In Silico Characterization of ADAR1: Structure, Dynamics, and Functional Implications
Abstract
1. Introduction
2. Methods
2.1. Sequence Retrieval
2.2. Template Search and Obtaining Template Structures
2.3. Homology Modeling
2.3.1. RosettaCM and Modeller
2.3.2. Improving Homology Modeling with AlphaFold
2.4. Molecular Dynamics Simulations-Based Model Refinement
2.5. Evaluation of Structural Changes and Dynamics
2.6. Model Quality Assessment
3. Results and Discussion
3.1. ADAR1 Structures
3.2. Template Search and De Novo Fragment Generation
3.3. Modeling with Robetta and Modeller
3.4. Modeling Improvement with AlphaFold DB
3.5. Evaluation of Model Quality
3.5.1. Structural Alignment Assessment
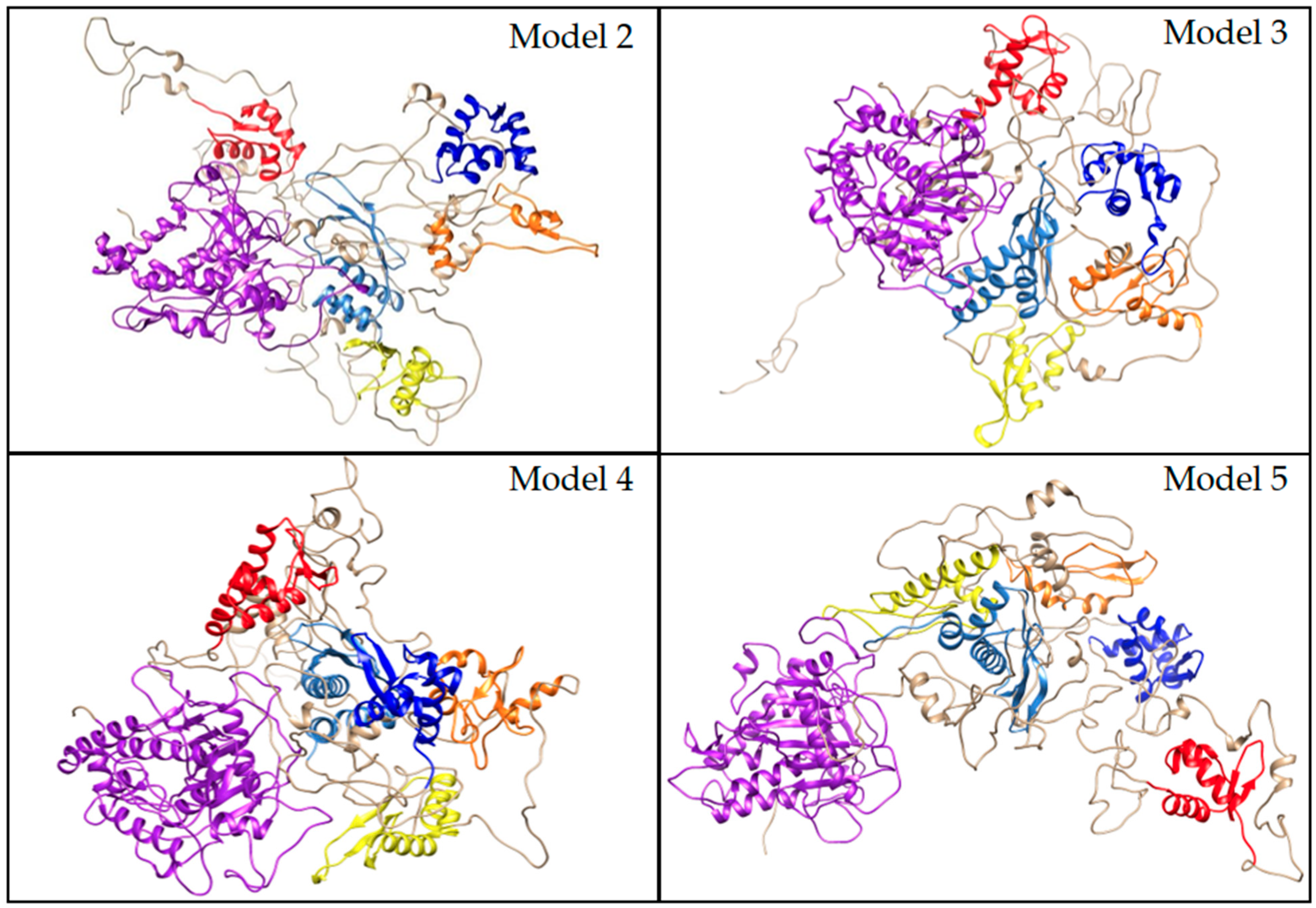
3.5.2. Protein Structure Analysis
3.5.3. Model Quality with SAVEs
3.6. Model Refinement with MD Simulations
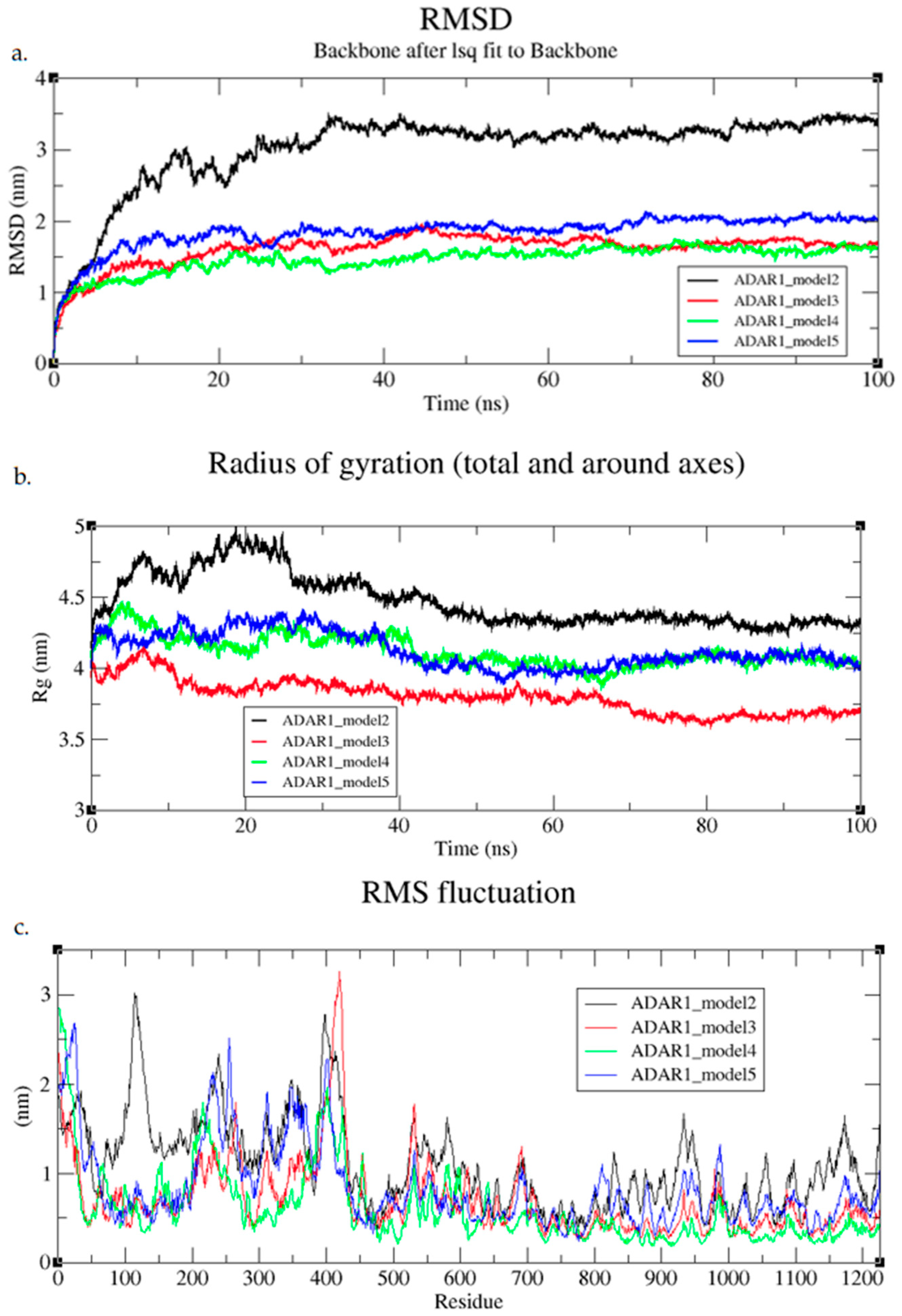
3.6.1. 100. ns MD Simulations
3.6.2. 800 ns MD Simulations
3.6.3. Post-MD Model Quality Analysis
3.6.4. MD Refinement of Initial Models
3.7. Principal Component Analysis
Principal Component Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Hundley, H.A.; Bass, B.L. ADAR Editing in Double-Stranded UTRs and Other Non-Coding RNA Sequences. Trends Biochem. Sci. 2010, 35, 383. [Google Scholar] [CrossRef]
- Li, X.; Overton, I.M.; Baines, R.A.; Keegan, L.P.; O’Connell, M.A. The ADAR RNA Editing Enzyme Controls Neuronal Excitability in Drosophila Melanogaster. Nucleic Acids Res. 2014, 42, 1139–1151. [Google Scholar] [CrossRef]
- Quin, J.; Sedmík, J.; Vukić, D.; Khan, A.; Keegan, L.P.; O’Connell, M.A. ADAR RNA Modifications, the Epitranscriptome and Innate Immunity. Trends Biochem. Sci. 2021, 46, 758–771. [Google Scholar] [CrossRef]
- Jacobs, M.M.; Fogg, R.L.; Emeson, R.B.; Stanwood, G.D. ADAR1 and ADAR2 Expression and Editing Activity During Forebrain Development. Dev. Neurosci. 2009, 31, 237. [Google Scholar] [CrossRef]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A-to-I RNA Editing Occurs at over a Hundred Million Genomic Sites, Located in a Majority of Human Genes. Genome Res. 2014, 24, 376. [Google Scholar] [CrossRef]
- Song, B.; Shiromoto, Y.; Minakuchi, M.; Nishikura, K. The Role of RNA Editing Enzyme ADAR1 in Human Disease. Wiley Interdiscip. Rev. RNA 2022, 13, e1665. [Google Scholar] [CrossRef]
