Genomic and Biological Insights of Bacteriophage ΦBc24 Targeting Bacillus cereus
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Phage Isolation and Purification
2.3. Phage Stock Preparation
2.4. Transmission Electron Microscopy
2.5. Determination of Host Range
2.6. Multiplicity of Infection
2.7. One-Step Growth Curve
2.8. Phage Stability
2.9. In Vitro Lytic Activity Assay
2.10. Genome Sequencing and Annotation
2.11. Bioinformatic and Proteomic Analysis
2.12. Statistical Analysis
3. Results
3.1. Phage Isolation
3.2. Phage Morphology
3.3. Host Range
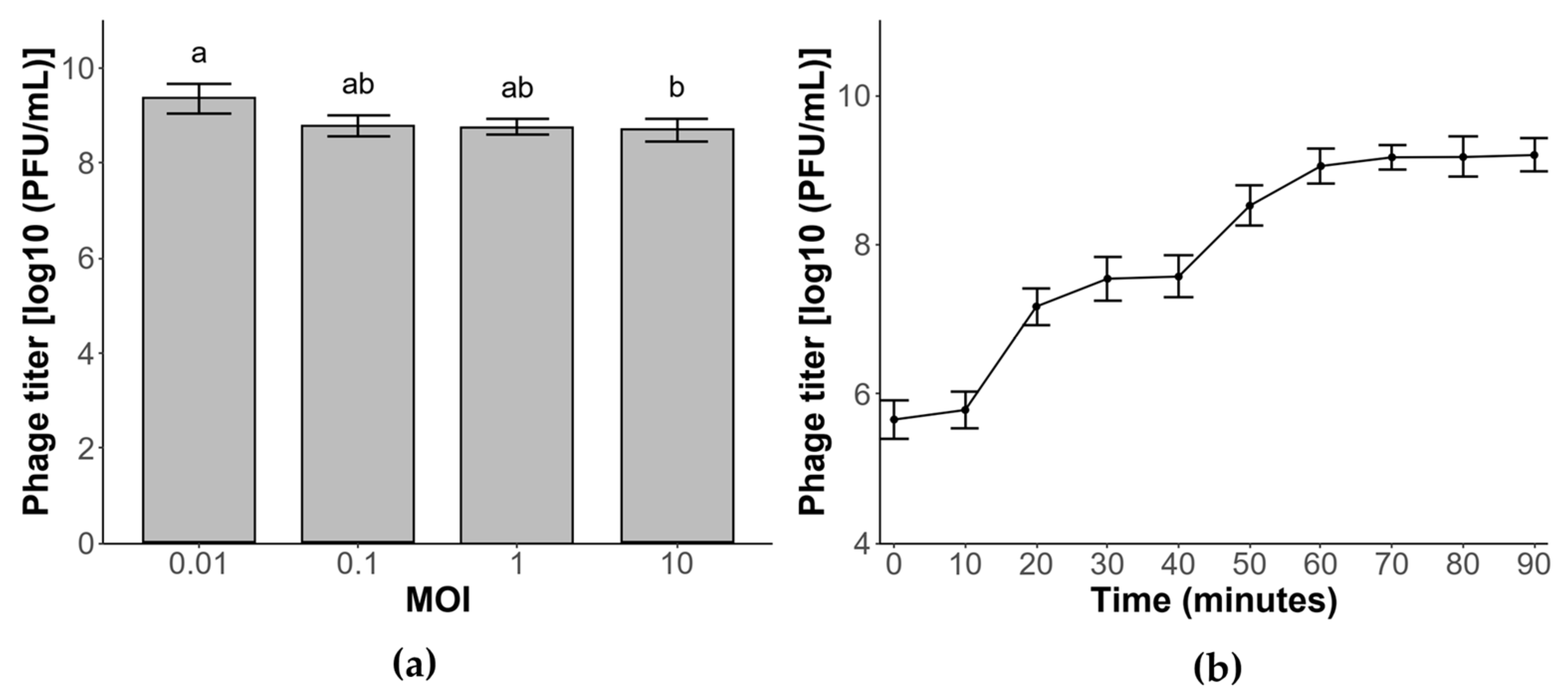
3.4. Optimal Multiplicity of Infection
3.5. One-Step Growth Curve
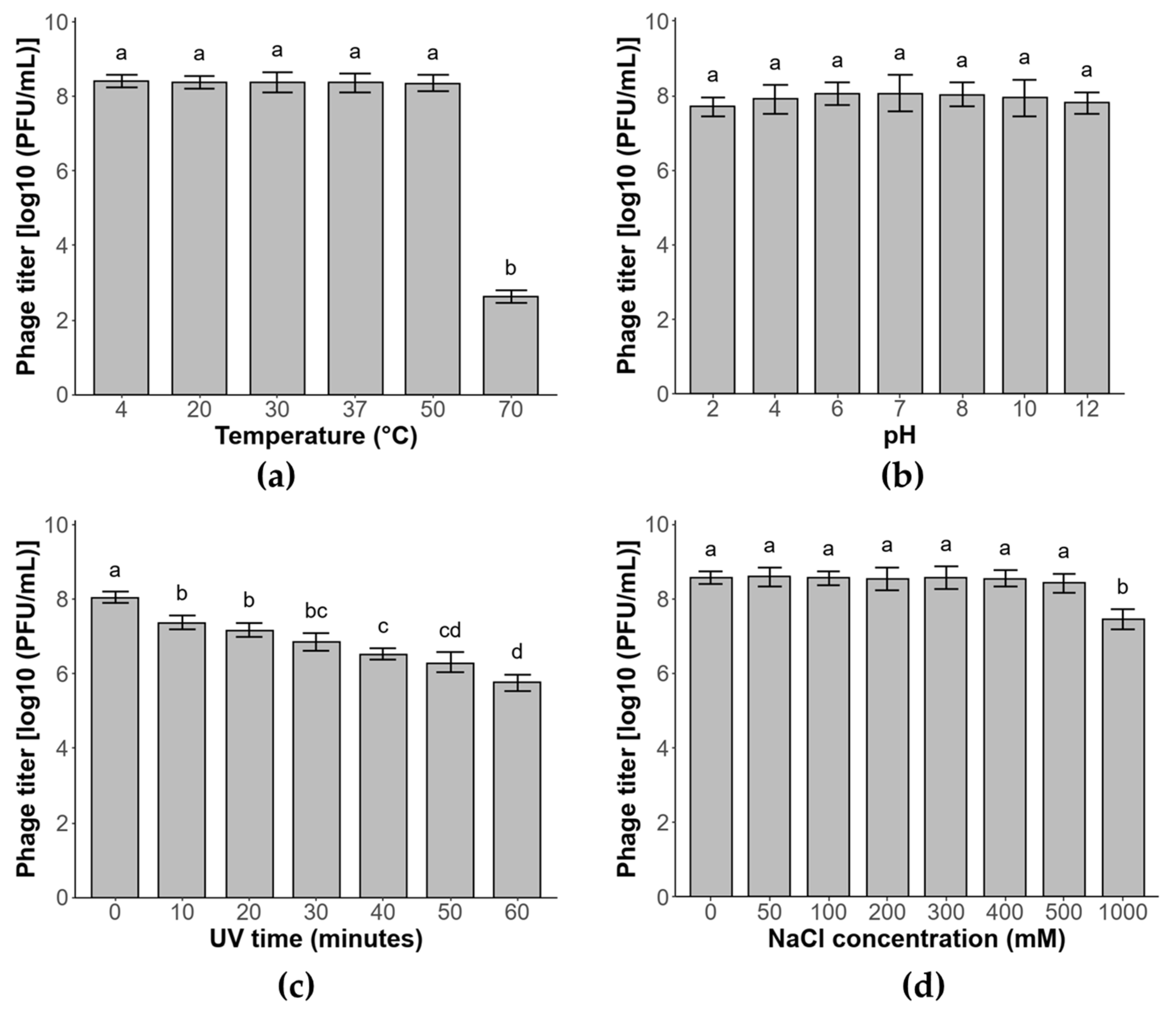
3.6. Phage Stability
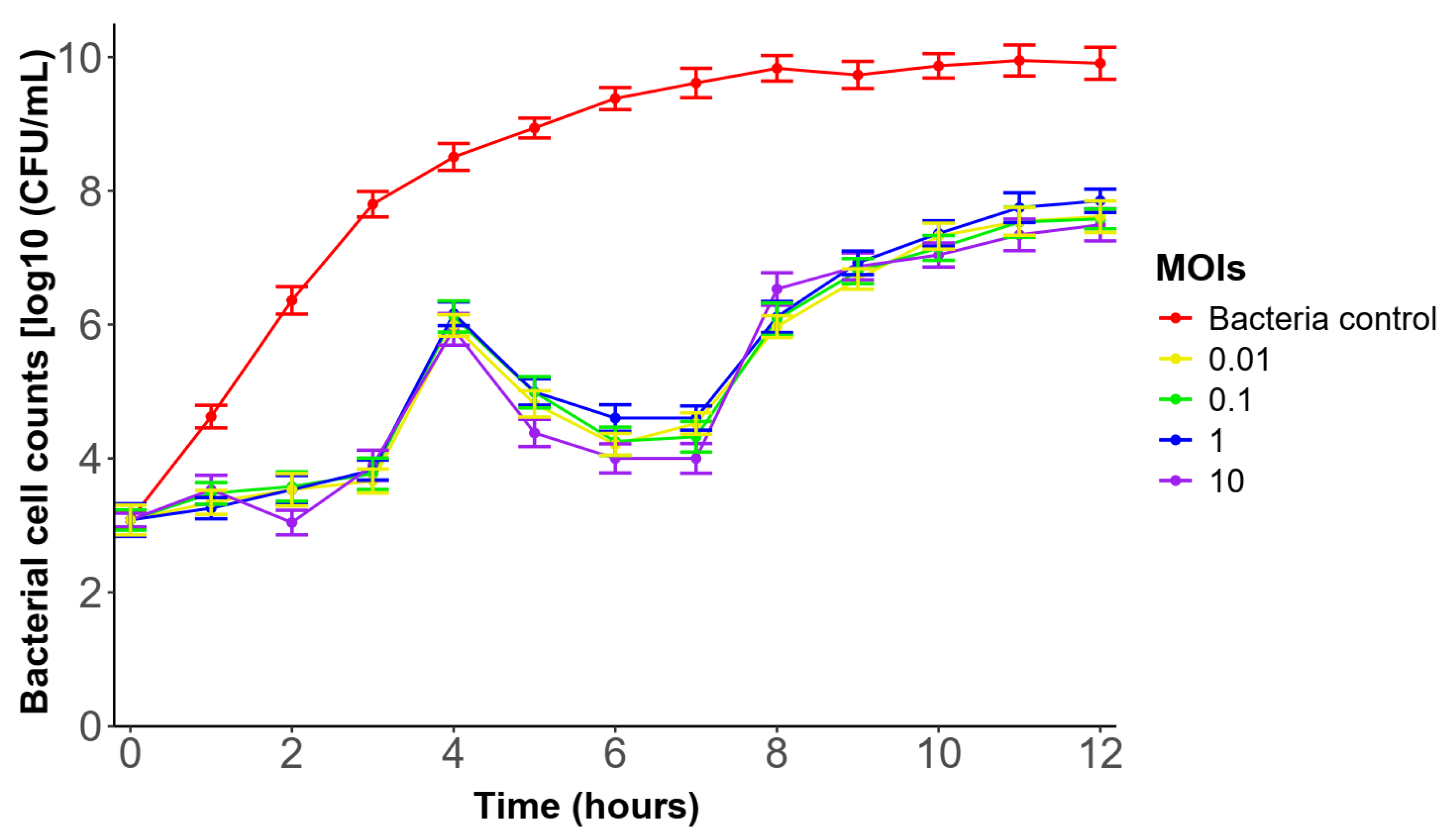
3.7. In Vitro Lytic Activity of Phage ΦBC24
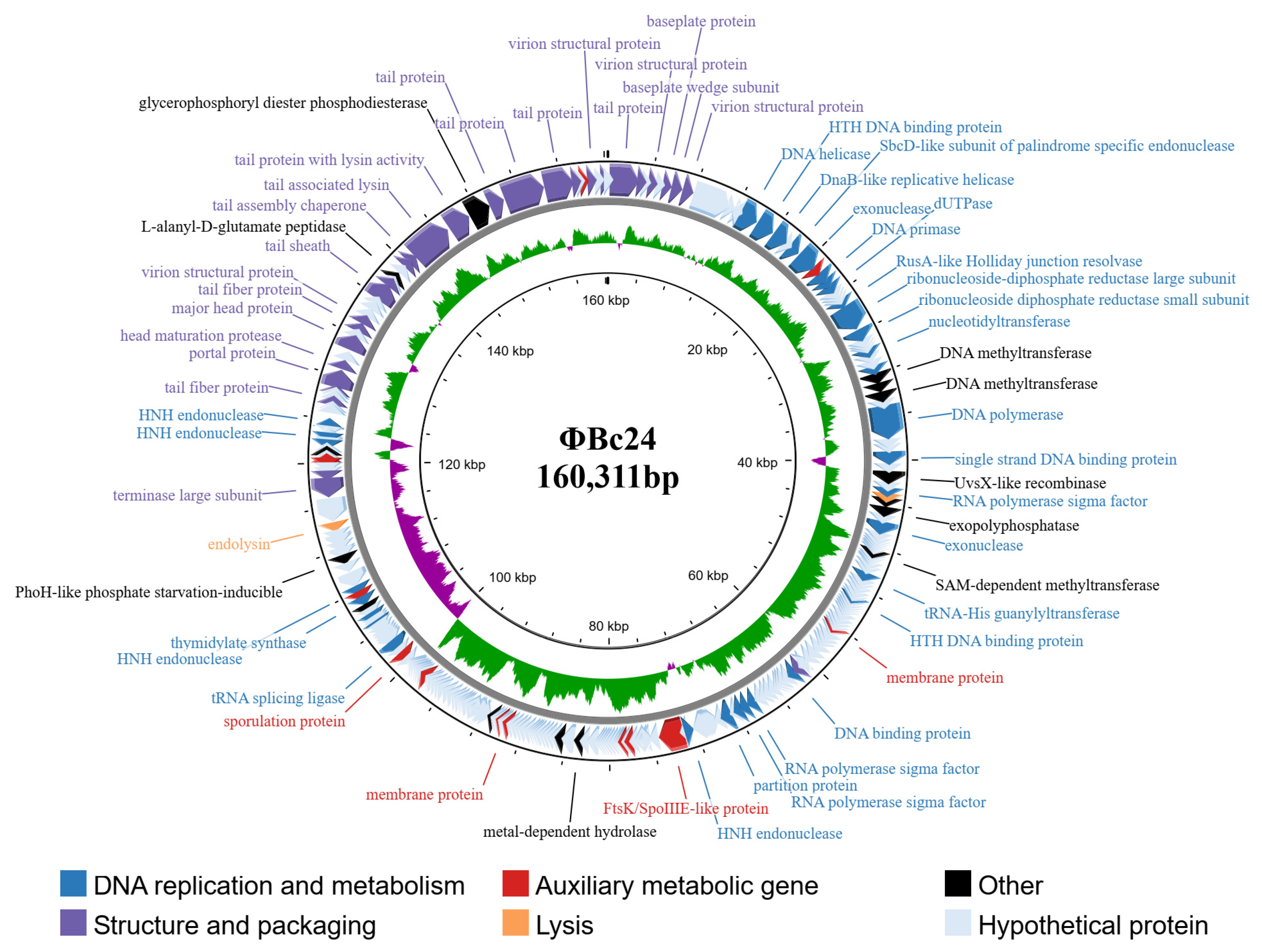
3.8. Characterization of Phage ΦBc24 Genome
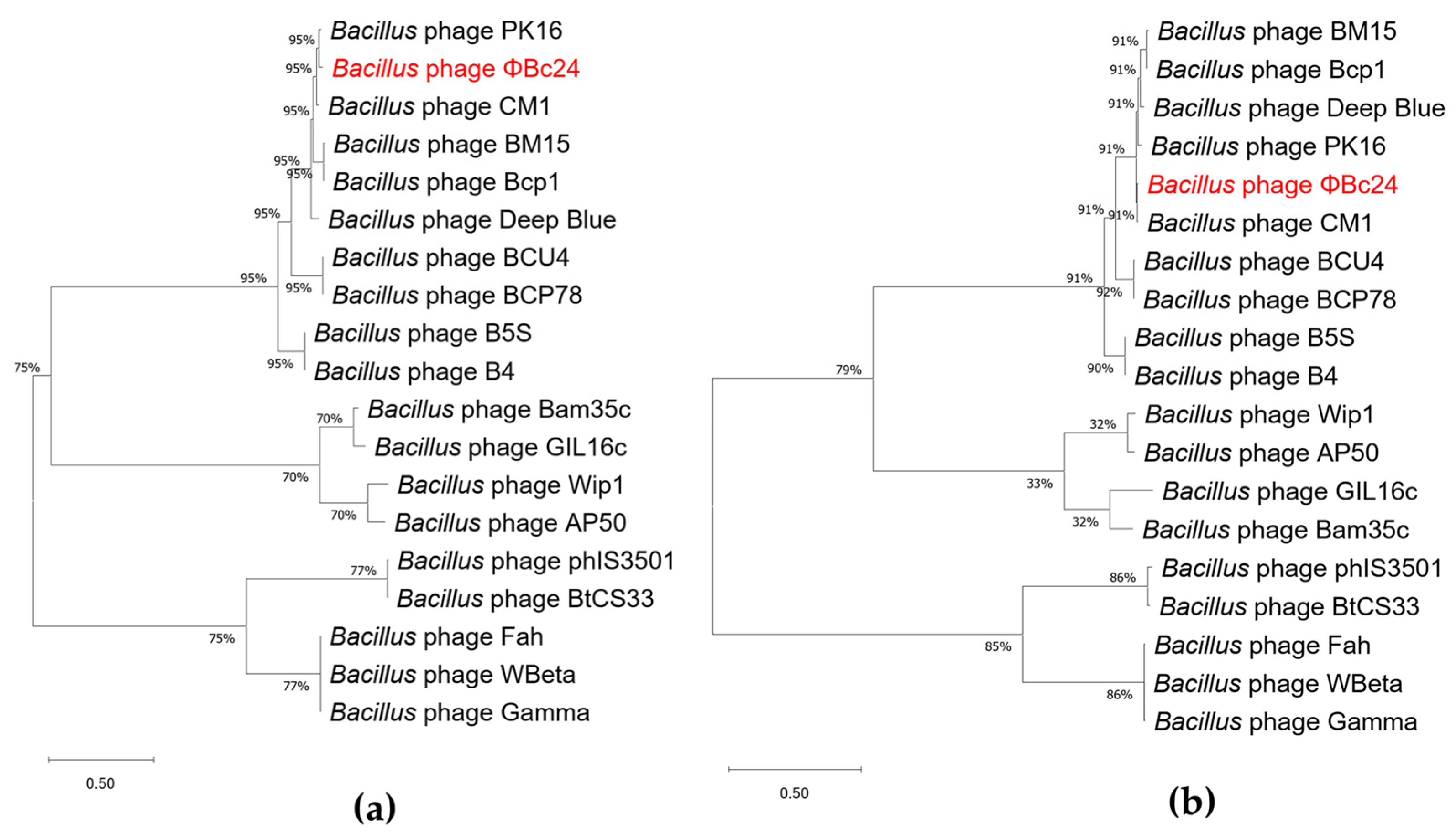
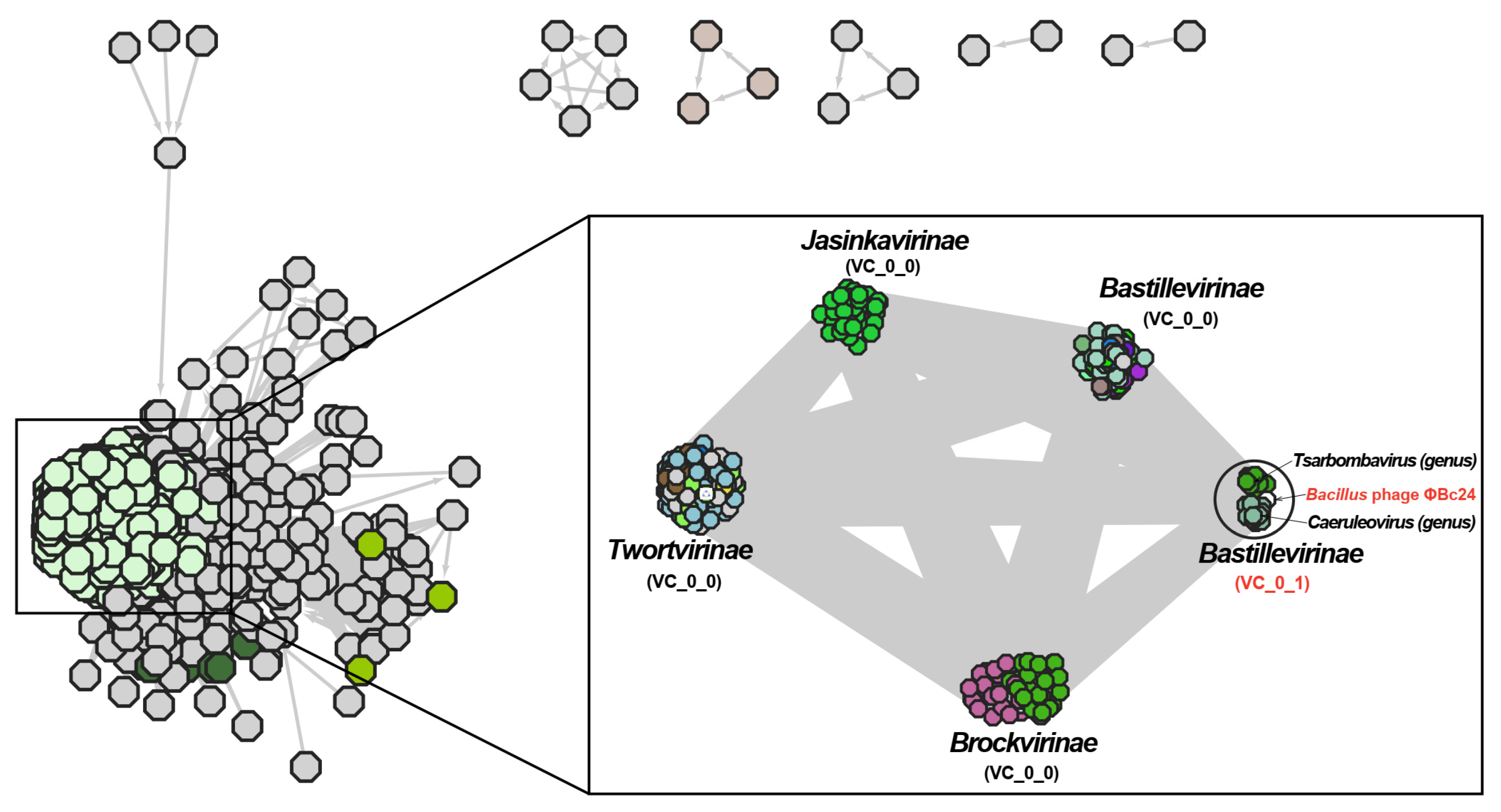
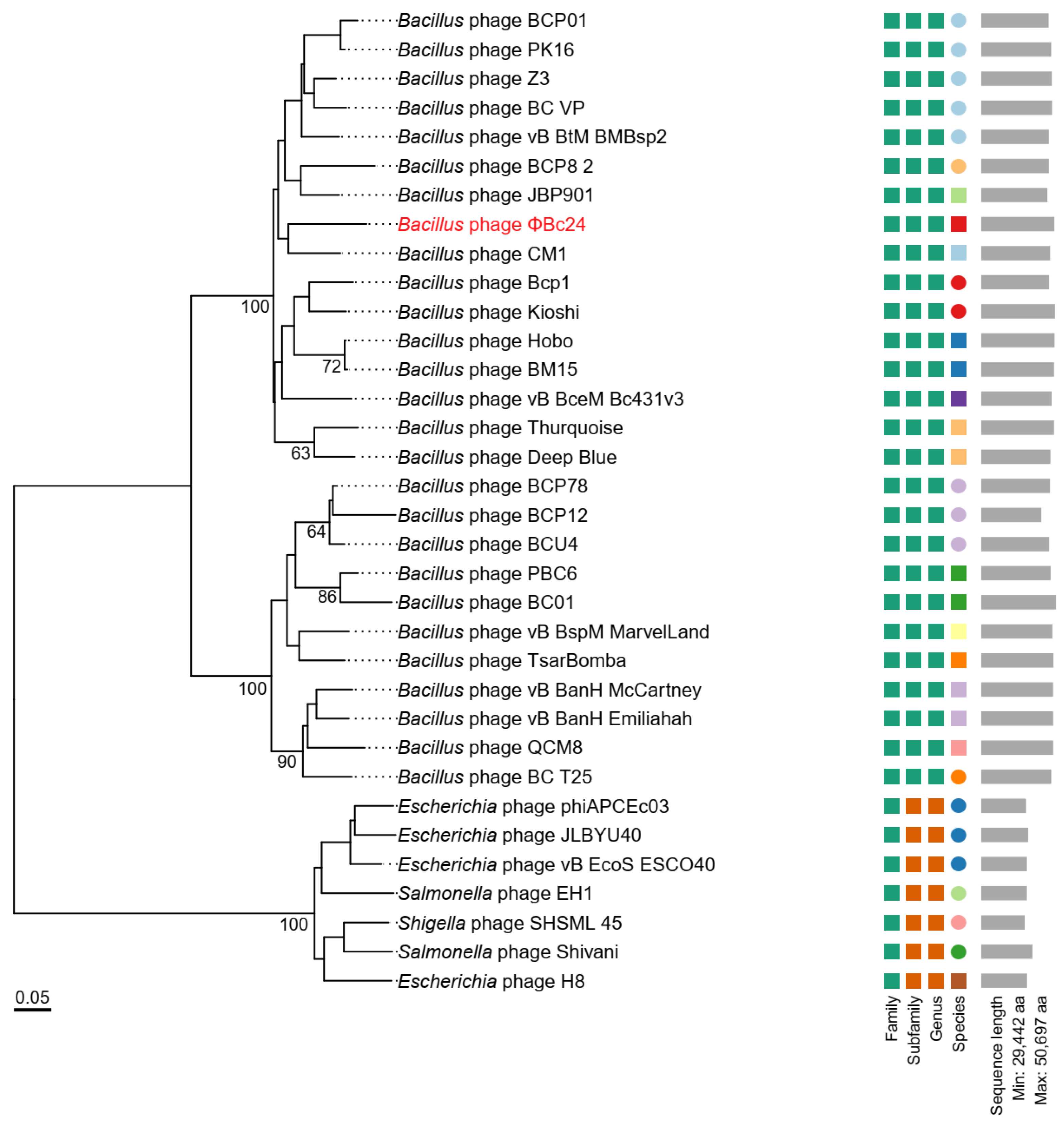
3.9. Phylogenetic Analyses of Phage ΦBc24
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic Acid |
| GC | Guanine-Cytosine |
| tRNA | Transfer ribonucleic acid |
| UV | Ultraviolet |
References
- Sun, H.; Xiang, X.; Li, Q.; Lin, H.; Wang, X.; Sun, J.; Luo, L.; Zheng, A. Comparative genome analysis of Bacillus thuringiensis strain HD521 and HS18-1. Sci. Rep. 2021, 11, 16590. [Google Scholar] [CrossRef]
- Xiao, Z.; Cheng, M.; Hu, X.; Xue, M.; Jiang, N.; Liu, W.; Fan, Y.; Meng, Y.; Xu, C.; Zhou, Y. Pathological changes of highly pathogenic Bacillus cereus on Pelodiscus sinensis. Vet. Q. 2023, 43, 1–10. [Google Scholar] [CrossRef]
- Kazantseva, O.A.; Piligrimova, E.G.; Shadrin, A.M. vB_BcM_Sam46 and vB_BcM_Sam112, members of a new bacteriophage genus with unusual small terminase structure. Sci. Rep. 2021, 11, 12173. [Google Scholar] [CrossRef]
- Huang, Z.; Yuan, X.; Zhu, Z.; Feng, Y.; Li, N.; Yu, S.; Li, C.; Chen, B.; Wu, S.; Gu, Q. Isolation and characterization of Bacillus cereus bacteriophage DZ1 and its application in foods. Food Chem. 2024, 431, 137128. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, J.; Ornelis, V.F.; Madder, A.; Rajkovic, A. Bacillus cereus food intoxication and toxicoinfection. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3719–3761. [Google Scholar] [CrossRef]
- Choi, W.; Kim, S.-S. Outbreaks, germination, and inactivation of Bacillus cereus in food products: A review. J. Food Prot. 2020, 83, 1480–1487. [Google Scholar] [CrossRef] [PubMed]
- Jessberger, N.; Dietrich, R.; Granum, P.E.; Märtlbauer, E. The Bacillus cereus food infection as multifactorial process. Toxins 2020, 12, 701. [Google Scholar]
- Thery, M.; Cousin, V.L.; Tissieres, P.; Enault, M.; Morin, L. Multi-organ failure caused by lasagnas: A case report of Bacillus cereus food poisoning. Front. Pediatr. 2022, 10, 978250. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yu, P.; Wang, J.; Li, C.; Guo, H.; Liu, C.; Kong, L.; Yu, L.; Wu, S.; Lei, T. A study on prevalence and characterization of Bacillus cereus in ready-to-eat foods in China. Front. Microbiol. 2020, 10, 3043. [Google Scholar] [CrossRef]
