Genome-Wide Identification, Phylogeny and Expression Analysis of the Magnesium Release Gene Family in Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening and Identification of MGR Family Members in Wheat
2.2. Construction of the Phylogenetic Tree and Analysis of Protein Conserved Motifs in MGR Family
2.3. Chromosomal Localization and Gene Structure Analysis
2.4. Gene Duplication Events and Correlation Analysis
2.5. Cis-Element Analysis
2.6. Analysis of Gene Expression Patterns
3. Results
3.1. Screening and Identification of MGR Family Members in Wheat
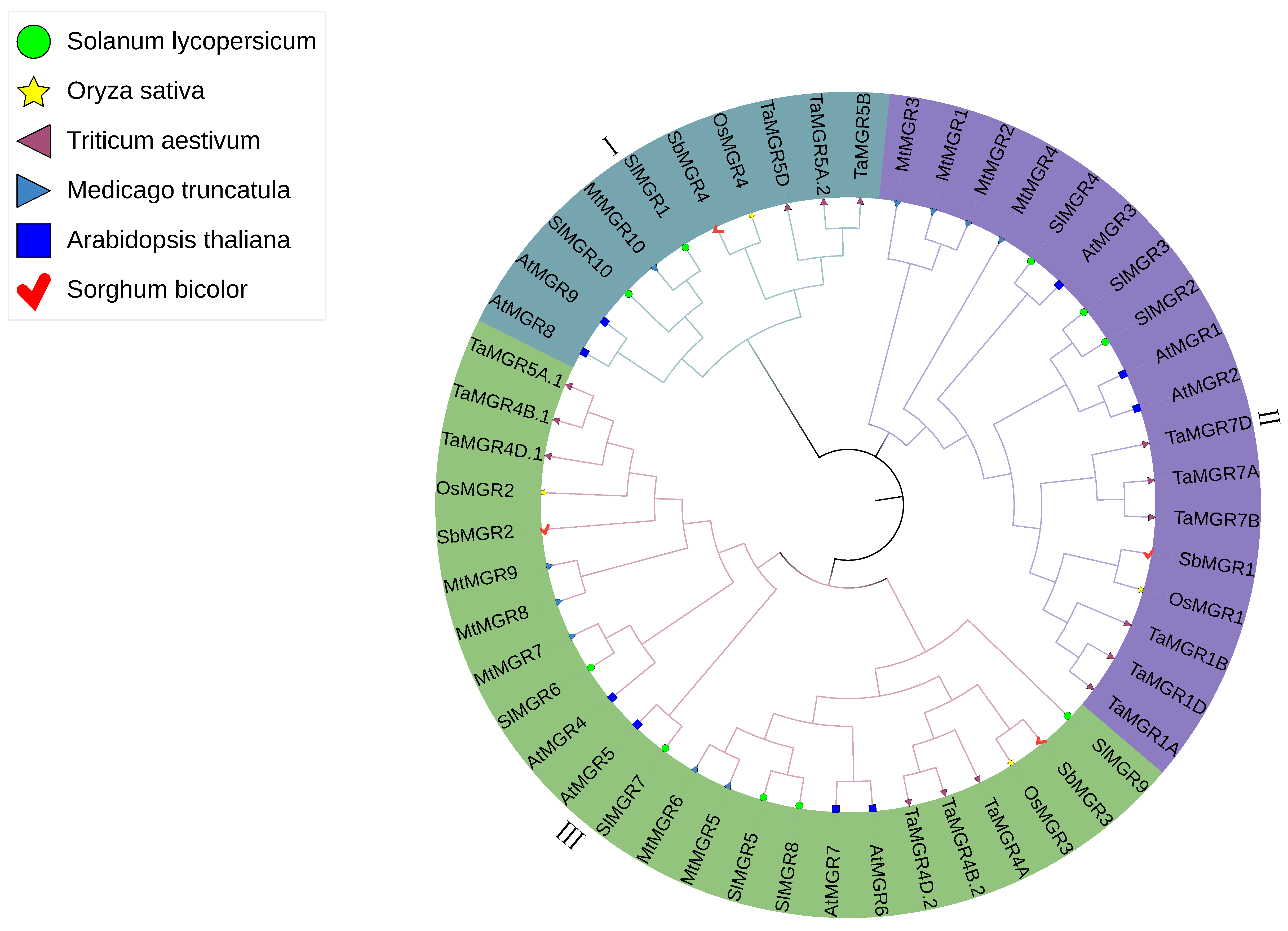
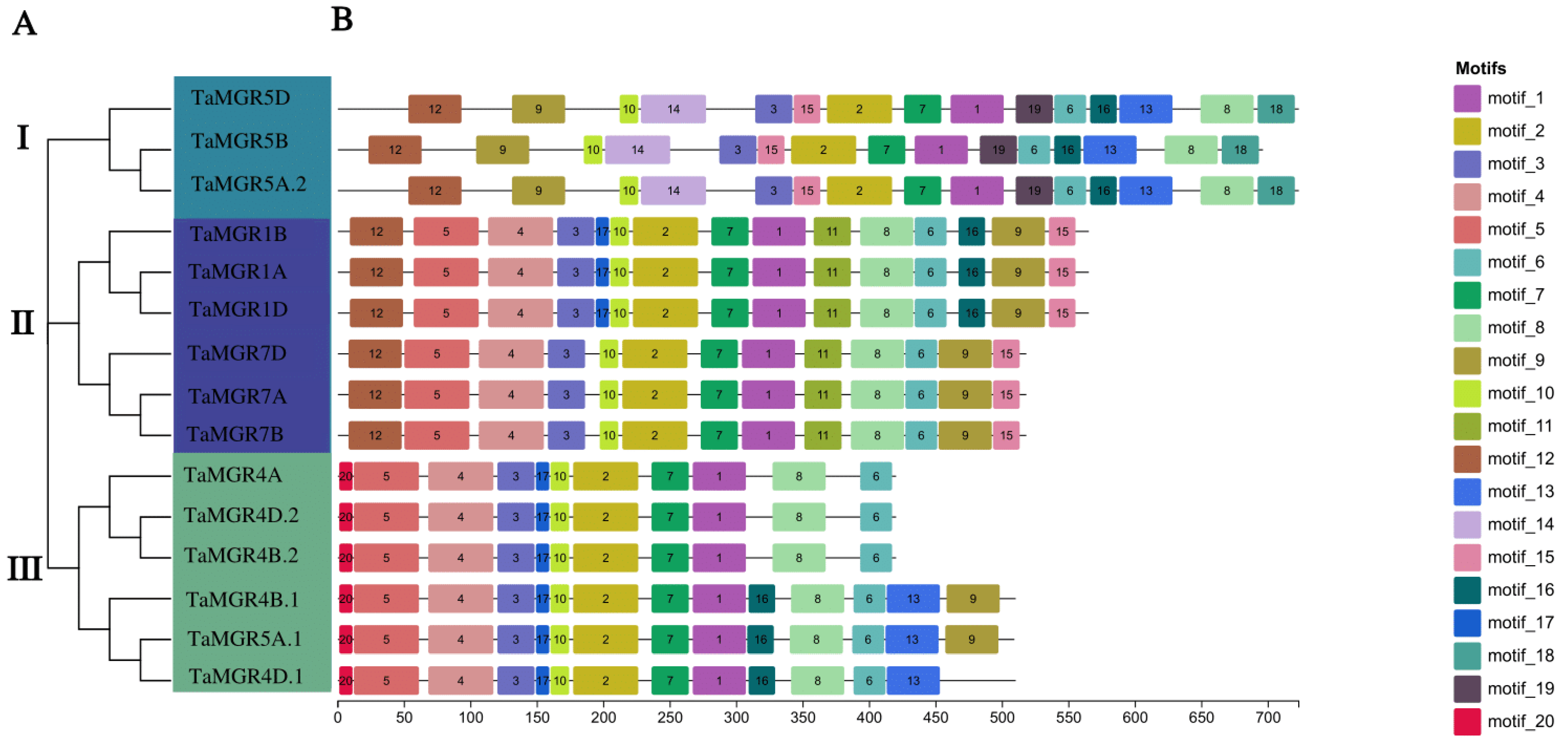
3.2. Construction of the Phylogenetic Tree and Analysis of Protein Conserved Motifs in the Wheat MGR Family
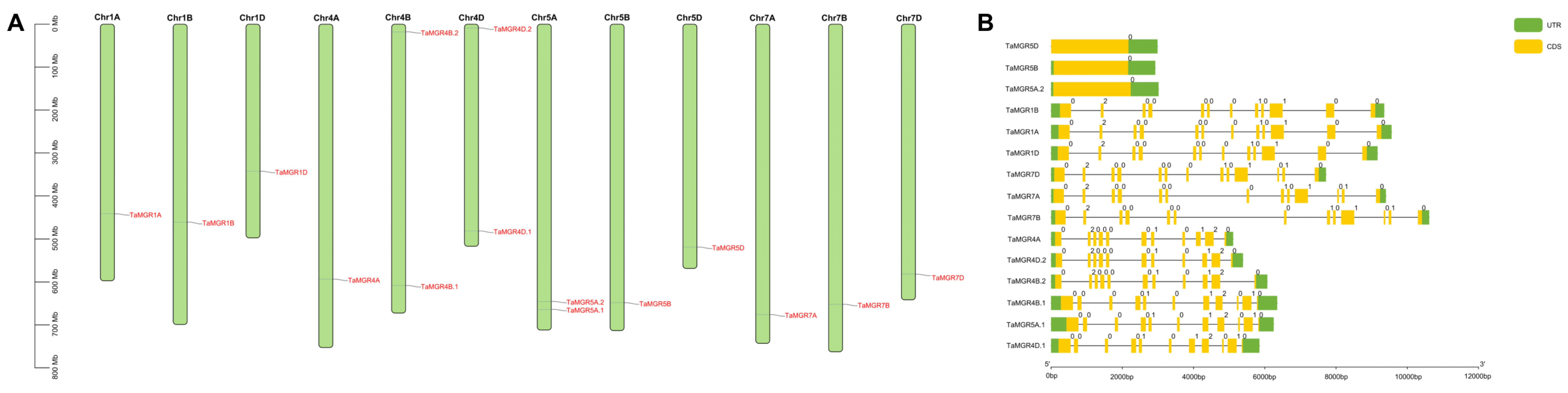
3.3. Chromosomal Localization and Gene Structure Analysis
3.4. Gene Duplication Events and Synteny Analysis
3.5. Analysis of Cis-Acting Elements
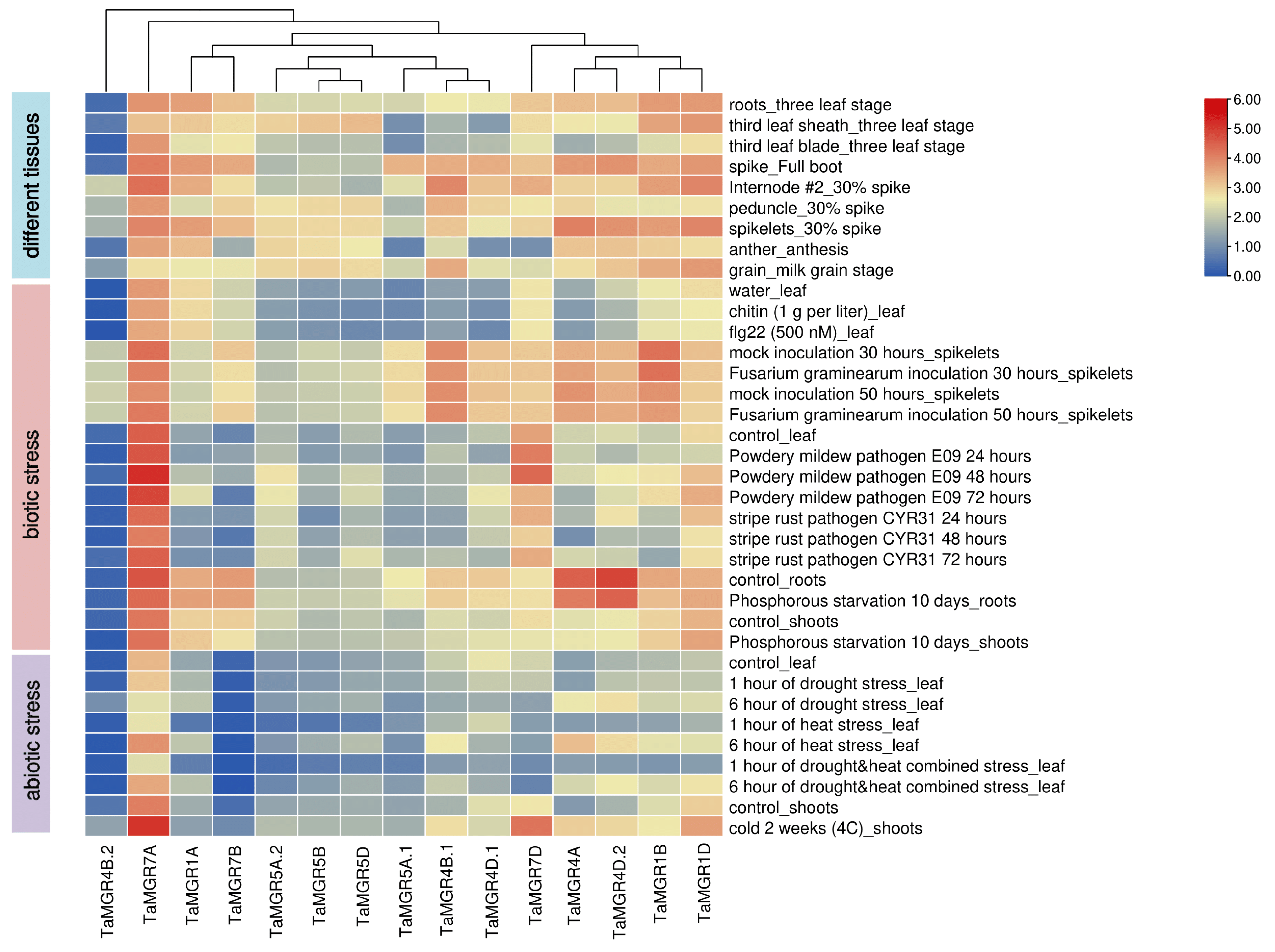
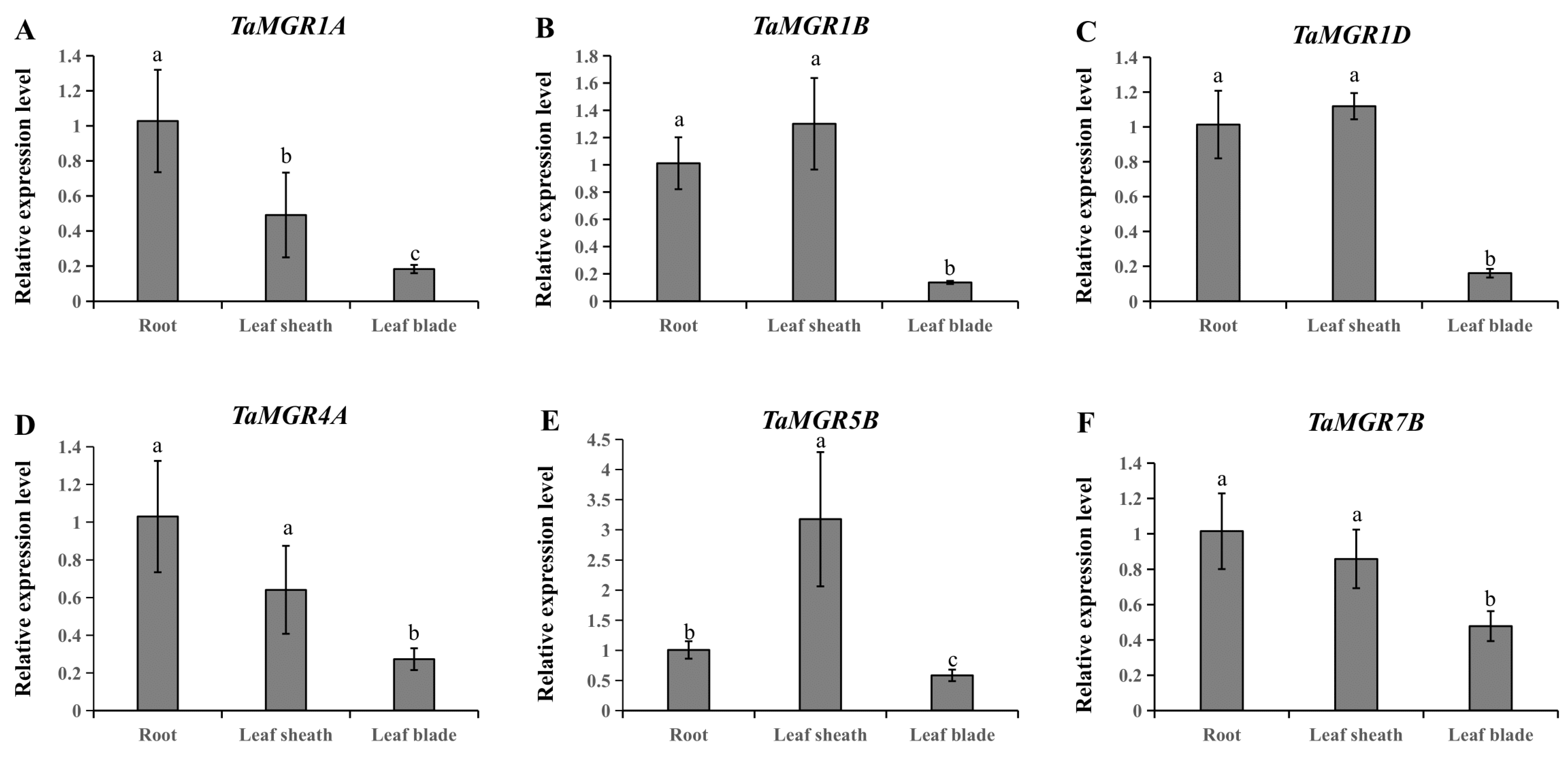
3.6. Analysis of TaMGRs Expression Patterns
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kobayashi, N.I.; Tanoi, K. Critical issues in the study of magnesium transport systems and magnesium deficiency symptoms in plants. Int. J. Mol. Sci. 2015, 16, 23076–23093. [Google Scholar] [CrossRef]
- Williams, L.; Salt, D.E. The plant ionome coming into focus. Curr. Opin. Plant Biol. 2009, 12, 247. [Google Scholar] [CrossRef]
- Cowan, J. Structural and catalytic chemistry of magnesium-dependent enzymes. Biometals 2002, 15, 225–235. [Google Scholar] [CrossRef]
- Horlitz, M.; Klaff, P. Gene-specific trans-regulatory functions of magnesium for chloroplast mRNA stability in higher plants. J. Biol. Chem. 2000, 275, 35638–35645. [Google Scholar] [CrossRef]
