Genetic Trends in General Combining Ability for Maize Yield-Related Traits in Northeast China
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials for the Test
2.2. Experimental Methods
2.3. Genome-Wide Association Analysis
2.4. Statistical Analysis
3. Results
3.1. Analysis of Yield-Related Traits in Major Maize Inbred Lines from Northeast China with Tester PH4CV and PH6WC
3.2. GCA Analysis of Major Maize Inbred Lines Traits in Northeast China
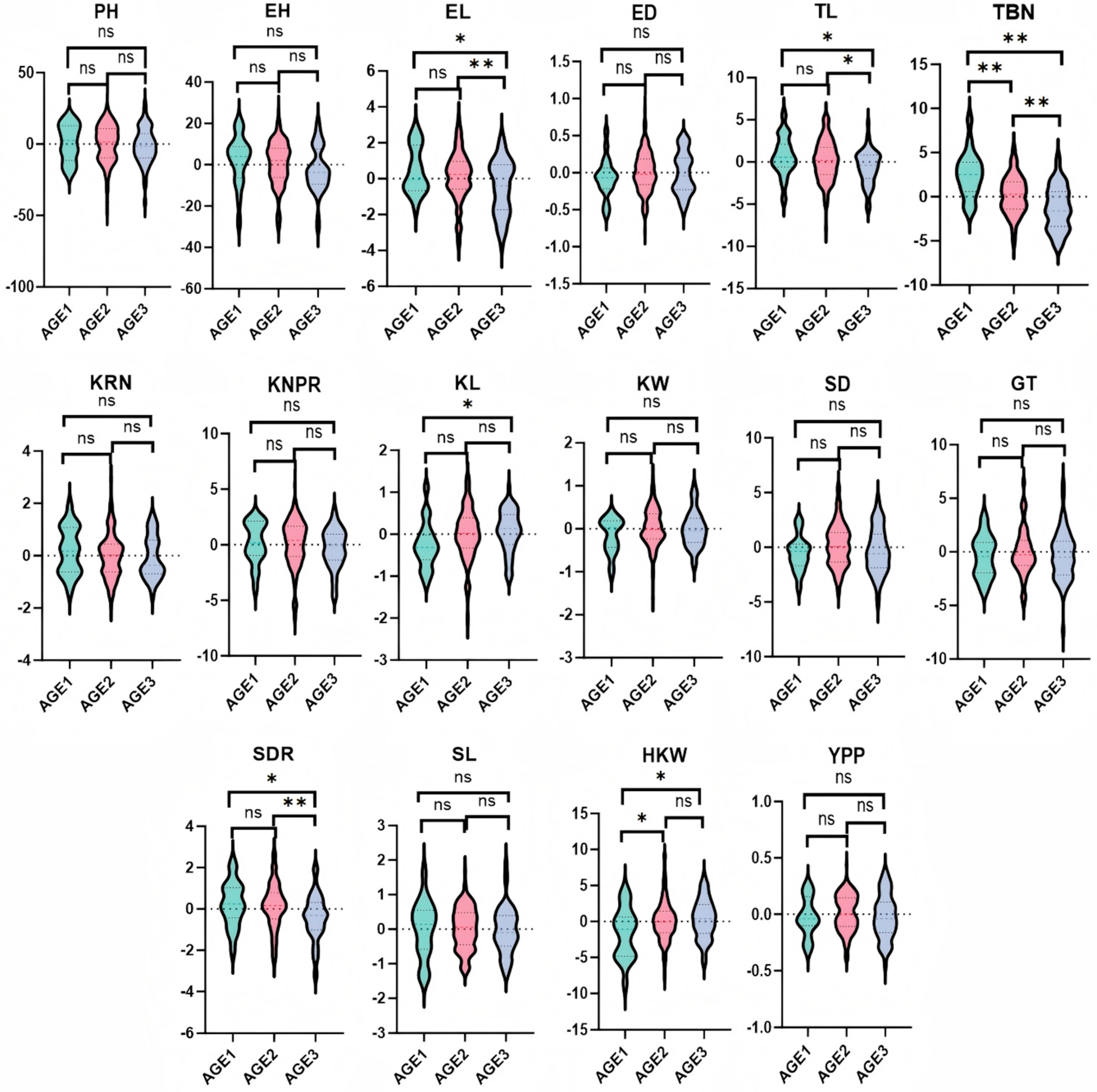
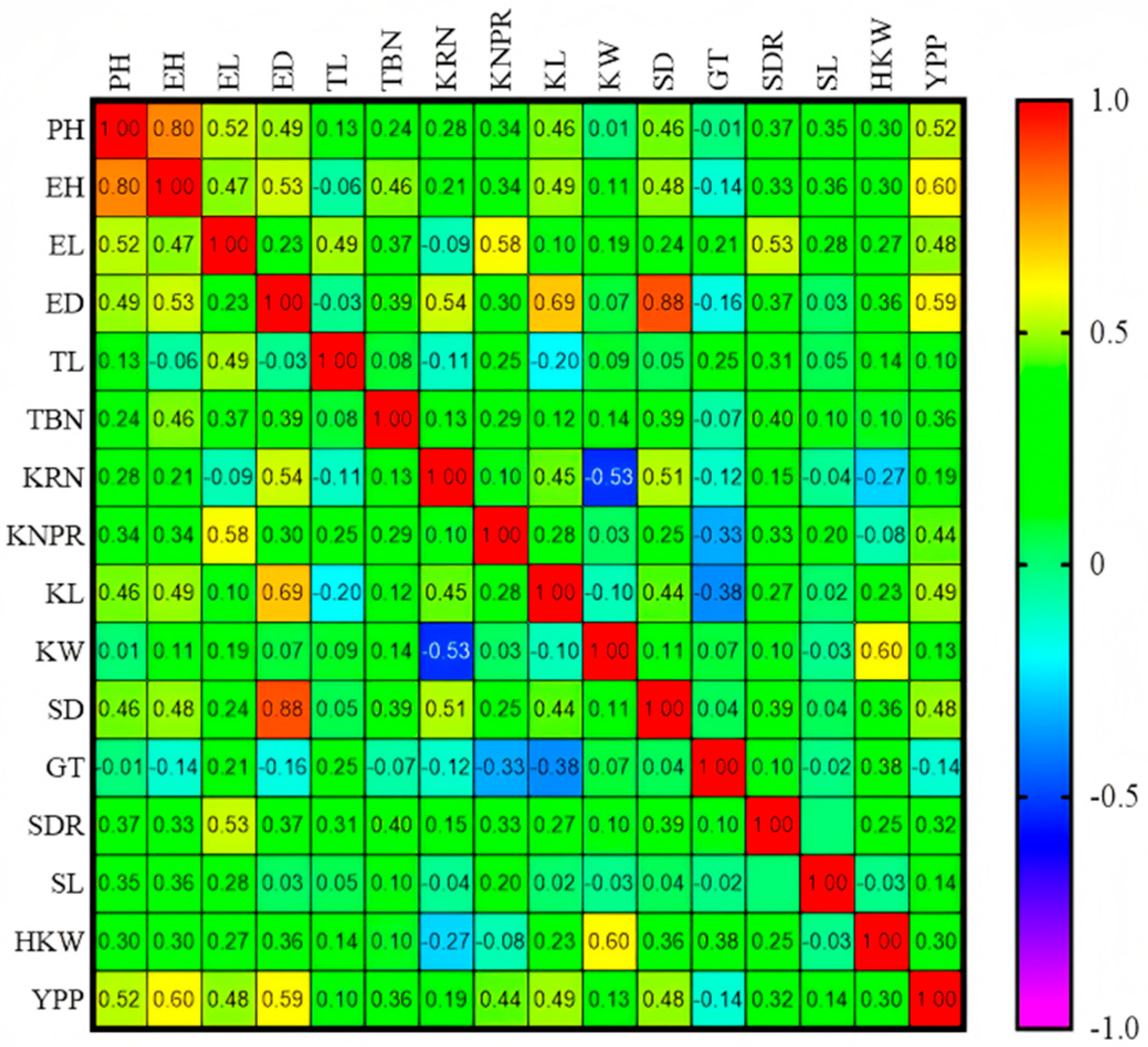
3.3. Trends of GCA and Correlations for Yield-Related Traits
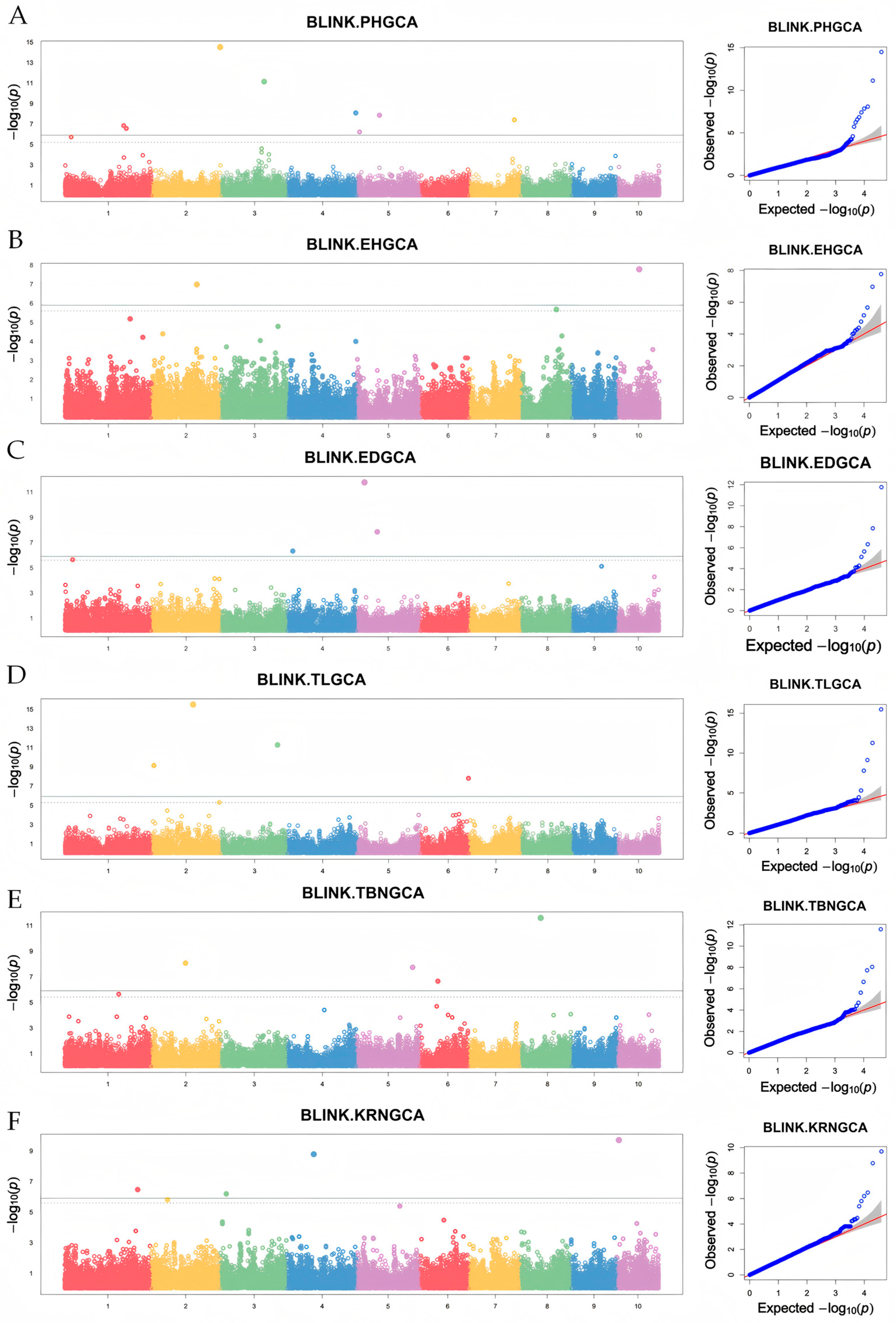
3.4. GWAS for GCA of Yield-Related Traits in Maize from Northeast China
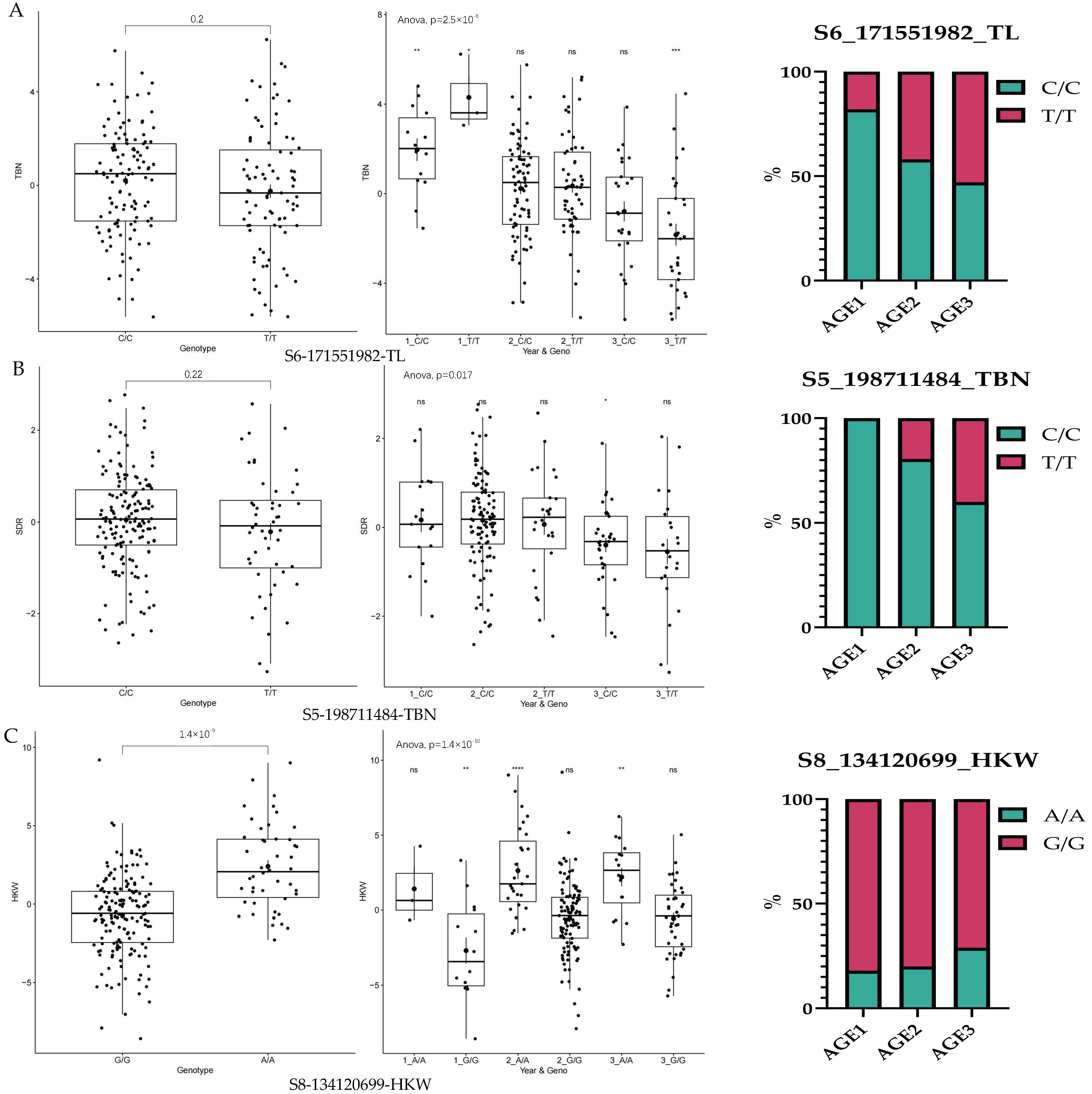
3.5. Trends in Elite Alleles of GCA During Breeding Stage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, K.; Tan, J.; Zhang, Q.; Zhang, Q.; Bai, T.; Zhou, S.; Hao, J.; Yu, X.; Zang, Z.; Zhang, D. Research on the Genetic Improvement Effects of Lodging Resistance-Related Traits in Maize Core Germplasm. Agronomy 2025, 15, 17. [Google Scholar] [CrossRef]
- Editorial Board and Compilation and Publication Team. China Statistical Yearbook 2024; Editorial Board and Compilation and Publication Team: China, Beijing, 2024; pp. 4–5. [Google Scholar]
- Yang, H.; Wang, H.; Han, X.; Zheng, F. Comparative Advantages and Spatial Distribution of Maize-Growing Regions in China: An Analysis Based on Provincial Data (1996–2015). Res. Agric. Mod. 2017, 38, 921–929. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Z.; Shao, Y.; Jin, Z.; Gao, L.; Yu, Y.; Zhang, F.; Zhang, Y.; Nan, Y.; Li, M.; et al. Adaptation of Diverse Maize Germplasm to Spring Season Conditions in Northeast China. Agronomy 2025, 15, 170. [Google Scholar] [CrossRef]