- Lamers, M.M.; van den Hoogen, B.G.; Haagmans, B.L. ADAR1: “Editor-in-Chief” of Cytoplasmic Innate Immunity. Front. Immunol. 2019, 10, 1763. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, F.; Zhang, Y.; Liu, J.; Zhao, B. ADAR1-Mediated RNA Editing and Its Role in Cancer. Front. Cell Dev. Biol. 2022, 10, 956649. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, J.J.; Manguso, R.T.; Cheruiyot, C.K.; Bi, K.; Panda, A.; Iracheta-Vellve, A.; Miller, B.C.; Du, P.P.; Yates, K.B.; Dubrot, J.; et al. Loss of ADAR1 in Tumours Overcomes Resistance to Immune Checkpoint Blockade. Nature 2019, 565, 43–48. [Google Scholar] [CrossRef]
- Maurano, M.; Snyder, J.M.; Connelly, C.; Henao-Mejia, J.; Sidrauski, C.; Stetson, D.B. Protein Kinase R and the Integrated Stress Response Drive Immunopathology Caused by Mutations in the RNA Deaminase ADAR1. Immunity 2021, 54, 1948–1960.e5. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wiley, C.A.; Steinman, R.A.; Sheng, Y.; Ji, B.; Wang, J.; Zhang, L.; Wang, T.; Zenatai, M.; Billiar, T.R.; et al. Aicardi-Goutières Syndrome-Associated Mutation at ADAR1 Gene Locus Activates Innate Immune Response in Mouse Brain. J. Neuroinflamm. 2021, 18, 169. [Google Scholar] [CrossRef]
- Inoue, M.; Nakahama, T.; Yamasaki, R.; Shibuya, T.; Kim, J.I.; Todo, H.; Xing, Y.; Kato, Y.; Morii, E.; Kawahara, Y. An Aicardi-Goutières Syndrome–Causative Point Mutation in Adar1 Gene Invokes Multiorgan Inflammation and Late-Onset Encephalopathy in Mice. J. Immunol. 2021, 207, 3016–3027. [Google Scholar] [CrossRef]
- Tang, Q.; Rigby, R.E.; Young, G.R.; Hvidt, A.K.; Davis, T.; Tan, T.K.; Bridgeman, A.; Townsend, A.R.; Kassiotis, G.; Rehwinkel, J. Adenosine-to-Inosine Editing of Endogenous Z-Form RNA by the Deaminase ADAR1 Prevents Spontaneous MAVS-Dependent Type I Interferon Responses. Immunity 2021, 54, 1961–1975.e5. [Google Scholar] [CrossRef]
- Jarmoskaite, I.; Li, J.B. Multifaceted Roles of RNA Editing Enzyme ADAR1 in Innate Immunity. RNA 2024, 30, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Booth, B.J.; Nourreddine, S.; Katrekar, D.; Savva, Y.; Bose, D.; Long, T.J.; Huss, D.J.; Mali, P. RNA Editing: Expanding the Potential of RNA Therapeutics. Mol. Ther. 2023, 31, 1533–1549. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Ding, S.; Zhou, S.; Du, F.; Liu, S.; Cui, X.; Dong, J.; Huang, X.; Tang, Z. Enhancing RNA Editing Efficiency and Specificity with Engineered ADAR2 Guide RNAs. Mol. Ther. Nucleic Acids 2025, 36, 102447. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; Cukura, A.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New Features and Functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhatt, R.; Bhikadiya, C.; Bi, C.; Biester, A.; Biswas, P.; Bittrich, S.; Blaumann, S.; Brown, R.; Chao, H.; et al. Updated Resources for Exploring Experimentally-Determined PDB Structures and Computed Structure Models at the RCSB Protein Data Bank. Nucleic Acids Res. 2025, 53, D564–D574. [Google Scholar] [CrossRef]
- Schwartz, T.; Rould, M.A.; Lowenhaupt, K.; Herbert, A.; Rich, A. Crystal Structure of the Zα Domain of the Human Editing Enzyme ADAR1 Bound to Left-Handed Z-DNA. Science 1999, 284, 1841–1845. [Google Scholar] [CrossRef]
- Schade, M.; Turner, C.J.; Kühne, R.; Schmieder, P.; Lowenhaupt, K.; Herbert, A.; Rich, A.; Oschkinat, H. The Solution Structure of the Za Domain of the Human RNA Editing Enzyme ADAR1 Reveals a Prepositioned Binding Surface for Z-DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 12465–12470. [Google Scholar] [CrossRef]
- Placido, D.; Brown, B.A.; Lowenhaupt, K.; Rich, A.; Athanasiadis, A. A Left-Handed RNA Double Helix Bound by the Zα Domain of the RNA-Editing Enzyme ADAR1. Structure 2007, 15, 395–404. [Google Scholar] [CrossRef]
- Athanasiadis, A.; Placido, D.; Maas, S.; Brown, B.A.; Lowenhaupt, K.; Rich, A. The Crystal Structure of the Zβ Domain of the RNA-Editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-Domains. J. Mol. Biol. 2005, 351, 496–507. [Google Scholar] [CrossRef]
- Sung, C.H.; Lowenhaupt, K.; Rich, A.; Kim, Y.G.; Kyeong, K.K. Crystal Structure of a Junction Between B-DNA and Z-DNA Reveals Two Extruded Bases. Nature 2005, 437, 1183–1186. [Google Scholar] [CrossRef]
- Ha, S.C.; Choi, J.; Hwang, H.Y.; Rich, A.; Kim, Y.G.; Kim, K.K. The Structures of Non-CG-Repeat Z-DNAs Co-Crystallized with the Z-DNA-Binding Domain, HZαADAR1. Nucleic Acids Res. 2009, 37, 637. [Google Scholar] [CrossRef] [PubMed]