- Rissler, H.M.; Collakova, E.; DellaPenna, D.; Whelan, J.; Pogson, B.J. Chlorophyll biosynthesis. Expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiol. 2002, 128, 770–779. [Google Scholar] [CrossRef]
- White, P.J.; Broadley, M.R. Biofortification of crops with seven mineral elements often lacking in human diets–iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 2009, 182, 49–84. [Google Scholar] [CrossRef]
- Gerendás, J.; Führs, H. The significance of magnesium for crop quality. Plant Soil. 2013, 368, 101–128. [Google Scholar] [CrossRef]
- Tanoi, K.; Kobayashi, N.I. Leaf senescence by magnesium deficiency. Plants 2015, 4, 756–772. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.L.; Chen, S.N.; Hussain, N.; Cong, Y.X.; Liang, Z.S.; Chen, K.M. Magnesium stress signaling in plant: Just a beginning. Plant Signal. Behav. 2015, 10, e992287. [Google Scholar] [CrossRef] [PubMed]
- Dukic, E.; Van Maldegem, K.A.; Shaikh, K.M.; Fukuda, K.; Töpel, M.; Solymosi, K.; Hellsten, J.; Hansen, T.H.; Husted, S.; Higgins, J. Chloroplast magnesium transporters play essential but differential roles in maintaining magnesium homeostasis. Front. Plant Sci. 2023, 14, 1221436. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.J.; Yang, Y.; Yan, Y.W.; Mao, D.D.; Yuan, H.M.; Wang, C.; Zhao, F.G.; Luan, S. Two transporters mobilize magnesium from vacuolar stores to enable plant acclimation to magnesium deficiency. Plant Physiol. 2022, 190, 1307–1320. [Google Scholar] [CrossRef]
- Silver, S. Active transport of magnesium in Escherichia coli. Proc. Natl. Acad. Sci. USA 1969, 62, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Franken, G.; Huynen, M.; Martínez-Cruz, L.; Bindels, R.; de Baaij, J. Structural and functional comparison of magnesium transporters throughout evolution. Cell Mol. Life Sci. 2022, 79, 418. [Google Scholar] [CrossRef]