- Xue, Y.; Zhao, Y.; Zhang, Y.; Wang, R.; Li, X.; Liu, Z.; Wang, W.; Zhu, S.; Fan, Y.; Xu, L.; et al. Insights into the genomic divergence of maize heterotic groups in China. J. Integr. Plant Biol. 2025, 67, 1467–1486. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. Study on Heterotic Groups and Heterotic Patterns of Maize in Northern China; Jilin Agricultural University: Changchun, China, 2014. [Google Scholar]
- Luo, P.; Yang, R.; Zhang, L.; Yang, J.; Wang, H.; Yong, H.; Zhang, R.; Li, W.; Wang, F.; Li, M.; et al. Genomic Prediction of Kernel Water Content in a Hybrid Population for Mechanized Harvesting in Maize in Northern China. Agronomy 2024, 14, 2795. [Google Scholar] [CrossRef]
- Lu, X.; Zhou, Z.; Yuan, Z.; Zhang, C.; Hao, Z.; Wang, Z.; Li, M.; Zhang, D.; Yong, H.; Han, J.; et al. Genetic Dissection of the General Combining Ability of Yield-Related Traits in Maize. Front. Plant Sci. 2020, 11, 788. [Google Scholar] [CrossRef] [PubMed]
- Yong, H.; Jin, Z.; Gao, L.; Zhang, L.; Liu, X.; Zhang, F.; Zhang, X.; Zhang, D.; Li, M.; Weng, J.; et al. Breeding potential of maize germplasm populations to improve yield and predominant heterotic pattern in Northeast China. Euphytica 2017, 213, 219. [Google Scholar] [CrossRef]
- Lewis, M.; Goodman, M. Incorporation of tropical maize germplasm into inbred lines derived from temperate × temperate-adapted tropical line crosses: Agronomic and molecular assessment. Theor. Appl. Genet. 2003, 107, 793–805. [Google Scholar] [CrossRef]
- Wang, N.; Wang, Z.; Wang, Y.; Zhang, X.; Lu, L. Utilization Patterns of Heterosis in Major Maize Germplasm in China. Sci. Agric. Sin. 1997, 4, 17–25. [Google Scholar] [CrossRef]
- Shi, G. Current Status and Fundamental Research of Maize Germplasm: A Genomic and Phenomic Perspective. Heilongjiang Agric. Sci. 2002, 2, 35–37. [Google Scholar] [CrossRef]
- Prasanna, B. Diversity in global maize germplasm: Characterization and utilization. J. Biosci. 2012, 37, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Wu, X. Genomic Characterization of Important Maize Inbred Lines and Association Analysis with Plant Type Traits; Sichuan Agricultural University: Ya’an, China, 2013. [Google Scholar]