- Woh, P.; Ng, C. Bacillus cereus in rice: A review on food poisoning, antimicrobial resistance, and control measures. Trop. Biomed. 2024, 41, 298–309. [Google Scholar] [CrossRef]
- Noppakuadrittidej, P.; Charlermroj, R.; Makornwattana, M.; Kaew-Amdee, S.; Waditee-Sirisattha, R.; Vilaivan, T.; Praneenararat, T.; Karoonuthaisiri, N. Development of peptide nucleic acid-based bead array technology for Bacillus cereus detection. Sci. Rep. 2023, 13, 12482. [Google Scholar] [CrossRef] [PubMed]
- Glasset, B.; Herbin, S.; Guillier, L.; Cadel-Six, S.; Vignaud, M.-L.; Grout, J.; Pairaud, S.; Michel, V.; Hennekinne, J.-A.; Ramarao, N. Bacillus cereus-induced food-borne outbreaks in France, 2007 to 2014: Epidemiology and genetic characterisation. Eurosurveillance 2016, 21, 30413. [Google Scholar]
- Carroll, L.M.; Wiedmann, M.; Mukherjee, M.; Nicholas, D.C.; Mingle, L.A.; Dumas, N.B.; Cole, J.A.; Kovac, J. Characterization of emetic and diarrheal Bacillus cereus strains from a 2016 foodborne outbreak using whole-genome sequencing: Addressing the microbiological, epidemiological, and bioinformatic challenges. Front. Microbiol. 2019, 10, 144. [Google Scholar] [CrossRef]
- Duan, S.; Yu, Y.; Guo, Y.; Lu, D.; Li, N.; Liu, Z.; Liang, J.; Jiang, Y.; Wang, S.; Fu, P. Epidemiological Evaluation of Bacillus cereus-Induced Foodborne Outbreaks—China, 2010–2020. China CDC Wkly 2023, 5, 737. [Google Scholar]
- Rusnan, A.N.; Nordin, N.; Radu, S.; Abdul-Mutalib, N.A. Pathogenic Bacillus cereus, an Overlooked Food Contaminants in Southeast Asia. Pertanika J. Trop. Agric. Sci. 2020, 43, 1–17. [Google Scholar]
- Garcia, P.; Martinez, B.; Obeso, J.; Rodriguez, A. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 2008, 47, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.S.; Kim, Y.; Oh, S.; Jeon, W.M.; Frank, J.F.; Kim, S.H. Susceptibility of Listeria monocytogenes biofilms and planktonic cultures to hydrogen peroxide in food processing environments. Biosci. Biotechnol. Biochem. 2012, 76, 2008–2013. [Google Scholar] [CrossRef]
- Han, Q.; Song, X.; Zhang, Z.; Fu, J.; Wang, X.; Malakar, P.K.; Liu, H.; Pan, Y.; Zhao, Y. Removal of foodborne pathogen biofilms by acidic electrolyzed water. Front. Microbiol. 2017, 8, 988. [Google Scholar] [CrossRef]
- Meireles, A.; Ferreira, C.; Melo, L.; Simões, M. Comparative stability and efficacy of selected chlorine-based biocides against Escherichia coli in planktonic and biofilm states. Food Res. Int. 2017, 102, 511–518. [Google Scholar] [CrossRef]
- Artawinata, P.C.; Lorraine, S.; Waturangi, D.E. Isolation and characterization of bacteriophages from soil against food spoilage and foodborne pathogenic bacteria. Sci. Rep. 2023, 13, 9282. [Google Scholar] [CrossRef]
- Oh, H.; Seo, D.J.; Jeon, S.B.; Park, H.; Jeong, S.; Chun, H.S.; Oh, M.; Choi, C. Isolation and characterization of Bacillus cereus bacteriophages from foods and soil. Food Environ. Virol. 2017, 9, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Sulakvelidze, A. Using lytic bacteriophages to eliminate or significantly reduce contamination of food by foodborne bacterial pathogens. J. Sci. Food Agric. 2013, 93, 3137–3146. [Google Scholar] [CrossRef]