- Li, L.; Tutone, A.F.; Drummond, R.S.; Gardner, R.C.; Luan, S. A novel family of magnesium transport genes in Arabidopsis. Plant Cell 2001, 13, 2761–2775. [Google Scholar] [CrossRef]
- Deng, W.; Luo, K.; Li, D.; Zheng, X.; Wei, X.; Smith, W.; Thammina, C.; Lu, L.; Li, Y.; Pei, Y. Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance. J. Exp. Bot. 2006, 57, 4235–4243. [Google Scholar] [CrossRef]
- Drummond, R.; Tutone, A.; Li, Y.C.; Gardner, R. A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci. 2006, 170, 78–89. [Google Scholar] [CrossRef]
- Li, L.G.; Sokolov, L.N.; Yang, Y.H.; Li, D.P.; Ting, J.; Pandy, G.K.; Luan, S. A mitochondrial magnesium transporter functions in Arabidopsis pollen development. Mol. Plant 2008, 1, 675–685. [Google Scholar] [CrossRef]
- Yan, Y.W.; Mao, D.D.; Yang, L.; Qi, J.L.; Zhang, X.X.; Tang, Q.L.; Li, Y.P.; Tang, R.J.; Luan, S. Magnesium transporter MGT6 plays an essential role in maintaining magnesium homeostasis and regulating high magnesium tolerance in Arabidopsis. Front. Plant. Sci. 2018, 9, 274. [Google Scholar] [CrossRef]
- Mao, D.D.; Tian, L.F.; Li, L.G.; Chen, J.; Deng, P.Y.; Li, D.P.; Luan, S. AtMGT7: An Arabidopsis gene encoding a low-affinity magnesium transporter. J. Integr. Plant Biol. 2008, 50, 1530–1538. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.C.; Yamaji, N.; Horie, T.; Che, J.; Li, J.; An, G.; Ma, J.F. A magnesium transporter OsMGT1 Plays a critical role in salt tolerance in rice. Plant Physiol. 2017, 174, 1837–1849. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yokosho, K.; Liu, S.; Cao, H.R.; Yamaji, N.; Zhu, X.G.; Liao, H.; Ma, J.F.; Chen, Z.C. Diel magnesium fluctuations in chloroplasts contribute to photosynthesis in rice. Nat. Plants 2020, 6, 848–859. [Google Scholar] [CrossRef]