- Wang, B.; Lin, Z.; Li, X.; Zhao, Y.; Zhao, B.; Wu, G.; Ma, X.; Wang, H.; Xie, Y.; Li, Q.; et al. Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 2020, 52, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Gao, R. Genetic Research on Improved Maize Germplasm; Jilin Agricultural University: Changchun, China, 2016. [Google Scholar]
- Zhang, X. Genetic Variation Analysis of Major Maize Germplasm Populations; Chinese Academy of Agricultural Sciences: Beijing, China, 2013. [Google Scholar]
- Shu, G.; Cao, G.; Li, N.; Wang, A.; Wei, F.; Li, T.; Yi, L.; Xu, Y.; Wang, Y. Genetic variation and population structure in China summer maize germplasm. Sci. Rep. 2021, 11, 8012. [Google Scholar] [CrossRef]
- Shiri, M.; Choukan, R.; Estakhr, A.; Fareghi, S.; Afarinesh, A. Temperate testers’ efficiency in the screening of tropical and subtropical maize germplasm. Cereal Res. Commun. 2021, 49, 639–647. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding Technologies to Increase Crop Production in a Changing World. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.; Eizenga, G.; Wright, M.; Ali, L.; Price, A.; Norton, G.; Islam, R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Dababat, A.; Arif, M.; Toktay, H.; Atyia, O.; Shokat, S.; Orakci, G.; Imren, M.; Singh, S. A GWAS to identify the cereal cyst nematode resistance loci in diverse wheat prebreeding lines. J. Appl. Genet. 2021, 62, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–976. [Google Scholar] [CrossRef]
- Chen, H.; Xie, W.; He, H.; Yu, H.; Chen, W.; Li, J.; Yu, R.; Yao, Y.; Zhang, W.; He, Y.; et al. A High-Density SNP Genotyping Array for Rice Biology and Molecular Breeding. Mol. Plant 2014, 7, 541–553. [Google Scholar] [CrossRef]
- Wu, H.; Matheson, A. General and specific combining ability from partial diallels of radiata pine: Implications for utility of SCA in breeding and deployment populations. Theor. Appl. Genet. 2004, 108, 1503–1512. [Google Scholar] [CrossRef]