- Mboukou, A.; Rajendra, V.; Messmer, S.; Catala, M.; Tisné, C.; Jantsch, M.F.; Barraud, P. Dimerization of ADAR1 Modulates Site-Specificity of RNA Editing. bioRxiv 2023, 15, 570066. [Google Scholar] [CrossRef] [PubMed]
- Barraud, P.; Banerjee, S.; Mohamed, W.I.; Jantsch, M.F.; Allain, F.H.T. A Bimodular Nuclear Localization Signal Assembled via an Extended Double-Stranded RNA-Binding Domain Acts as an RNA-Sensing Signal for Transportin 1. Proc. Natl. Acad. Sci. USA 2014, 111, E1852–E1861. [Google Scholar] [CrossRef]
- Deng, X.; Sun, L.; Zhang, M.; Basavaraj, R.; Wang, J.; Weng, Y.-L.; Gao, Y. Biochemical Profiling and Structural Basis of ADAR1-Mediated RNA Editing. Mol. Cell 2025, 85, 1381–1394.e6. [Google Scholar] [CrossRef]
- Park, S.H.; Doherty, E.E.; Xie, Y.; Padyana, A.K.; Fang, F.; Zhang, Y.; Karki, A.; Lebrilla, C.B.; Siegel, J.B.; Beal, P.A. High-Throughput Mutagenesis Reveals Unique Structural Features of Human ADAR1. Nat. Commun. 2020, 11, 5130. [Google Scholar] [CrossRef]
- Song, Y.; DiMaio, F.; Wang, R.Y.-R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef]
- Fiser, A.; Do, R.K.G.; Šali, A. Modeling of Loops in Protein Structures. Protein Sci. 2000, 9, 1753–1773. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5–6. [Google Scholar] [CrossRef]
- Jamroz, M.; Kolinski, A. Modeling of Loops in Proteins: A Multi-Method Approach. BMC Struct. Biol. 2010, 10, 5. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.O.; He, Y. Benchmarking the Accuracy of AlphaFold 2 in Loop Structure Prediction. Biomolecules 2022, 12, 985. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-chain Torsion Potentials for the Amber Ff99SB Protein Force Field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Robertson, M.J.; Tirado-Rives, J.; Jorgensen, W.L. Improved Peptide and Protein Torsional Energetics with the OPLS-AA Force Field. J. Chem. Theory Comput. 2015, 11, 3499–3509. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. GROMACS: High Performance Molecular Simulations Through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Allu, A.S.; Tiriveedhi, V. Cancer Salt Nostalgia. Cells 2021, 10, 1285. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-Align: A Protein Structure Alignment Algorithm Based on the TM-Score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Maisuradze, G.G.; Liwo, A.; Scheraga, H.A. Principal Component Analysis for Protein Folding Dynamics. J. Mol. Biol. 2009, 385, 312–329. [Google Scholar] [CrossRef]
