptosis in vitro and suppress tumor growth in vivo. Nutr. Cancer 2005, 53, 232–243. [Google Scholar] [CrossRef]
- Nas, J.S. Exploring the binding affinity and non-covalent interactions of anthocyanins with aging-related enzymes through molecular docking. J. Health Res. 2020, 24, 9–19. [Google Scholar]
- Pervaiz, T.; Songtao, J.; Faghihi, F.; Haider, M.S.; Fang, J. Naturally Occurring Anthocyanin, Structure, Functions and Biosynthetic Pathway in Fruit Plants. J. Plant Biochem. Physiol. 2017, 5, 1–9. [Google Scholar] [CrossRef]
- Alappat, B.; Alappat, J. Anthocyanin Pigments: Beyond Aesthetics. Molecules 2020, 25, 5500. [Google Scholar] [CrossRef]
- Ichikawa, H.; Ichiyanagi, T.; Xu, B.; Yoshii, Y.; Nakajima, M.; Konishi, T. Antioxidant Activity of Anthocyanin Extract from Purple Black Rice. J. Med. Food 2001, 4, 211–218. [Google Scholar] [CrossRef]
- Bajpai, M.; Pande, A.; Tewari, S.K.; Prakash, D. Phenolic contents and antioxidant activity of some food and medicinal plants. Int. J. Food Sci. Nutr. 2005, 56, 287–291. [Google Scholar] [CrossRef]
- Nozzolillo, C.; Isabelle, P.; Andersen, Ø.M.; Abou-Zaid, M. Anthocyanins of jack pine (Pinus banksiana) seedlings. Can. J. Bot. 2002, 80, 796–801. [Google Scholar] [CrossRef]
- Lev-Yadun, S.; Gould, K.S. Role of anthocyanins in plant defence. In Anthocyanins: Biosynthesis, Functions, and Applications; Springer: New York, NY, USA, 2009; pp. 22–28. [Google Scholar]
- Xie, X.; Gu, Y.; Fox, T.; Coll, J.T.; Fleming, M.A.; Markland, W.; Caron, P.R.; Wilson, K.P.; Su, S.S. Crystal structure of JNK3: A kinase implicated in neuronal apoptosis. Structure 1998, 6, 983–991. [Google Scholar] [CrossRef]
- King, A.J.; Patrick, D.R.; Batorsky, R.S.; Ho, M.L.; Do, H.T.; Zhang, S.Y.; Kumar, R.; Rusnak, D.W.; Takle, A.K.; Wilson, D.M.; et al. Demonstration of a Genetic Therapeutic Index for Tumors Expressing Oncogenic BRAF by the Kinase Inhibitor SB-590885. Cancer Res. 2006, 66, 11100–11105. [Google Scholar] [CrossRef]
- Fujino, A.; Fukushima, K.; Kubota, T.; Kosugi, T.; Takimoto-Kamimura, M. Crystal structure of human cyclin-dependent kinase-2 complex with MK2 inhibitor TEI-I01800: Insight into the selectivity. J. Synchrotron Radiat. 2013, 20 Pt. 6, 905–909. [Google Scholar] [CrossRef]
- Haroon; Li, Y.X.; Ye, C.X.; Su, J.; Nabi, G.; Su, X.H.; Xing, L.X. De Novo Transcriptome Assembly and Analysis of Longevity Genes Using Subterranean Termite (Reticulitermes chinensis) Castes. Int. J. Mol. Sci. 2022, 23, 13660. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M.; Forli, S. Using AutoDock 4 and AutoDock vina with AutoDockTools: A tutorial. Scripps Res. Inst. Mol. Graph. Lab. 2012, 10550, 1000. [Google Scholar]
- Mobley, D.L.; Graves, A.P.; Chodera, J.D.; McReynolds, A.C.; Shoichet, B.K.; Dill, K.A. Predicting absolute ligand binding free energies to a simple model site. J. Mol. Biol. 2007, 371, 1118–1134. [Google Scholar] [CrossRef] [PubMed]
- Azam, S.S.; Abbasi, S.W. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor. Biol. Med. Model. 2013, 10, 1–16. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Hernández-Rodríguez, M.C.; Rosales-Hernández, M.; Mendieta-Wejebe, J.E.; Martínez-Archundia, M.; Correa Basurto, J. Current tools and methods in molecular dynamics (MD) simulations for drug design. Curr. Med. Chem. 2016, 23, 3909–3924. [Google Scholar] [CrossRef]