- Sun, J.; Gao, J.; Yu, X.; Liu, J.; Su, Z.; Feng, Y.; Wang, D. Combining Ability of Sixteen USA Maize Inbred Lines and Their Outbreeding Prospectsin China. Agronomy 2018, 8, 281. [Google Scholar] [CrossRef]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Y.; Feng, Q.; Gong, H.; Li, W.; Zhan, Q.; Cheng, B.; Xia, J.; et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef]
- Li, Y.; Li, C.; Bradbury, P.; Liu, X.; Lu, F.; Romay, C.; Glaubitz, J.; Wu, X.; Peng, B.; Shi, Y.; et al. Identification of genetic variants associated with maize flowering time using an extremely large multi-genetic background population. Plant J. 2016, 86, 391–402. [Google Scholar] [CrossRef]
- Hu, G.; Li, Z.; Lu, Y.; Li, C.; Gong, S.; Yan, S.; Li, G.; Wang, M.; Ren, H.; Guan, H.; et al. Genome-wide association study Identified multiple Genetic Loci on Chilling Resistance During Germination in Maize. Sci. Rep. 2017, 7, 10840. [Google Scholar] [CrossRef]
- Lv, Z.; Zhang, H.; Zhang, Z.; Tao, Y.; Yue, B.; Zheng, Y. Conversion of the Statistical Combining Ability into a Genetic Concept. J. Integr. Agric. 2012, 11, 43–52. [Google Scholar] [CrossRef]
- Qi, H.; Huang, J.; Zheng, Q.; Huang, Y.; Shao, R.; Zhu, L.; Zhang, Z.; Qiu, F.; Zhou, G.; Zheng, Y.; et al. Identification of combining ability loci for five yield-related traits in maize using a set of testcrosses with introgression lines. Theor. Appl. Genet. 2013, 126, 369–377. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, C.; Lu, X.; Wang, L.; Hao, Z.; Li, M.; Zhang, D.; Yong, H.; Zhu, H.; Weng, J.; et al. Dissecting the Genetic Basis Underlying Combining Ability of Plant Height Related Traits in Maize. Front. Plant Sci. 2018, 9, 1117. [Google Scholar] [CrossRef]
- Adebayo, A.; Menkir, A.; Hearne, S.; Kolawole, A. Gene action controlling normalized difference vegetation index in crosses of elite maize inbred lines. Cereal Res. Commun. 2017, 45, 675–686. [Google Scholar] [CrossRef]