- Palma, J.; Pierdominici-Sottile, G. On the Uses of PCA to Characterise Molecular Dynamics Simulations of Biological Macromolecules: Basics and Tips for an Effective Use. ChemPhysChem 2023, 24, e202200491. [Google Scholar] [CrossRef]
- Lemkul, J.A. Introductory Tutorials for Simulating Protein Dynamics with GROMACS. J. Phys. Chem. B 2024, 128, 9418–9435. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of Protein Models with Three-Dimensional Profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Morris, A.L.; MacArthur, M.W.; Hutchinson, E.G.; Thornton, J.M. Stereochemical Quality of Protein Structure Coordinates. Proteins 1992, 12, 345–364. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Nayeem, A.; Sitkoff, D.; Krystek, S. A Comparative Study of Available Software for High-accuracy Homology Modeling: From Sequence Alignments to Structural Models. Protein Sci. 2006, 15, 808–824. [Google Scholar] [CrossRef]
- Fiser, A. Template-Based Protein Structure Modeling. Methods Mol. Biol. 2014, 2010, 73–94. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Strehblow, A.; Hallegger, M.; Jantsch, M.F. Nucleocytoplasmic Distribution of Human RNA-Editing Enzyme ADAR1 Is Modulated by Double-Stranded RNA-Binding Domains, a Leucine-Rich Export Signal, and a Putative Dimerization Domain. Mol. Biol. Cell 2002, 13, 3822–3835. [Google Scholar] [CrossRef] [PubMed]
- Patra, D.; Paul, J.; Rai, U.; P. S., A.; Deshmukh, M.V. Conformational Plasticity in DsRNA-Binding Domains Drives Functional Divergence in RNA Recognition. J. Am. Chem. Soc. 2025, 147, 17088–17100. [Google Scholar] [CrossRef]
- Sinigaglia, K.; Cherian, A.; Du, Q.; Lacovich, V.; Vukić, D.; Melicherová, J.; Linhartova, P.; Zerad, L.; Stejskal, S.; Malik, R.; et al. An ADAR1 DsRBD3-PKR Kinase Domain Interaction on DsRNA Inhibits PKR Activation. Cell Rep. 2024, 43, 114618. [Google Scholar] [CrossRef]
- Ashley, C.N.; Broni, E.; Miller, W.A. ADAR Family Proteins: A Structural Review. Curr. Issues Mol. Biol. 2024, 46, 3919–3945. [Google Scholar] [CrossRef]