- Li, C.; Yuan, X.; Li, N.; Wang, J.; Yu, S.; Zeng, H.; Zhang, J.; Wu, Q.; Ding, Y. Isolation and characterization of Bacillus cereus phage vB_BceP-DLc1 reveals the largest member of the Φ29-like phages. Microorganisms 2020, 8, 1750. [Google Scholar] [CrossRef]
- Nacharaju, D.; Tapley, A.; Karmarkar, E.N.; Fang, F.C.; Johnson, N.J. Vancomycin-Resistant Bacillus cereus Pneumonia and Bacteremia After Nonfatal Freshwater Drowning. Ann. Intern. Med. Clin. Cases 2024, 3, e240226. [Google Scholar] [CrossRef]
- Abdelaziz, M.N.S.; Zayda, M.G.; Maung, A.T.; El-Telbany, M.; Mohammadi, T.N.; Lwin, S.Z.C.; Linn, K.Z.; Wang, C.; Yuan, L.; Masuda, Y.; et al. Genetic Characterization, Antibiotic Resistance, and Virulence Genes Profiling of Bacillus cereus Strains from Various Foods in Japan. Antibiotics 2024, 13, 774. [Google Scholar] [CrossRef] [PubMed]
- Kutateladze, M.; Adamia, R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 2010, 28, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Seol, H.; Kim, B.S.; Kim, M. Control of Bacillus cereus in rice cake and food contact surfaces with novel Becedseptimavirus genus phage BCC348 and its partial SPOR domain-containing endolysin LysBCC348. LWT 2024, 213, 117034. [Google Scholar] [CrossRef]
- Henry, M.; Debarbieux, L. Tools from viruses: Bacteriophage successes and beyond. Virology 2012, 434, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Elbreki, M.; Ross, R.P.; Hill, C.; O′ Mahony, J.; McAuliffe, O.; Coffey, A. Bacteriophages and their derivatives as biotherapeutic agents in disease prevention and treatment. J. Viruses 2014, 2014, 382539. [Google Scholar] [CrossRef]
- Abraha, H.B.; Kim, K.P.; Sbhatu, D.B. Bacteriophages for detection and control of foodborne bacterial pathogens—The case of Bacillus cereus and their phages. J. Food Saf. 2023, 43, e12906. [Google Scholar]
- Lee, W.J.; Billington, C.; Hudson, J.A.; Heinemann, J.A. Isolation and characterization of phages infecting Bacillus cereus. Lett. Appl. Microbiol. 2011, 52, 456–464. [Google Scholar] [CrossRef]
- Bandara, N.; Jo, J.; Ryu, S.; Kim, K.-P. Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol. 2012, 31, 9–16. [Google Scholar] [CrossRef]
- Van Quyen, D.; Lanh, P.T.; Thu, H.T. Isolation and characterization of beneficial bacteria from Apis Cerana honeybees from Hanoi, Vietnam. Vietnam J. Biotechnol. 2024, 22, 367–381. [Google Scholar] [CrossRef]
- Van Quyen, D.; Lanh, P.T.; Oanh, N.K.; Khang, T.N. Isolation and characterization of Salmonella enterica associated with diarrhea in chickens and ducks in Hai Duong province. Vietnam J. Biotechnol. 2024, 22, 425–436. [Google Scholar] [CrossRef]
- Pham, T.L.; Nguyen, K.O. Isolation and characterization of Escherichia coli associated with diarrhea in chickens and ducks in Hai Phong province. Acad. J. Biol. 2024, 46, 17–26. [Google Scholar] [CrossRef]
- Van Quyen, D.; Lanh, P.T.; Gahyun, L.; Hoa, N.T.; Phuoc, M.H. Isolation and phylogenetic analysis of Staphylococcus aureus strains isolated from meat in traditional markets in Ha Noi. Acad. J. Biol. 2025, 47, 73–84. [Google Scholar] [CrossRef]