- Luo, P.; Wang, H.; Ni, Z.; Yang, R.; Wang, F.; Yong, H.; Zhang, L.; Zhou, Z.; Song, W.; Li, M.; et al. Genomic prediction of yield performance among single-cross maize hybrids using a partial diallel cross design. Crop J. 2023, 11, 1884–1892. [Google Scholar] [CrossRef]
- Bates, D.; Machler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Amani, A.; Gamal, O. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 2017, 13, 1. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Weber, J.; Aldana, R.; Gallagher, B.; Edwards, J. Sentieon DNA pipeline for variant detection-Software-only solution, over 20 faster than GATK 3.3 with identical results. PeerJ Prepr. 2016, 4, 1672. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. Available online: http://www.genome.org/cgi/doi/10.1101/gr.107524.110 (accessed on 15 September 2020).
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, 164. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Giga Sci. 2019, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Lei, Y.; Xu, Z.; Cheng, Z.; Liu, M.; Tai, Y.; Han, X.; Hao, Z.; Li, M.; Zhang, D.; et al. Natural variations in the promoter of ZmDeSI2 encoding a deSUMOylating isopeptidase controls kernel methionine content in maize. Mol. Plant 2025, 18, 872–891. [Google Scholar] [CrossRef] [PubMed]
- Griffing, B. Concept of general and specific combining ability in relation to diallel cross systems. Aust. J. Biol. Sci. 1956, 9, 463–493. [Google Scholar] [CrossRef]
- Yu, K.; Wang, H.; Liu, X.; Xu, C.; Li, Z.; Xu, X.; Liu, J.; Wang, Z.; Xu, Y. Large-Scale Analysis of Combining Ability and Heterosis for Development of Hybrid Maize Breeding Strategies Using Diverse Germplasm Resources. Front. Plant Sci. 2020, 11, 660. [Google Scholar] [CrossRef]
- Zhang, N. Genetic Basis and Combining Ability Analysis of 125 Foreign Maize Inbred Lines; Jilin Agricultural University: Changchun, China, 2024; Available online: https://link.cnki.net/doi/10.27163/d.cnki.gjlnu.2024.000434 (accessed on 3 February 2025).