- Mascali, F.C.; Crespo, R.; Tabares, L.C.; Rasia, R.M. Conserved Linker Length in Double DsRBD Proteins from Plants Restricts Interdomain Motion. J. Magn. Reson. Open 2023, 16–17, 100109. [Google Scholar] [CrossRef]
- Banerjee, S.; Barraud, P. Functions of Double-Stranded RNA-Binding Domains in Nucleocytoplasmic Transport. RNA Biol. 2014, 11, 1232. [Google Scholar] [CrossRef]
- Van Quyen, D.; Ha, S.C.; Lowenhaupt, K.; Rich, A.; Kim, K.K.; Kim, Y.-G. Characterization of DNA-Binding Activity of Zα Domains from Poxviruses and the Importance of the β-Wing Regions in Converting B-DNA to Z-DNA. Nucleic Acids Res. 2007, 35, 7714–7720. [Google Scholar] [CrossRef][Green Version]
- Kaufmann, K.W.; Lemmon, G.H.; DeLuca, S.L.; Sheehan, J.H.; Meiler, J. Practically Useful: What the Rosetta Protein Modeling Suite Can Do for You. Biochemistry 2010, 49, 2987–2998. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, R.; Strauss, C.E.M.; Rohl, C.A.; Chivian, D.; Bradley, P.; Malmström, L.; Robertson, T.; Baker, D. De Novo Prediction of Three-Dimensional Structures for Major Protein Families. J. Mol. Biol. 2002, 322, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Mark, A.E. Refinement of Homology-based Protein Structures by Molecular Dynamics Simulation Techniques. Protein Sci. 2004, 13, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 Forms a Complex with Dicer to Promote MicroRNA Processing and RNA-Induced Gene Silencing. Cell 2013, 153, 589. [Google Scholar] [CrossRef]
- Liang, Z.; Walkley, C.R.; Heraud-Farlow, J.E. A-to-I RNA Editing and Hematopoiesis. Exp. Hematol. 2024, 139, 104621. [Google Scholar] [CrossRef]
- Samuel, C.E. Adenosine Deaminase Acting on RNA (ADAR1), a Suppressor of Double-Stranded RNA–Triggered Innate Immune Responses. J. Biol. Chem. 2019, 294, 1710–1720. [Google Scholar] [CrossRef]
- Eggington, J.M.; Greene, T.; Bass, B.L. Predicting Sites of ADAR Editing in Double-Stranded RNA. Nat. Commun. 2011, 2, 319. [Google Scholar] [CrossRef]
- Thuy-Boun, A.S.; Thomas, J.M.; Grajo, H.L.; Palumbo, C.M.; Park, S.; Nguyen, L.T.; Fisher, A.J.; Beal, P.A. Asymmetric Dimerization of Adenosine Deaminase Acting on RNA Facilitates Substrate Recognition. Nucleic Acids Res. 2020, 48, 7958–7972. [Google Scholar] [CrossRef]
- Schwartz, T.; Lowenhaupt, K.; Kim, Y.-G.; Li, L.; Brown, B.A.; Herbert, A.; Rich, A. Proteolytic Dissection of Zab, the Z-DNA-Binding Domain of Human ADAR1. J. Biol. Chem. 1999, 274, 2899–2906. [Google Scholar] [CrossRef]