- Lanh, P.T.; Duong, B.T.; Thu, H.T.; Hoa, N.T.; Van Quyen, D. Comprehensive analysis of the microbiome in Apis cerana honey highlights honey as a potential source for the isolation of beneficial bacterial strains. PeerJ 2024, 12, e17157. [Google Scholar] [CrossRef] [PubMed]
- Lukman, C.; Yonathan, C.; Magdalena, S.; Waturangi, D.E. Isolation and characterization of pathogenic Escherichia coli bacteriophages from chicken and beef offal. BMC Res. Notes 2020, 13, 8. [Google Scholar] [CrossRef]
- Khan, M.A.S.; Islam, Z.; Barua, C.; Sarkar, M.M.H.; Ahmed, M.F.; Rahman, S.R. Phenotypic characterization and genomic analysis of a Salmonella phage L223 for biocontrol of Salmonella spp. in poultry. Sci. Rep. 2024, 14, 15347. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; Van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef]
- Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Humana Press: Totowa, NJ, USA, 2009; pp. 15–21. [Google Scholar]
- Hong, Y.; Pan, Y.; Ebner, P. Development of bacteriophage treatments to reduce Escherichia coli O157: H7 contamination of beef products and produce. J. Anim. Sci. 2014, 92, 1366–1377. [Google Scholar] [CrossRef]
- Hien, V.T.; Lanh, P.T.; Pham, T.T.P.; Tran, K.N.; Duy, N.D.; Hoa, N.T.; Canh, N.X.; Nguyen, Q.H.; Kim, S.; Van Quyen, D. Isolation and characterization of a novel lytic bacteriophage Pv27 with biocontrol potential against Vibrio parahaemolyticus infections in shrimp. PeerJ 2025, 13, e19421. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Pharokka: A fast scalable bacteriophage annotation tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Li, X.; Guo, R.; Zou, X.; Yao, Y.; Lu, L. The First Cbk-Like Phage Infecting Erythrobacter, Representing a Novel Siphoviral Genus. Front. Microbiol. 2022, 13, 861793. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Cook, R.; Brown, N.; Redgwell, T.; Rihtman, B.; Barnes, M.; Clokie, M.; Stekel, D.J.; Hobman, J.; Jones, M.A.; Millard, A. INfrastructure for a PHAge REference database: Identification of large-scale biases in the current collection of cultured phage genomes. Phage 2021, 2, 214–223. [Google Scholar] [CrossRef]
- Bin Jang, H.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Brister, J.R.; Kropinski, A.M.; Krupovic, M.; Lavigne, R. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A. Complete genome sequence of DSM 30083 T, the type strain (U5/41 T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Fokas, R.; Kotsiri, Z.; Vantarakis, A. Can Bacteriophages Be Effectively Utilized for Disinfection in Animal-Derived Food Products? A Systematic Review. Pathogens 2025, 14, 291. [Google Scholar] [CrossRef]
- Endersen, L.; Coffey, A. The use of bacteriophages for food safety. Curr. Opin. Food Sci. 2020, 36, 1–8. [Google Scholar] [CrossRef]
- Bumunang, E.W.; Zaheer, R.; Niu, D.; Narvaez-Bravo, C.; Alexander, T.; McAllister, T.A.; Stanford, K. Bacteriophages for the targeted control of foodborne pathogens. Foods 2023, 12, 2734. [Google Scholar] [CrossRef]