- Wang, C. Combining Ability and Heterotic Group Analysis of 14 Maize Inbred Lines; Gansu Agricultural University: Lanzhou, China, 2016. [Google Scholar]
- Li, Q.; Zhang, D.; Yang, S.; Liu, J.; Sun, K.; Wu, X. Diversity Analysis of 326 Maize Germplasm Accessions. J. Gansu Agric. Univ. 2024, 1–16. Available online: https://link.cnki.net/urlid/62.1055.S.20240702.1713.006 (accessed on 15 March 2025).
- Imdad, U.; Tang, W.; He, J.; Khan, S.; Hong, D. Association analysis uncovers the genetic basis of general combining ability of 11 yield-related traits in parents of hybrid rice. AoB Plants 2019, 11, 77. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, H.; Xie, W.; Xia, D.; Gao, G.; Zhang, Q.; Wang, G.; Lian, X.; Xiao, J.; He, Y. Genome-wide association analyses reveal the genetic basis of combining ability in rice. Plant Biotechnol. J. 2019, 20, 2211–2222. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Guan, H.; Jing, X.; Li, Y.; Wang, B.; Li, Y.; Liu, X.; Zhang, D.; Liu, C.; Xie, X.; et al. Genomic insights into historical improvement of heterotic groups during modern hybrid maize breeding. Nat. Plants 2022, 8, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Austin, D.; Lee, M.; Veldboom, L. Genetic mapping in maize with hybrid progeny across testers and generations: Plant height and flowering. Theor. Appl. Genet. 2001, 102, 163–176. [Google Scholar] [CrossRef]
- Elisabetta, F.; Angela, M.; Mario, E.; Pea, G.; Morgante, M.; Landi, P. QTL detection in maize testcross progenies as affected by related and unrelated testers. Theor. Appl. Genet. 2009, 118, 993–1004. [Google Scholar] [CrossRef]
| Traits | Blo | Loc | Cro | Blo × Loc | Blo × Cro | Loc × Cro | Blo × Loc × Cro |
|---|---|---|---|---|---|---|---|
| DF | 3 | 2 | 217 | 6 | 651 | 434 | 1302 |
| PH | 431.93 | 170,653.76 ** | 1935.44 * | 205.38 | 181.73 | 314.35 | 155.10 |
| EH | 386.79 * | 45,063.38 ** | 1182.96 ** | 426.61 ** | 80.10 | 106.09 | 78.52 |
| EL | 25.05 | 56.27 * | 23.44 * | 3.78 | 3.91 | 4.47 | 3.51 |
| ED | 0.03 | 2.02 ** | 0.59 ** | 0.34 * | 0.09 | 0.14 | 0.09 |
| TL | 120.97 * | 1724.24 ** | 58.43 | 44.38 | 9.38 | 11.32 | 7.77 |
| TBN | 10.42 | 78.74 | 42.48 | 30.84 | 6.10 | 6.75 | 5.86 |
| KRN | 1.36 | 6.26 * | 5.40 * | 0.14 | 1.86 | 2.91 | 1.36 |
| KNPR | 75.67 | 1116.03 ** | 40.12 | 200.56 * | 19.58 | 33.47 | 18.61 |
| KL | 3.45 * | 13.67 ** | 2.73 * | 1.95 | 0.42 | 0.62 | 0.38 |
| KW | 62.86 ** | 84.56 ** | 1.55 | 55.82 | 0.54 ** | 0.94 | 0.74 |
| SD | 62.25 ** | 949.66 ** | 27.96 * | 84.11 ** | 3.86 | 4.48 | 3.35 |
| GT | 15.45 | 870.07 ** | 50.81 * | 46.66 * | 7.64 | 15.71 | 7.90 |
| SDR | 20.59 ** | 4370.51 ** | 14.82 ** | 103.28 ** | 6.11 * | 5.40 | 4.74 |
| SL | 164.35 ** | 1034.33 ** | 4.77 | 114.14 ** | 2.13 | 2.84 | 2.26 |
| HKW | 6.22 | 1440.90 ** | 65.85 | 8.62 | 7.80 | 16.02 | 7.62 |
| YPP | 0.08 | 0.93 ** | 0.34 * | 0.10 | 0.07 | 0.08 | 0.05 |
| Traits | Blo | Loc | Cro | Blo × Loc | Blo × Cro | Loc × Cro | Blo × Loc × Cro |
|---|---|---|---|---|---|---|---|
| DF | 3 | 2 | 217 | 6 | 651 | 434 | 1302 |
| PH | 1180.91 | 131,570.40 ** | 1862.35 * | 115.34 | 167.99 | 335.17 | 128.80 |
| EH | 79.21 | 29,114.31 ** | 1060.37 ** | 842.73 * | 81.17 | 102.29 | 69.36 |