- Mackereth, C.D.; Sattler, M. Dynamics in Multi-Domain Protein Recognition of RNA. Curr. Opin. Struct. Biol. 2012, 22, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Tarus, B.; Chevalier, C.; Richard, C.-A.; Delmas, B.; Di Primo, C.; Slama-Schwok, A. Molecular Dynamics Studies of the Nucleoprotein of Influenza A Virus: Role of the Protein Flexibility in RNA Binding. PLoS ONE 2012, 7, e30038. [Google Scholar] [CrossRef] [PubMed]





| PDB | ||||||
|---|---|---|---|---|---|---|
| 1QBJ | 1XMK | 2MDR | 9B83 | 9B89 | ||
| AF-P55265-F1 | TM-score | 0.94350 | 0.79996 | 0.86573 | 0.98091 | 0.84275 |
| RMSD Value (Å) | 0.77 | 1.84 | 1.55 | 1.08 | 3.07 | |
| Aligned Length | 66 | 71 | 90 | 385 | 422 | |
| Model 1 | TM-score | 0.91895 | 0.83429 | 0.74950 | 0.98121 | 0.86723 |
| RMSD Value (Å) | 1.40 | 1.77 | 1.85 | 1.13 | 2.86 | |
| Aligned Length | 66 | 72 | 81 | 385 | 432 | |
| Model 2 | TM-score | 0.91366 | 0.84227 | 0.80269 | 0.97891 | 0.84385 |
| RMSD Value (Å) | 1.58 | 1.64 | 2.54 | 1.30 | 3.01 | |
| Aligned Length | 66 | 72 | 93 | 385 | 421 | |
| Model 3 | TM-score | 0.95466 | 0.81490 | 0.84983 | 0.98802 | 0.85914 |
| RMSD Value (Å) | 0.65 | 1.79 | 1.83 | 0.90 | 3.06 | |
| Aligned Length | 66 | 71 | 90 | 385 | 430 | |
| Model 4 | TM-score | 0.93571 | 0.82709 | 0.84068 | 0.98879 | 0.83827 |
| RMSD Value (Å) | 0.79 | 1.81 | 2.12 | 0.92 | 1.80 | |
| Aligned Length | 66 | 72 | 91 | 385 | 403 | |
| Model 5 | TM-score | 0.94630 | 0.83280 | 0.77372 | 0.98141 | 0.86549 |
| RMSD Value (Å) | 0.75 | 1.76 | 2.54 | 1.29 | 3.19 | |
| Aligned Length | 66 | 72 | 90 | 385 | 436 | |
| AlphaFold-included Models Pre-MD Simulation | |||||||
| Model | Molpdf | ERRAT | VERIFY3D | PROCHECK | |||
| Favored | Allowed | Generously Allowed | Disallowed | ||||
| AF-P55265-F1 | 95.3737 | 61.42% | 78.6% | 12.6% | 2.4% | 6.5% | |
| Model 1 | 95,629.57031 | 52.9412 | 67.58% | 82.9% | 12.8% | 3.6% | 0.7% |
| Model 2 | 96,981.09375 | 54.4348 | 71.23% | 82.7% | 13.1% | 2.6% | 1.7% |
| Model 3 | 89,784.28125 | 54.491 | 71.12% | 84.1% | 11.9% | 2.1% | 1.9% |
| Model 4 | 85,828.26562 | 61.7058 | 68.61% | 83.1% | 12.4% | 2.6% | 1.9% |
| Model 5 | 92,985.09375 | 56.4957 | 67.81% | 85.3% | 11.3% | 2.0% | 1.3% |
