Pathogenic FANCC Variants Are Associated with Accessory Breasts in a Sub-Saharan African Multiplex Family
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection, DNA Processing, and Whole Exome Sequencing
2.3. Bioinformatic Analysis of Whole Exome Sequencing Dataset
2.4. Sanger Sequencing to Confirm Pathogenic Variants
2.5. Gene Expression and Interactome Analyses
3. Results
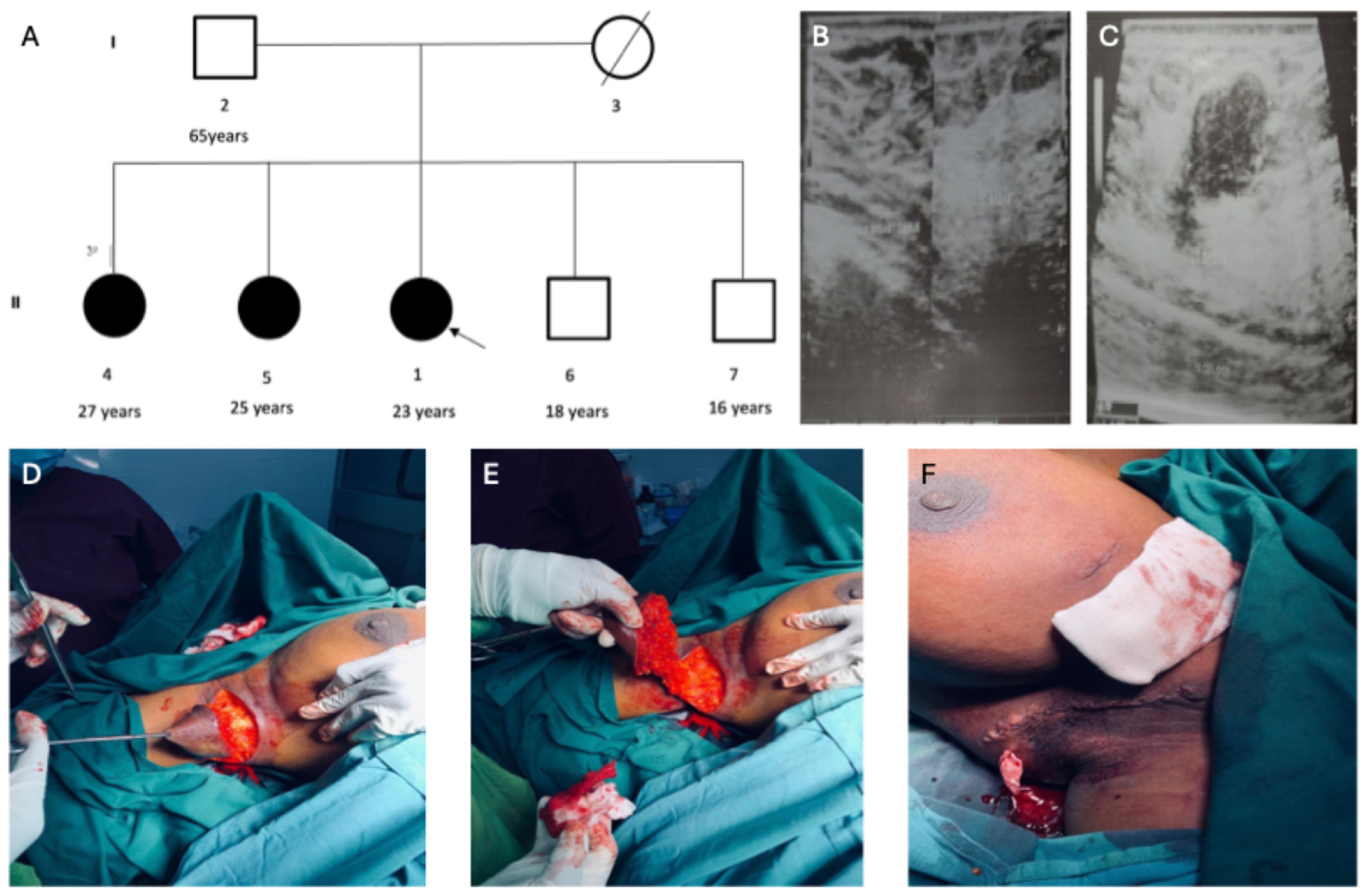
3.1. Clinical Characteristics of the Accessory Breast Phenotype in the Multiplex Family
3.2. Detection of Etiologic Variants for Accessory Breast Phenotype
3.3. Rare Genetic Variants for Co-Occurring Conditions in Individuals with Polymastia
3.4. ACMG Secondary Finding Genes
4. Discussion
4.1. Clinical Presentations and Diagnosis of the Accessory Breasts
4.2. Genetic Aetiology of the Accessory Breast Phenotype in the Multiplex Family
4.3. Other Likely Pathogenic Variants Segregating with the Accessory Breasts
4.4. Clinically Actionable Secondary Findings Observed in Family Members
4.5. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laor, T.; Collins, M.H.; Emery, K.H.; Donnelly, L.F.; Bove, K.E.; Ballard, E.T. MRI appearance of accessory breast tissue: A diagnostic consideration for an axillary mass in a peripubertal or pubertal girl. Am. J. Roentgenol. 2004, 183, 1779–1781. [Google Scholar] [CrossRef]
- Bellahsene-Bendib, S.; Achir, Y.; Aiche, D.; Aimeur, C. Polymastia: What you Should Know. From One Man Case Discovered by Chance. Clin. Nurs. 2023, 2, 520–2644. [Google Scholar]
- Rho, J.Y.; Juhng, S.K.; Yoon, K.J. Carcinoma originating from aberrant breast tissue of the right upper anterior chest wall: A case report. J. Korean Med. Sci. 2001, 16, 519. [Google Scholar] [CrossRef]
- Bakker, J.R.; Sataloff, D.M.; Haupt, H.M. Breast cancer presenting in aberrant axillary breast tissue. Community Oncol. Elsevier Oncol. 2005, 2, 117–120. [Google Scholar] [CrossRef]
- Hong, J.H.; Oh, M.J.; Hur, J.Y.; Lee, J.K. Accessory breast tissue presenting as a vulvar mass in an adolescent girl. Arch. Gynecol. Obstet. 2009, 280, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Youn, H.J.; Jung, S.H. Accessory Breast Carcinoma. Breast Care 2009, 4, 104. [Google Scholar] [CrossRef] [PubMed]
- Schoenwolf, G.C.; Bleyl, S.B.; Brauer, P.R.; Francis-West, P.H. Larsen’s Human Embryology. Larsen’s Hum. Embryol. 2014, 5, 155–171. [Google Scholar]
- Goyal, S.; Puri, T.; Gupta, R.; Julka, P.; Rath, G. Accessory breast tissue in axilla masquerading as breast cancer recurrence. J. Cancer Res. Ther. 2008, 4, 95–96. [Google Scholar] [CrossRef]
- Sahu, S.K.; Husain, M.; Sachan, P.K. Bilateral Accessory Breast. Internet J. Surg. 2007, 17, 1–4. [Google Scholar]