| EL | 3.79 | 26.86 * | 18.44 * | 4.03 | 2.04 | 3.40 | 2.17 |
| ED | 0.18 | 2.79 * | 0.64 | 0.33 | 0.07 | 0.09 | 0.06 |
| TL | 104.25 * | 1814.83 ** | 51.73 | 38.85 | 7.53 | 8.12 | 6.17 |
| TBN | 24.81 | 45.00 | 77.70 | 34.84 | 5.70 | 7.99 | 7.02 |
| KRN | 5.57 | 2.35 | 6.67 | 2.83 | 1.79 | 3.26 | 1.64 |
| KNPR | 133.93 | 122.37 * | 50.85 * | 99.99 | 20.14 | 38.46 | 22.38 |
| KL | 1.02 | 37.20 * | 2.99 | 0.30 | 0.45 | 0.76 | 0.43 |
| KW | 88.05 ** | 115.81 ** | 1.98 * | 72.78 ** | 0.65 | 0.92 | 0.83 |
| SD | 52.08 * | 408.12 ** | 32.90 * | 105.91 ** | 4.18 | 5.59 | 3.91 |
| GT | 11.53 | 153.31 * | 34.83 | 32.19 | 6.46 | 10.29 | 5.82 |
| SDR | 23.45 * | 4229.27 ** | 12.68 | 141.79 ** | 4.06 | 3.65 | 3.83 |
| SL | 84.67 ** | 994.61 ** | 4.18 | 78.70 ** | 1.72 | 2.78 | 1.95 |
| HKW | 22.10 | 2112.79 ** | 75.27 * | 16.99 | 7.69 | 18.79 | 8.21 |
| YPP | 0.05 | 1.67 ** | 0.27 | 0.07 | 0.05 | 0.07 | 0.04 |
| Traits | Loc | Blo | Line | Line × loc | Tester | Tester × Loc | Line × Tester | Line × Tester × Loc |
|---|---|---|---|---|---|---|---|---|
| DF | 2 | 3 | 217 | 434 | 1 | 2 | 217 | 434 |
| PH | 288,784.40 ** | 1308.83 ** | 15,899.80 ** | 1288.49 ** | 2914.74 ** | 450.09 ** | 807.09 ** | 198.51 ** |
| EH | 69,987.21 ** | 315.91 * | 416.74 * | 839.93 ** | 1916.82 ** | 116.10 ** | 268.90 ** | 88.66 * |
| EL | 15.47 ** | 7.97 | 321.93 ** | 97.97 ** | 27.98 ** | 4.87 ** | 12.57 ** | 3.39 * |
| ED | 7.36 ** | 0.02 | 1.41 ** | 1.19 ** | 1.01 ** | 0.13 ** | 0.19 ** | 0.11 ** |
| TL | 3356.62 ** | 227.59 ** | 463.13 ** | 11.93 | 85.38 ** | 10.22 ** | 22.63 ** | 9.48 ** |
| TBN | 119.97 ** | 4.65 | 5418.17 ** | 8.88 | 102.96 ** | 8.49 ** | 15.27 ** | 6.61 |
| KRN | 6.03 * | 4.42 | 5.16 | 7.00 * | 11.11 ** | 4.52 ** | 1.66 | 1.72 |
| KNPR | 1113.90 ** | 324.60 ** | 7065.20 ** | 820.96 ** | 58.76 ** | 45.77 ** | 29.58 ** | 25.05 ** |
| KL | 42.87 ** | 2.91 ** | 134.02 ** | 7.93 ** | 4.23 ** | 0.93 ** | 1.24 ** | 0.52 ** |
| KW | 398.90 ** | 119.45 ** | 1.45 | 0.1 | 2.81 ** | 1.18 * | 1.12 | 0.9 |
| SD | 1231.03 ** | 9.8 | 29.17 ** | 62.15 ** | 52.22 ** | 5.58 ** | 7.70 ** | 5.10 ** |
| GT | 883.52 ** | 3.3 | 18,593.25 ** | 228.38 ** | 66.30 ** | 17.59 ** | 18.35 ** | 11.96 ** |
| SDR | 8397.87 ** | 17.25 * | 0.82 | 0.15 | 20.24 ** | 5.09 | 6.90 ** | 3.95 |
| SL | 1787.86 ** | 246.36 ** | 95.58 ** | 51.05 ** | 6.20 ** | 3.20 ** | 2.85 * | 2.47 |
| HKW | 3841.97 ** | 31.95 * | 8472.96 ** | 42.48 ** | 108.12 ** | 25.30 ** | 24.34 ** | 11.24 ** |
| YPP | 3.62 ** | 0.09 | 2.72 ** | 0.14 | 0.40 ** | 0.09 ** | 0.19 ** | 0.07 ** |
| Trait | SNPs | Chromosome | Position (bp) | p Value | MAF |
|---|---|---|---|---|---|
| PH | S1_208497876 | 1 | 208497876 | 1.459878 × 10−7 | 0.0926829 |
| PH | S1_217817403 | 1 | 217817403 | 2.732274 × 10−7 | 0.4731707 |
| PH | S2_242867385 | 2 | 242867385 | 3.079791 × 10−15 | 0.0658536 |
| PH | S3_154176114 | 3 | 154176114 | 7.300432 × 10−12 | 0.1560975 |
| PH | S4_242979954 | 4 | 242979954 | 8.332607 × 10−9 | 0.2707317 |
| PH | S5_9690315 | 5 | 9690315 | 6.014419 × 10−7 | 0.4341463 |
| PH | S5_80232841 | 5 | 80232841 | 1.405806 × 10−8 | 0.2463414 |
| PH | S7_160382018 | 7 | 160382018 | 3.959368 × 10−8 | 0.2146341 |