- Goodridge, L.D.; Bisha, B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage 2011, 1, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.B.; Bhaskar, R.; Han, S.S. Bacteriophages: Natural antimicrobial bioadditives for food preservation in active packaging. Int. J. Biol. Macromol. 2024, 276, 133945. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, X. The application and research progress of bacteriophages in food safety. J. Appl. Microbiol. 2022, 133, 2137–2147. [Google Scholar] [CrossRef]
- Brovko, L.Y.; Anany, H.; Griffiths, M.W. Bacteriophages for detection and control of bacterial pathogens in food and food-processing environment. Adv. Food Nutr. Res. 2012, 67, 241–288. [Google Scholar]
- Singh, V. Bacteriophage-mediated biosensors for detection of foodborne pathogens. In Microbial Bioprospecting for Sustainable Development; Springer: Singapore, 2018; pp. 353–384. [Google Scholar]
- Zinke, M.; Schröder, G.F.; Lange, A. Major tail proteins of bacteriophages of the order Caudovirales. J. Biol. Chem. 2022, 298, 101472. [Google Scholar] [CrossRef]
- Li, N.; Yuan, X.; Li, C.; Chen, N.; Wang, J.; Chen, B.; Yu, S.; Yu, P.; Zhang, J.; Zeng, H. A novel Bacillus cereus bacteriophage DLn1 and its endolysin as biocontrol agents against Bacillus cereus in milk. Int. J. Food Microbiol. 2022, 369, 109615. [Google Scholar] [CrossRef]
- Na, H.; Kong, M.; Ryu, S. Characterization of LysPBC4, a novel Bacillus cereus-specific endolysin of bacteriophage PBC4. FEMS Microbiol. Lett. 2016, 363, fnw092. [Google Scholar] [CrossRef]
- Hock, L.; Gillis, A.; Mahillon, J. Complete genome sequence of bacteriophage Deep-Purple, a novel member of the family Siphoviridae infecting Bacillus cereus. Arch. Virol. 2018, 163, 2555–2559. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Chen, H.; Huang, S.; Zhu, B.; Wu, J.; Chen, M.; Zhang, J.; Wang, J.; Ding, Y.; Wu, Q. Characterization of the novel phage vB_BceP_LY3 and its potential role in controlling Bacillus cereus in milk and rice. Int. J. Food Microbiol. 2024, 421, 110778. [Google Scholar] [CrossRef]
- Peng, Q.; Yuan, Y. Characterization of a novel phage infecting the pathogenic multidrug-resistant Bacillus cereus and functional analysis of its endolysin. Appl. Microbiol. Biotechnol. 2018, 102, 7901–7912. [Google Scholar] [CrossRef]
- Chen, B.; Huang, Z.; Yuan, X.; Li, C.; Wang, J.; Chen, M.; Xue, L.; Zhang, J.; Wu, Q.; Ding, Y. Isolation and characterization of two phages against emetic Bacillus cereus and their potential applications. Food Front. 2024, 5, 2305–2318. [Google Scholar] [CrossRef]
- Tan, C.W.; Rukayadi, Y.; Hasan, H.; Abdul-Mutalib, N.-A.; Jambari, N.N.; Hara, H.; Thung, T.Y.; Lee, E.; Radu, S. Isolation and characterization of six Vibrio parahaemolyticus lytic bacteriophages from seafood samples. Front. Microbiol. 2021, 12, 616548. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.; Niu, X.; Xiong, G.; Chen, G.; Wu, H.; Ma, Z.; Zhu, K.; Liu, Y.; Wang, G. Phenotypic and genotypic characterization of the new Bacillus cereus phage SWEP1. Arch. Virol. 2021, 166, 3183–3188. [Google Scholar] [CrossRef] [PubMed]
- Tey, B.T.; Ooi, S.T.; Yong, K.C.; Ng, M.Y.T.; Ling, T.C.; Tan, W.S. Production of fusion m13 phage bearing the di-sulphide constrained peptide sequence (C-WSFFSNI-C) that interacts with hepatitis B core antigen. Afr. J. Biotechnol. 2009, 8, 268. [Google Scholar]