- Gowans, L.J.J.; Oseni, G.; Mossey, P.A.; Adeyemo, W.L.; Eshete, M.A.; Busch, T.D.; Donkor, P.; Obiri-Yeboah, S.; Plange-Rhule, G.; Oti, A.A.; et al. Novel GREM1 variations in Sub-Saharan African patients with cleft lip and/or cleft palate. Cleft Palate-Craniofacial J. 2018, 55, 736–742. [Google Scholar] [CrossRef]
- Alade, A.; Peter, T.; Busch, T.; Awotoye, W.; Anand, D.; Abimbola, O.; Aladenika, E.; Olujitan, M.; Rysavy, O.; Nguyen, P.F.; et al. Shared genetic risk between major orofacial cleft phenotypes in an African population. Genet. Epidemiol. 2024, 48, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Feng, X.; Ping, J.; Gao, S.; Han, D.; Song, W.; Li, X.; Tao, Y.; Wang, L. Novel JAG1 variants leading to Alagille syndrome in two Chinese cases. Sci. Rep. 2014, 14, 1812. [Google Scholar] [CrossRef]
- Kendig, K.I.; Baheti, S.; Bockol, M.A.; Drucker, T.M.; Hart, S.N.; Heldenbrand, J.R.; Hernaez, M.; Hudson, M.E.; Kalmbach, M.T.; Klee, E.W.; et al. Sentieon DNASeq Variant Calling Workflow Demonstrates Strong Computational Performance and Accuracy. Front. Genet. 2019, 10, 736. [Google Scholar] [CrossRef]
- Mahmoud, M.; Huang, Y.; Garimella, K.; Audano, P.A.; Wan, W.; Prasad, N.; Handsaker, R.E.; Hall, S.; Pionzio, A.; Schatz, M.C.; et al. Utility of long-read sequencing for All of Us. Nat. Commun. 2024, 15, 837. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.J.; Biesecker, L.G. Databases of genomic variation and phenotypes: Existing resources and future needs. Hum. Mol. Genet. 2013, 22, R27. [Google Scholar] [CrossRef]
- Fairley, S.; Lowy-Gallego, E.; Perry, E.; Flicek, P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 2020, 48, D941–D947. [Google Scholar] [CrossRef]
- Gudmundsson, S.; Singer-Berk, M.; Watts, N.A.; Phu, W.; Goodrich, J.K.; Solomonson, M.; Rehm, H.L.; MacArthur, D.G.; O’Donnell-Luria, A. Variant interpretation using population databases: Lessons from gnomAD. Hum. Mutat. 2022, 43, 1012–1030. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. dbNSFP v4: A comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Stein, T.I.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Plaschkes, I.; Oz-Levi, D.; Alkelai, A.; Olender, T.; Zimmerman, S.; Twik, M.; Belinky, F.; Fishilevich, S.; Nudel, R.; et al. VarElect: The phenotype-based variation prioritizer of the GeneCards Suite. BMC Genom. 2016, 17, 444. [Google Scholar] [CrossRef]