- Li, H.; Du, H.; Huang, K.; Chen, X.; Liu, T.; Gao, S.; Liu, H.; Tang, Q.; Rong, T.; Zhang, S. Identification, and functional and expression analyses of the CorA/MRS2/MGT-type magnesium transporter family in maize. Plant Cell Physiol. 2016, 57, 1153–1168. [Google Scholar] [CrossRef]
- Li, H.Y.; Wang, N.; Ding, J.; Liu, C.; Du, H.; Huang, K.; Cao, M.; Lu, Y.; Gao, S.; Zhang, S. The maize CorA/MRS2/MGT-type magnesium transporter, ZmMGT10, responses to magnesium deficiency and confers low magnesium tolerance in transgenic Arabidopsis. Plant Mol. Biol. 2017, 95, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Dölger, J.L.; Sagervanshi, A.; Pitann, B.; Mühling, K.H. The magnesium-specific uptake and translocation transporters ZmMGT10 and MGR6 are upregulated not only by magnesium deficiency but also by high potassium concentrations in maize. Plant Physiol. Bioch. 2025, 224, 109977. [Google Scholar] [CrossRef]
- Cao, H.R.; Peng, W.T.; Nie, M.M.; Bai, S.; Chen, C.Q.; Liu, Q.; Guo, Z.L.; Liao, H.; Chen, Z.C. Carbon-nitrogen trading in symbiotic nodules depends on magnesium import. Curr. Biol. 2022, 32, 4337–4349.e4335. [Google Scholar] [CrossRef]
- Anwar, A.; Akhtar, J.; Aleem, S.; Aleem, M.; Razzaq, M.K.; Alamri, S.; Raza, Q.; Sharif, I.; Iftikhar, A.; Naseer, S. Genome-wide identification of MGT gene family in soybean (Glycine max) and their expression analyses under magnesium stress conditions. BMC Plant Biol. 2025, 25, 83. [Google Scholar] [CrossRef]
- Tang, R.J.; Meng, S.F.; Zheng, X.J.; Zhang, B.; Yang, Y.; Wang, C.; Fu, A.G.; Zhao, F.G.; Lan, W.Z.; Luan, S. Conserved mechanism for vacuolar magnesium sequestration in yeast and plant cells. Nat. Plants 2022, 8, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Shi, J.D.; Yang, P.; Kumar, P.G.; Li, Q.Z.; Run, Q.G.; Su, Y.C.; Scott, H.S.; Kao, K.J.; She, J.X. Molecular cloning and characterization of a novel gene family of four ancient conserved domain proteins (ACDP). Gene 2003, 306, 37–44. [Google Scholar] [CrossRef]