- Nakai, T.; Park, S.C. Bacteriophage therapy of infectious diseases in aquaculture. Res. Microbiol. 2002, 153, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Jarrell, K.F.; Vydykhan, T.; Lee, P.; Agnew, M.D.; Thomas, N.A. Isolation and characterization of bacteriophage BCJA1, a novel temperate bacteriophage active against the alkaliphilic bacterium, Bacillus clarkii. Extremophiles 1997, 1, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Sae-Ueng, U.; Bunsuwansakul, C.; Showpanish, K.; Phironrit, N.; Thadajarassiri, J.; Nehls, C. Nanomechanical resilience and thermal stability of RSJ2 phage. Sci. Rep. 2024, 14, 19389. [Google Scholar] [CrossRef]
- Hardy, J.M.; Dunstan, R.A.; Grinter, R.; Belousoff, M.J.; Wang, J.; Pickard, D.; Venugopal, H.; Dougan, G.; Lithgow, T.; Coulibaly, F. The architecture and stabilisation of flagellotropic tailed bacteriophages. Nat. Commun. 2020, 11, 3748. [Google Scholar] [CrossRef]
- Węglewska, M.; Barylski, J.; Wojnarowski, F.; Nowicki, G.; Łukaszewicz, M. Genome, biology and stability of the Thurquoise phage—A new virus from the Bastillevirinae subfamily. Front. Microbiol. 2023, 14, 1120147. [Google Scholar] [CrossRef]
- Kong, M.; Ryu, S. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl. Environ. Microbiol. 2015, 81, 2274–2283. [Google Scholar] [CrossRef]
- Alrafaie, A.M.; Stafford, G.P. Enterococcal bacteriophage: A survey of the tail associated lysin landscape. Virus Res. 2023, 327, 199073. [Google Scholar] [CrossRef]
- Mursalin, M.H.; Astley, R.; Coburn, P.S.; Bagaruka, E.; Hunt, J.J.; Fischetti, V.A.; Callegan, M.C. Therapeutic potential of Bacillus phage lysin PlyB in ocular infections. Msphere 2023, 8, e00044-00023. [Google Scholar] [CrossRef]
- Kaliniene, L.; Noreika, A.; Kaupinis, A.; Valius, M.; Jurgelaitis, E.; Lazutka, J.; Meškienė, R.; Meškys, R. Analysis of a novel bacteriophage vB_AchrS_AchV4 highlights the diversity of achromobacter viruses. Viruses 2021, 13, 374. [Google Scholar] [CrossRef]
- Ge, F.; Guo, R.; Liang, Y.; Chen, Y.; Shao, H.; Sung, Y.Y.; Mok, W.J.; Wong, L.L.; McMinn, A.; Wang, M. Characterization and genomic analysis of Stutzerimonas stutzeri phage vB_PstS_ZQG1, representing a novel viral genus. Virus Res. 2023, 336, 199226. [Google Scholar] [CrossRef]
- Xu, H.; Bao, X.; Hong, W.; Wang, A.; Wang, K.; Dong, H.; Hou, J.; Govinden, R.; Deng, B.; Chenia, H.Y. Biological Characterization and Evolution of Bacteriophage T7-Δholin During the Serial Passage Process. Front. Microbiol. 2021, 12, 705310. [Google Scholar] [CrossRef]
- Kornienko, M.; Bespiatykh, D.; Gorodnichev, R.; Abdraimova, N.; Shitikov, E. Transcriptional Landscapes of Herelleviridae Bacteriophages and Staphylococcus aureus during Phage Infection: An Overview. Viruses 2023, 15, 1427. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Mikoulinskaia, G.V.; Prokhorov, D.A.; Chernyshov, S.V.; Sitnikova, D.S.; Arakelian, A.G.; Uversky, V.N. Conservative Tryptophan Residue in the Vicinity of an Active Site of the M15 Family l, d-Peptidases: A Key Element in the Catalysis. Int. J. Mol. Sci. 2023, 24, 13249. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.A.; Rodríguez-Ramos, J.A.; Shaffer, M.; Flynn, R.M.; Daly, R.A.; Wrighton, K.C.; Lennon, J.T. Human-gut phages harbor sporulation genes. mBio 2023, 14, e00182-23. [Google Scholar] [CrossRef]