| AlphaFold-included Models Post-MD Simulations | |||||||
| Model | ERRAT | VERIFY3D | PROCHECK | ||||
| Favored | Allowed | Generously Allowed | Disallowed | ||||
| Model 2 | 85.6275 | 62.56% | 88.6% | 8.8% | 2.1% | 0.6% | |
| Model 3 | 86.9822 | 63.13% | 88.6% | 9.3% | 1.6% | 0.5% | |
| Model 4 | 88.9872 | 61.53% | 88.0% | 10.3% | 1.4% | 0.3% | |
| Model 5 | 85.7282 | 57.65% | 87.9% | 9.8% | 1.6% | 0.7% | |
| PDB | ||||||
|---|---|---|---|---|---|---|
| 1QBJ | 1XMK | 2MDR | 9B83 | 9B89 | ||
| Model 2 | TM-score | 0.82375 | 0.70837 | 0.64477 | 0.85329 | 0.79438 |
| RMSD Value (Å) | 2.03 | 2.75 | 3.15 | 3.22 | 4.20 | |
| Aligned Length | 65 | 67 | 93 | 376 | 440 | |
| Model 3 | TM-score | 0.79966 | 0.78393 | 0.82476 | 0.89278 | 0.78305 |
| RMSD Value (Å) | 1.89 | 1.82 | 1.87 | 2.98 | 4.21 | |
| Aligned Length | 65 | 72 | 92 | 382 | 428 | |
| Model 4 | TM-score | 0.80826 | 0.74380 | 0.81220 | 0.88156 | 0.77782 |
| RMSD Value (Å) | 1.69 | 2.24 | 2.24 | 3.11 | 3.92 | |
| Aligned Length | 66 | 70 | 89 | 383 | 422 | |
| Model 5 | TM-score | 0.82185 | 0.72882 | 0.73847 | 0.86625 | 0.73615 |
| RMSD Value (Å) | 1.77 | 2.30 | 2.20 | 3.16 | 3.65 | |
| Aligned Length | 66 | 67 | 84 | 378 | 392 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashley, C.N.; Broni, E.; Wood, C.M.; Miller, W.A., III. In Silico Characterization of ADAR1: Structure, Dynamics, and Functional Implications. Curr. Issues Mol. Biol. 2025, 47, 958. https://doi.org/10.3390/cimb47110958
Ashley CN, Broni E, Wood CM, Miller WA III. In Silico Characterization of ADAR1: Structure, Dynamics, and Functional Implications. Current Issues in Molecular Biology. 2025; 47(11):958. https://doi.org/10.3390/cimb47110958
Chicago/Turabian StyleAshley, Carolyn N., Emmanuel Broni, ChaNyah M. Wood, and Whelton A. Miller, III. 2025. "In Silico Characterization of ADAR1: Structure, Dynamics, and Functional Implications" Current Issues in Molecular Biology 47, no. 11: 958. https://doi.org/10.3390/cimb47110958
APA StyleAshley, C. N., Broni, E., Wood, C. M., & Miller, W. A., III. (2025). In Silico Characterization of ADAR1: Structure, Dynamics, and Functional Implications. Current Issues in Molecular Biology, 47(11), 958. https://doi.org/10.3390/cimb47110958