- de Baaij, J.H.; Stuiver, M.; Meij, I.C.; Lainez, S.; Kopplin, K.; Venselaar, H.; Müller, D.; Bindels, R.J.; Hoenderop, J.G. Membrane topology and intracellular processing of cyclin M2 (CNNM2). J. Biol. Chem. 2012, 287, 13644–13655. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.F.; Zhang, B.; Tang, R.J.; Zheng, X.J.; Chen, R.; Liu, C.G.; Jing, Y.P.; Ge, H.M.; Zhang, C.; Chu, Y.L.; et al. Four plasma membrane-localized MGR transporters mediate xylem Mg2+ loading for root-to-shoot Mg2+ translocation in Arabidopsis. Mol. Plant 2022, 15, 805–819. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, C.; Tang, R.; Zheng, X.; Zhao, F.; Fu, A.; Lan, W.; Luan, S. Two magnesium transporters in the chloroplast inner envelope essential for thylakoid biogenesis in Arabidopsis. New Phytol. 2022, 236, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Dubcovsky, J.; Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef]
- Gransee, A.; Führs, H. Magnesium mobility in soils as a challenge for soil and plant analysis, magnesium fertilization and root uptake under adverse growth conditions. Plant Soil 2013, 368, 5–21. [Google Scholar] [CrossRef]
- Hawkesford, M.J.; Cakmak, I.; Coskun, D.; De Kok, L.J.; Lambers, H.; Schjoerring, J.K.; White, P.J. Functions of macronutrients. In Marschner’s Mineral Nutrition of Plants; Elsevier: Amsterdam, The Netherlands, 2023; pp. 201–281. [Google Scholar]
- Chaudhry, A.H.; Nayab, S.; Hussain, S.B.; Ali, M.; Pan, Z. Current understandings on magnesium deficiency and future outlooks for sustainable agriculture. Int. J. Mol. Sci. 2021, 22, 1819. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yang, X.; Li, H.; Shuai, Y.; Chen, W.; Ma, D.; Lü, Z. Uncovering the role of wheat magnesium transporter family genes in abiotic responses. Front. Plant Sci. 2023, 14, 1078299. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I. InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.e.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Ødum, M.T.; Teufel, F.; Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Winther, O.; Nielsen, H. DeepLoc 2.1: Multi-label membrane protein type prediction using protein language models. Nucleic Acids Res. 2024, 52, W215–W220. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ramírez-González, R.; Borrill, P.; Lang, D.; Harrington, S.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; Van Ex, F.; Pasha, A. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef]
- Paolacci, A.R.; Tanzarella, O.A.; Porceddu, E.; Ciaffi, M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 2009, 10, 11. [Google Scholar] [CrossRef]
- Zhu, T.; Liu, Y.; Ma, L.; Wang, X.; Zhang, D.; Han, Y.; Ding, Q.; Ma, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol. 2020, 20, 420. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Fukao, T.; Bailey-Serres, J. Rop’rheostat’management of carbohydrate consumption under hypoxia. Trends Plant Sci. 2004, 9, 449–456. [Google Scholar] [CrossRef]
- Seo, P.J.; Xiang, F.N.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef]
- Birnbaum, K.; Shasha, D.E.; Wang, J.Y.; Jung, J.W.; Lambert, G.M.; Galbraith, D.W.; Benfey, P.N. A gene expression map of the Arabidopsis root. Science 2003, 302, 1956–1960. [Google Scholar] [CrossRef] [PubMed]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Gubler, F.; Kalla, R.; Roberts, J.K.; Jacobsen, J.V. Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 1995, 7, 1879–1891. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Kobayashi, N.I.; Tanoi, K.; Iwata, N.; Suzuki, H.; Iwata, R.; Nakanishi, T.M. Expression and functional analysis of the CorA-MRS2-ALR-type magnesium transporter family in rice. Plant Cell Physiol. 2013, 54, 1673–1683. [Google Scholar] [CrossRef]