- Butala, M.; Dragoš, A. Unique relationships between phages and endospore-forming hosts. Trends Microbiol. 2023, 31, 498–510. [Google Scholar] [CrossRef]
- Cattoni, D.I.; Thakur, S.; Godefroy, C.; Le Gall, A.; Lai-Kee-Him, J.; Milhiet, P.-E.; Bron, P.; Nöllmann, M. Structure and DNA-binding properties of the Bacillus subtilis SpoIIIE DNA translocase revealed by single-molecule and electron microscopies. Nucleic Acids Res. 2014, 42, 2624–2636. [Google Scholar] [CrossRef]
- Chan, H.; Mohamed, A.M.; Grainge, I.; Rodrigues, C.D. FtsK and SpoIIIE, coordinators of chromosome segregation and envelope remodeling in bacteria. Trends Microbiol. 2022, 30, 480–494. [Google Scholar] [CrossRef]
- Brown, K.L.; Hughes, K.T. The role of anti-sigma factors in gene regulation. Mol. Microbiol. 1995, 16, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Walkinshaw, M.; Taylor, P.; Sturrock, S.; Atanasiu, C.; Berge, T.; Henderson, R.M.; Edwardson, J.; Dryden, D. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell 2002, 9, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Xie, N.; Zhang, X.; Yang, S.; Lv, S. Computational insights into the dynamic structural features and binding characteristics of recombinase UvsX compared with RecA. Molecules 2023, 28, 3363. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Ma, Y.; Wang, J. Isolation and characteristic of Bacillus cereus phage Z3 and its application in rice and milk. LWT 2024, 198, 116022. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Brister, J.R. How to name and classify your phage: An informal guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A roadmap for genome-based phage taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Barylski, J.; Enault, F.; Dutilh, B.E.; Schuller, M.B.; Edwards, R.A.; Gillis, A.; Klumpp, J.; Knezevic, P.; Krupovic, M.; Kuhn, J.H. Analysis of spounaviruses as a case study for the overdue reclassification of tailed phages. Syst. Biol. 2020, 69, 110–123. [Google Scholar] [CrossRef]
- Koonin, E.V.; Yutin, N. The crAss-like phage group: How metagenomics reshaped the human virome. Trends Microbiol. 2020, 28, 349–359. [Google Scholar] [CrossRef]
- de Jonge, P.A.; van den Born, B.-J.H.; Zwinderman, A.H.; Nieuwdorp, M.; Dutilh, B.E.; Herrema, H. Phylogeny and disease associations of a widespread and ancient intestinal bacteriophage lineage. Nat. Commun. 2024, 15, 6346, Correction in Nat. Commun. 2024, 15, 9724. [Google Scholar] [CrossRef]
- Sada, T.S.; Alemayehu, D.H.; Tafese, K.M.; Tessema, T.S. Genomic analysis and characterization of lytic bacteriophages that target antimicrobial resistant Escherichia coli in Addis Ababa, Ethiopia. Heliyon 2024, 10, e40342. [Google Scholar] [CrossRef]
- Rohwer, F.; Edwards, R. The Phage Proteomic Tree: A genome-based taxonomy for phage. J. Bacteriol. 2002, 184, 4529–4535. [Google Scholar] [CrossRef] [PubMed]