| EH | S2_160298536 | 2 | 160298536 | 1.038463 × 10−7 | 0.0902439 |
| EH | S10_79122567 | 10 | 79122567 | 1.669485 × 10−8 | 0.0487804 |
| ED | S4_20634817 | 4 | 20634817 | 4.724012 × 10−7 | 0.3390244 |
| ED | S5_27726473 | 5 | 27726473 | 1.693935 × 10−12 | 0.3560976 |
| ED | S5_72936373 | 5 | 72936373 | 1.417658 × 10−8 | 0.3292683 |
| TL | S2_8474932 | 2 | 8474932 | 7.390562 × 10−10 | 0.0878048 |
| TL | S2_147092451 | 2 | 147092451 | 3.235304 × 10−16 | 0.3975609 |
| TL | S3_201614406 | 3 | 201614406 | 5.281157 × 10−12 | 0.4317073 |
| TL | S6_171551982 | 6 | 171551982 | 1.589827 × 10−8 | 0.4268292 |
| TBN | S2_121462400 | 2 | 121462400 | 8.794725 × 10−9 | 0.3731707 |
| TBN | S5_198711484 | 5 | 198711484 | 1.853238 × 10−8 | 0.2414634 |
| TBN | S6_64593869 | 6 | 64593869 | 2.240302 × 10−7 | 0.2926829 |
| KRN | S1_259340649 | 1 | 259340649 | 3.391704 × 10−7 | 0.3536585 |
| KRN | S3_21754072 | 3 | 21754072 | 6.418560 × 10−7 | 0.4682927 |
| KRN | S4_95867722 | 4 | 95867722 | 1.648411 × 10−9 | 0.1609756 |
| KRN | S10_9329687 | 10 | 9329687 | 1.973185 × 10−10 | 0.3975610 |
| KW | S10_91130327 | 10 | 91130327 | 1.104042 × 10−7 | 0.3487805 |
| SD | S2_102852731 | 2 | 102852731 | 1.045193 × 10−7 | 0.0756097 |
| SD | S2_150753314 | 2 | 150753314 | 2.675444 × 10−7 | 0.2951219 |
| SD | S3_199474950 | 3 | 199474950 | 5.598493 × 10−7 | 0.1853658 |
| SD | S5_27726473 | 5 | 27726473 | 3.455339 × 10−10 | 0.3560975 |
| SD | S6_87803583 | 6 | 87803583 | 6.698875 × 10−15 | 0.1756097 |
| SD | S8_157428343 | 8 | 157428343 | 1.370149 × 10−9 | 0.4048780 |
| GT | S2_71773878 | 2 | 71773878 | 2.280263 × 10−10 | 0.1560976 |
| GT | S10_6348231 | 10 | 6348231 | 5.224577 × 10−7 | 0.0902439 |
| SDR | S3_124027250 | 3 | 124027250 | 1.641799 × 10−8 | 0.1243902 |
| SDR | S8_115429920 | 8 | 115429920 | 6.130413 × 10−7 | 0.0585365 |
| SL | S8_105193919 | 8 | 105193919 | 4.301483 × 10−11 | 0.2682927 |
| HKW | S8_134120699 | 8 | 134120699 | 2.863774 × 10−14 | 0.2219512 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Zhang, X.; Weng, J.; Li, M.; Hao, Z.; Zhang, D.; Yong, H.; Han, J.; Zhou, Z.; Li, X. Genetic Trends in General Combining Ability for Maize Yield-Related Traits in Northeast China. Curr. Issues Mol. Biol. 2025, 47, 877. https://doi.org/10.3390/cimb47110877
Wang H, Zhang X, Weng J, Li M, Hao Z, Zhang D, Yong H, Han J, Zhou Z, Li X. Genetic Trends in General Combining Ability for Maize Yield-Related Traits in Northeast China. Current Issues in Molecular Biology. 2025; 47(11):877. https://doi.org/10.3390/cimb47110877
Chicago/Turabian StyleWang, Haochen, Xiaocong Zhang, Jianfeng Weng, Mingshun Li, Zhuanfang Hao, Degui Zhang, Hongjun Yong, Jienan Han, Zhiqiang Zhou, and Xinhai Li. 2025. "Genetic Trends in General Combining Ability for Maize Yield-Related Traits in Northeast China" Current Issues in Molecular Biology 47, no. 11: 877. https://doi.org/10.3390/cimb47110877
APA StyleWang, H., Zhang, X., Weng, J., Li, M., Hao, Z., Zhang, D., Yong, H., Han, J., Zhou, Z., & Li, X. (2025). Genetic Trends in General Combining Ability for Maize Yield-Related Traits in Northeast China. Current Issues in Molecular Biology, 47(11), 877. https://doi.org/10.3390/cimb47110877