- Salo-Ahen, O.M.; Alanko, I.; Bhadane, R.; Bonvin, A.M.; Honorato, R.V.; Hossain, S.; Juffer, A.H.; Kabedev, A.; Lahtela-Kakkonen, M.; Larsen, A.S.; et al. Molecular dynamics simulations in drug discovery and pharmaceutical development. Processes 2020, 9, 71. [Google Scholar] [CrossRef]
- Lu, Y.; Zhao, R.; Wang, C.; Zhang, X.; Wang, C. Deciphering the non-covalent binding patterns of three whey proteins with rosmarinic acid by multi-spectroscopic, molecular docking and molecular dynamics simulation approaches. Food Hydrocoll. 2022, 132, 107895. [Google Scholar] [CrossRef]
- Andrei, P.; Nogara; Aquino, R.D.; Saraiva; Diones; Bueno, C.; Juliana, L.; Lissner; Cristiane; Corte, L.D. Virtual screening of acetylcholinesterase inhibitors using the Lipinski’s rule of five and ZINC databank. BioMed Res. Int. 2015, 2015, 870389. [Google Scholar]
- Mannhold, R.; Kubinyi, H.; Folkers, G. Pharmacokinetics and Metabolism in Drug Design; John Wiley & Sons: Hoboken, NJ, USA, 2012; Volume 51. [Google Scholar]
- Andleeb, H.; Danish, L.; Munawar, S.; Ahmed, M.N.; Khan, I.; Ali, H.S.; Tahir, M.N.; Simpson, J.; Hameed, S. Theoretical and computational insight into the supramolecular assemblies of Schiff bases involving hydrogen bonding and C-H…π interactions: Synthesis, X-ray characterization, Hirshfeld surface analysis, anticancer activity and molecular docking analysis. J. Mol. Struct. 2021, 1235, 130223. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Goh, N.Y.; Yap, Y.H.Y.; Ng, C.L.; Kong, B.H.; Ng, S.T.; Tan, C.S.; Razif, M.F.M.; Fung, S.Y. Water-soluble compounds from Lignosus rhinocerus TM02®(xLr™) modulate ACE2 activity and inhibit its interaction with SARS-CoV-2 spike-protein. Food Biosci. 2024, 59, 104232. [Google Scholar] [CrossRef]
- Hospital, A.; Goñi, J.R.; Orozco, M.; Gelpí, J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. [Google Scholar] [PubMed]
- Sargsyan, K.; Grauffel, C.; Lim, C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 1–13. [Google Scholar] [CrossRef]
- Ishitani, K.; Maekawa, K. Ovarian development of female-female pairs in the termite, Reticulitermes speratus. J. Insect Sci. 2010, 10, 194. [Google Scholar] [CrossRef]
- Van Hiel, M.B.; Van Wielendaele, P.; Temmerman, L.; Van Soest, S.; Vuerinckx, K.; Huybrechts, R.; Broeck, J.V.; Simonet, G. Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol. Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef]
- Wang, B.; Tang, X.; Mao, B.; Zhang, Q.; Tian, F.; Zhao, J.; Cui, S.; Chen, W. Anti-aging effects and mechanisms of anthocyanins and their intestinal microflora metabolites. Crit. Rev. Food Sci. Nutr. 2024, 64, 2358–2374. [Google Scholar] [CrossRef]
- Li, Z.; Tian, J.; Cheng, Z.; Teng, W.; Zhang, W.; Bao, Y.; Wang, Y.; Song, B.; Chen, Y.; Li, B. Hypoglycemic bioactivity of anthocyanins: A review on proposed targets and potential signaling pathways. Crit. Rev. Food Sci. Nutr. 2022, 63, 7878–7895. [Google Scholar] [CrossRef]
- Choi, K.H.; Lee, H.A.; Park, M.H.; Han, J.S. Mulberry (Morus alba L.) Fruit Extract Containing Anthocyanins Improves Glycemic Control and Insulin Sensitivity via Activation of AMP-Activated Protein Kinase in Diabetic C57BL/Ksj-db/db Mice. J. Med. Food 2016, 19, 737. [Google Scholar] [CrossRef]