- Dewanggana, M.N.; Waturangi, D.E.; Yogiara. Genomic characterization of bacteriophage BI-EHEC infecting strains of Enterohemorrhagic Escherichia coli. BMC Res. Notes 2021, 14, 459. [Google Scholar] [CrossRef]
- Rizkinata, D.; Kusnadi, V.C.; Waturangi, D.E.; Yulandi, A. Isolation and molecular characterization of bacteriophages isolated from lake water and their application in foods against Bacillus cereus. BMC Res. Notes 2025, 18, 364. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Bandara, N.; Shin, E.; Ryu, S.; Kim, K.-p. Prevalence of Bacillus cereus bacteriophages in fermented foods and characterization of phage JBP901. Res. Microbiol. 2011, 162, 791–797. [Google Scholar] [CrossRef]
- Gillis, A.; Mahillon, J. Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: Past, present and future. Viruses 2014, 6, 2623–2672. [Google Scholar] [CrossRef]
- Wójcicki, M.; Sokołowska, B.; Górski, A.; Jończyk-Matysiak, E. Dual nature of bacteriophages: Friends or foes in minimally processed food products—A comprehensive review. Viruses 2025, 17, 778. [Google Scholar] [CrossRef]
- Ha, P.N.; Hanh, N.T.; Van, V.K.; Le Minh, T.; Trung, N.T. Isolation and characterization of Bacillus cereus strains isolated from a beef pizza food poisoning incident in hanoi. Health Risk Anal. 2025, 98–106. [Google Scholar] [CrossRef]
- Huy, N.Q.; Trang, P.Q.; Loi, N.T. The occurrence of potential pathogenic antibiotic-resistant bacteria in the marine environment in Ha Long Bay, Vietnam. Vietnam J. Sci. Technol. Eng. 2024, 66, 101–106. [Google Scholar]
- Amjad, N.; Naseer, M.S.; Imran, A.; Menon, S.V.; Sharma, A.; Islam, F.; Tahir, S.; Shah, M.A. A mini-review on the role of bacteriophages in food safety. CYTA-J. Food 2024, 22, 2357192. [Google Scholar]
- Dhulipalla, H.; Basavegowda, N.; Haldar, D.; Syed, I.; Ghosh, P.; Rana, S.S.; Somu, P.; Naidu, R.; Yadav, A.K.; Lee, M.-J. Integrating phage biocontrol in food production: Industrial implications and regulatory overview. Discov. Appl. Sci. 2025, 7, 314. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, N.K.; Lanh, P.T.; Trinh Thu, T.; Phuoc, M.H.; Duy, N.D.; Hien, V.T.; Quyen, D.V. Genomic and Biological Insights of Bacteriophage ΦBc24 Targeting Bacillus cereus. Curr. Issues Mol. Biol. 2025, 47, 906. https://doi.org/10.3390/cimb47110906
Tran NK, Lanh PT, Trinh Thu T, Phuoc MH, Duy ND, Hien VT, Quyen DV. Genomic and Biological Insights of Bacteriophage ΦBc24 Targeting Bacillus cereus. Current Issues in Molecular Biology. 2025; 47(11):906. https://doi.org/10.3390/cimb47110906
Chicago/Turabian StyleTran, Nam Khang, Pham Thi Lanh, Trang Trinh Thu, Man Hong Phuoc, Nguyen Dinh Duy, Vu Thi Hien, and Dong Van Quyen. 2025. "Genomic and Biological Insights of Bacteriophage ΦBc24 Targeting Bacillus cereus" Current Issues in Molecular Biology 47, no. 11: 906. https://doi.org/10.3390/cimb47110906
APA StyleTran, N. K., Lanh, P. T., Trinh Thu, T., Phuoc, M. H., Duy, N. D., Hien, V. T., & Quyen, D. V. (2025). Genomic and Biological Insights of Bacteriophage ΦBc24 Targeting Bacillus cereus. Current Issues in Molecular Biology, 47(11), 906. https://doi.org/10.3390/cimb47110906