- Pál, C.; Papp, B.; Hurst, L.D. Genomic function: Rate of evolution and gene dispensability. Nature 2003, 421, 496–497. [Google Scholar] [CrossRef]
- Levasseur, A.; Pontarotti, P. The role of duplications in the evolution of genomes highlights the need for evolutionary-based approaches in comparative genomics. Biol. Direct. 2011, 6, 11. [Google Scholar] [CrossRef]
- Deng, Q.Y.; Luo, J.T.; Zheng, J.M.; Tan, W.F.; Pu, Z.J.; Wang, F. Genome-wide systematic characterization of the NRT2 gene family and its expression profile in wheat (Triticum aestivum L.) during plant growth and in response to nitrate deficiency. BMC Plant Biol. 2023, 23, 353. [Google Scholar] [CrossRef]
- Melamed-Bessudo, C.; Shilo, S.; Levy, A.A. Meiotic recombination and genome evolution in plants. Curr. Opin. Plant Biol. 2016, 30, 82–87. [Google Scholar] [CrossRef]
- Roy, S.W.; Penny, D. Patterns of intron loss and gain in plants: Intron loss–dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol. Biol. Evol. 2007, 24, 171–181. [Google Scholar] [CrossRef]
- Reddy, A.S.N.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [PubMed]
- Mignone, F.; Gissi, C.; Liuni, S.; Pesole, G. Untranslated regions of mRNAs. Genome Biol. 2002, 3, reviews0004. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Ang, L.H.; Puente, P.; Deng, X.W.; Wei, N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 1998, 10, 673–683. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Zeng, W.Z.; Zhang, Y.J.; Tian, X.Y.; Li, W.Y.; Meng, H.; Zhou, Y.C.; Wang, Z.H.; Chen, Z.C.; Zhang, K.W.; Wang, M. Increased cytoplasmic Mg2+ level contributes to rice salicylic acid accumulation and broad-spectrum resistance. Plant Physiol. 2024, 195, 2515–2519. [Google Scholar] [CrossRef]