- Tian, J.L.; Wang, Y.H.; Si, X.; Gong, E.S.; Xie, X.; Liao, X.J.; Shu, C.; Ran, X.L.; Li, B. Correction to Identification of Cyanidin-3-arabinoside Extracted from Blueberry as a Selective Protein Tyrosine Phosphatase 1B Inhibitor. J. Agric. Food Chem. 2022, 70, 2060. [Google Scholar] [CrossRef] [PubMed]
- Gowd, V.; Jia, Z.; Chen, W. Anthocyanins as promising molecules and dietary bioactive components against diabetes—A review of recent advances. Trends Food Sci. Technol. 2017, 68, 1–13. [Google Scholar] [CrossRef]
- Oliveira, H.; Fernandes, A.; Brás, N.F.; Mateus, N.; de Freitas, V.; Fernandes, I. Anthocyanins as Antidiabetic Agents—In Vitro and In Silico Approaches of Preventive and Therapeutic Effects. Molecules 2020, 25, 3813. [Google Scholar] [CrossRef] [PubMed]
- Damián-Medina, K.; Salinas-Moreno, Y.; Milenkovic, D.; Figueroa-Yáñez, L.; Marino-Marmolejo, E.; Higuera-Ciapara, I.; Vallejo-Cardona, A.; Lugo-Cervantes, E. In silico analysis of antidiabetic potential of phenolic compounds from blue corn (Zea mays L.) and black bean (Phaseolus vulgaris L.). Heliyon 2020, 6, e03632. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 64, 4–17. [Google Scholar] [CrossRef]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drug ability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef]
- Gao, S.; Wu, W.; Mo, Y. The B-H⋯H-P dihydrogen bonding in ion pair complexes [(CF3)3BH−][HPH3−n(Me)n+] (n = 0 − 3) and its implication in H2 elimination and activation reactions. J. Phys. Chem. A 2009, 113, 8108–8117. [Google Scholar] [CrossRef]
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef]
- Paula, K.; Dirk, W. Physicochemical characteristics of structurally determined metabolite-protein and drug-protein binding events with respect to binding specificity. Front. Mol. Biosci. 2015, 2, 51. [Google Scholar]
- Copeland, R.A. Evaluation of enzyme inhibitors in drug discovery. A guide for medicinal chemists and pharmacologists. In Evaluation of Enzyme Inhibitors in Drug Discovery; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Klebe, G. Protein-Ligand Interactions as the Basis for Drug Action; Springer: Dordrecht, The Netherlands, 2015; pp. 83–92. [Google Scholar]
- Xiao, J.; Cao, H.; Wang, Y.; Yamamoto, K.; Wei, X. Structure-affinity relationship of flavones on binding to serum albumins: Effect of hydroxyl groups on ring A. Mol. Nutr. Food Res. 2010, 54, S253–S260. [Google Scholar] [CrossRef]














| Gene ID | Symbol | Primers Sequences |
|---|---|---|
| Beta-actin | Forward: CCCAACACAGCGTCTTACAA Reverse: CAGATGTCCTCAGCTTCACG | |
| Unigene 0082575 | PdK1 | Forward: TCCTCCTCCTGCTACTGCTGAAG Reverse: CGACATATGACGGAGTAGGTGGTG |
| Unigene 0092210 | Tsc2 | Forward: AGTGGTGCTAACATGCCTGC Reverse: ACCTTCCAGCTGCTCTGACA |
| Unigene 0011832 | EIF4E | Forward: GGATCTCGTCTTGGCCGTCATTG Reverse: AGCAACCTCGTGAGCCACTCC |
| Pdk1 | Tsc2 | EIF4E | |
|---|---|---|---|
| Number of amino acids | 290 | 65 | 296 |
| Molecular weight | 32,997.1 | 7229.54 | 33,978.5 |
| Theoretical pI | 9.58 | 10.77 | 8.93 |
| Negatively charged residues (Asp + Glu) | 6 | 3 | 16 |
| Positively charged residues (Arg + Lys) | 24 | 14 | 32 |
| Carbon (C) | 1519 | 313 | 1541 |
| Hydrogen (H) | 2354 | 543 | 2422 |
| Nitrogen (N) | 404 | 99 | 400 |
| Oxygen (O) | 378 | 90 | 402 |
| Sulfur (S) | 21 | 3 | 31 |
| Total number of atoms | 4676 | 1048 | 4796 |