- Chen, Z.C.; Yamaji, N.; Motoyama, R.; Nagamura, Y.; Ma, J.F. Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol. 2012, 159, 1624–1633. [Google Scholar] [CrossRef]
- Zhang, L.D.; Peng, Y.Y.; Li, J.; Tian, X.Y.; Chen, Z.C. OsMGT1 confers resistance to magnesium deficiency by enhancing the import of Mg in rice. Inter. J. Mol. Sci. 2019, 20, 207. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Zhang, W.; Zhao, F.; Liu, G.; Zhao, D.; Xu, J.; Wang, X.; Zong, X.; Zhang, J.; Ji, X.; et al. Genome-Wide Identification, Phylogeny and Expression Analysis of the Magnesium Release Gene Family in Wheat (Triticum aestivum L.). Curr. Issues Mol. Biol. 2025, 47, 882. https://doi.org/10.3390/cimb47110882
Chen Y, Zhang W, Zhao F, Liu G, Zhao D, Xu J, Wang X, Zong X, Zhang J, Ji X, et al. Genome-Wide Identification, Phylogeny and Expression Analysis of the Magnesium Release Gene Family in Wheat (Triticum aestivum L.). Current Issues in Molecular Biology. 2025; 47(11):882. https://doi.org/10.3390/cimb47110882
Chicago/Turabian StyleChen, Yuanxue, Weiwei Zhang, Fengjuan Zhao, Guolan Liu, Deyong Zhao, Jikun Xu, Xin Wang, Xuehui Zong, Jingmin Zhang, Xiaoqing Ji, and et al. 2025. "Genome-Wide Identification, Phylogeny and Expression Analysis of the Magnesium Release Gene Family in Wheat (Triticum aestivum L.)" Current Issues in Molecular Biology 47, no. 11: 882. https://doi.org/10.3390/cimb47110882
APA StyleChen, Y., Zhang, W., Zhao, F., Liu, G., Zhao, D., Xu, J., Wang, X., Zong, X., Zhang, J., Ji, X., Ma, J., Zhao, S., & Li, J. (2025). Genome-Wide Identification, Phylogeny and Expression Analysis of the Magnesium Release Gene Family in Wheat (Triticum aestivum L.). Current Issues in Molecular Biology, 47(11), 882. https://doi.org/10.3390/cimb47110882