| Ext. coefficient | 76,150 | 1615 | 40,935 |
| Abs 0.1% (=1 g/L) (Cys residues form cystines) | 2.308 | 0.223 | 1.205 |
| Ext. coefficient | 75,400 | 1490 | 39,310 |
| Abs 0.1% (=1 g/L) (Cys residues are reduced) | 2.285 | 0.206 | 1.157 |
| Instability index (II) | 29.21 | 59.09 | 39.26 |
| Aliphatic index | 105.14 | 91.54 | 106.25 |
| GRAVY | 0.292 | −0.309 | 0.307 |
| Pdk1 | Tsc2 | EIF4E | ||
|---|---|---|---|---|
| Metrics | Hydrophobic Fitness | −17 | −15 | −18 |
| Isoelectric Point | 8 | 8 | 5 | |
| Number of Residues | 275 | 191 | 211 | |
| Mass (Da) | 31,845 | 21,894 | 24,684 | |
| Mean Packing Density | 58 | 57 | 58 | |
| BUDE force field results | Total Energy | −4272 | - | - |
| Steric | 251 | - | - | |
| Desolvation | −2873 | - | - | |
| Charge | −1650 | - | - | |
| EvoEF2 energy function results-summary | Total Energy | −1227 | −934 | −1082 |
| Reference | −54 | −40 | −32 | |
| Intra-Residue | 598 | 391 | 435 | |
| Inter-Residue—Same Chain | −1771 | −1285 | −1380 | |
| Inter-Residue—Different Chains | 0 | 0 | −105 | |
| DFIRE2 energy function results | Total Energy | −523 | −343 | −404 |
| Rosetta energy function results | Total Energy | 81 | −175 | −276 |
| Reference | 72 | 72 | 66 | |
| VDW Attractive | −1697 | −1100 | −1259 | |
| VDW Repulsive | 366 | 76 | 146 | |
| VDW Repulsive Intra-Residue | 5 | 2 | 2 | |
| Electrostatics | −383 | −239 | −332 | |
| Solvation Isotropic | 995 | 670 | 752 | |
| Solvation Anisotropic Polar Atoms | −34 | −23 | −32 | |
| Solvation Isotropic Intra Residue | 62 | 38 | 53 | |
| HB Long Range Backbone | −46 | −51 | −64 | |
| HB Short Range Backbone | −85 | −46 | −63 | |
| HB Backbone Sidechain | −33 | −40 | −21 | |
| HB Sidechain Sidechain | −14 | −21 | −22 | |
| Disulfide Bridges | 0 | 0 | 0 | |
| Backbone Torsion Preference | 35 | 9 | 18 | |
| Amino Acid Propensity | −29 | −31 | −26 | |
| Dunbrack Rotamer | 781 | 381 | 434 | |
| Omega Penalty | 60 | 89 | 62 | |
| Open Proline Penalty | 25 | 40 | 11 | |
| Tyrosine χ3 Dihedral Angle Penalty | 0 | 0 | 0 | |
| Aggregation propensity results | Total Score | −227 | −103 | −181 |
| Average Score | −0.83 | −0.54 | −0.86 | |
| Minimum Score | −4.31 | −4.31 | −4.48 | |
| Maximum Score | 1.9 | 2.9 | 1.56 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haroon; Khan, Z.; Javaid, W.; Xing, L.-X. Anthocyanin-Binding Affinity and Non-Covalent Interactions with IIS-Pathway-Related Protein Through Molecular Docking. Curr. Issues Mol. Biol. 2025, 47, 87. https://doi.org/10.3390/cimb47020087
Haroon, Khan Z, Javaid W, Xing L-X. Anthocyanin-Binding Affinity and Non-Covalent Interactions with IIS-Pathway-Related Protein Through Molecular Docking. Current Issues in Molecular Biology. 2025; 47(2):87. https://doi.org/10.3390/cimb47020087
Chicago/Turabian StyleHaroon, Zahid Khan, Wasim Javaid, and Lian-Xi Xing. 2025. "Anthocyanin-Binding Affinity and Non-Covalent Interactions with IIS-Pathway-Related Protein Through Molecular Docking" Current Issues in Molecular Biology 47, no. 2: 87. https://doi.org/10.3390/cimb47020087
APA StyleHaroon, Khan, Z., Javaid, W., & Xing, L.-X. (2025). Anthocyanin-Binding Affinity and Non-Covalent Interactions with IIS-Pathway-Related Protein Through Molecular Docking. Current Issues in Molecular Biology, 47(2), 87. https://doi.org/10.3390/cimb47020087


_Kim.png)