- Stenson, P.D.; Ball, E.V.; Mort, M.; Phillips, A.D.; Shaw, K.; Cooper, D.N. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr. Protoc. Bioinform. 2012, 39, 1.13.1–1.13.20. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Chitipiralla, S.; Kaur, K.; Brown, G.; Chen, C.; Hart, J.; Hoffman, D.; Jang, W.; Liu, C.; Maddipatla, Z.; et al. ClinVar: Updates to support classifications of both germline and somatic variants. Nucleic Acids Res. 2025, 53, D1313–D1321. [Google Scholar] [CrossRef]
- Baldarelli, R.M.; Smith, C.L.; Ringwald, M.; Richardson, J.E.; Bult, C.J.; Anagnostopoulos, A.; Begley, D.A.; Bello, S.M.; Christie, K.; Finger, J.H.; et al. Mouse Genome Informatics: An integrated knowledgebase system for the laboratory mouse. Genetics 2024, 227, iyae031. [Google Scholar] [CrossRef]
- Del Valle, J.; Rofes, P.; Moreno-Cabrera, J.M.; López-Dóriga, A.; Belhadj, S.; Vargas-Parra, G.; Teulé, À.; Cuesta, R.; Muñoz, X.; Campos, O.; et al. Exploring the Role of Mutations in Fanconi Anemia Genes in Hereditary Cancer Patients. Cancers 2020, 12, 829. [Google Scholar] [CrossRef]
- Miller, D.T.; Lee, K.; Abul-Husn, N.S.; Amendola, L.M.; Brothers, K.; Chung, W.K.; Gollob, M.H.; Gordon, A.S.; Harrison, S.M.; Hershberger, R.E.; et al. ACMG SF v3.1 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2022, 24, 1407–1414. [Google Scholar] [CrossRef]
- Lakkawar, N.J.; Maran, G.; Srinivasan, S.; Rangaswamy, T. Accessory breast tissue in the axilla in a puerperal woman- case study. Acta Medica Median. 2010, 49, 45–48. [Google Scholar]
- Alex, A.; Bhandary, E.; McGuire, K.P. Anatomy and physiology of the breast during pregnancy and lactation. Adv. Exp. Med. Biol. 2020, 1252, 3–7. [Google Scholar] [PubMed]
- Schock, H.; Zeleniuch-Jacquotte, A.; Lundin, E.; Grankvist, K.; Lakso, H.Å.; Idahl, A.; Lehtinen, M.; Surcel, H.M.; Fortner, R.T. Hormone concentrations throughout uncomplicated pregnancies: A longitudinal study. BMC Pregnancy Childbirth 2016, 16, 146. [Google Scholar] [CrossRef]
- Jaiswal, G.; Thakur, G.S. An alternative yogic approach for cyclical mastalgia—A narrative review. J. Fam. Med. Prim. Care 2021, 10, 601–608. [Google Scholar] [CrossRef]
- Sivarajah, R.; Welkie, J.; Mack, J.; Casas, R.S.; Paulishak, M.; Chetlen, A.L. A review of breast pain: Causes, imaging recommendations, and treatment. J. Breast Imaging 2020, 2, 101–111. [Google Scholar] [CrossRef]
- Eren, T.; Aslan, A.; Ozemir, I.A.; Baysal, H.; Sagiroglu, J.; Ekinci, O.; Alimoglu, O. Factors effecting mastalgia. Breast Care 2016, 11, 188–193. [Google Scholar] [CrossRef]
- Farcy, D.A.; Rabinowitz, D.; Frank, M. Ectopic glandular breast tissue in a lactating young woman. J. Emerg. Med. 2011, 41, 627–629. [Google Scholar] [CrossRef]
- Darlington, A.J. Anatomy of the breast. In Digital Mammography: A Holistic Approach; Springer: Berlin/Heidelberg, Germany, 2015; Volume 1, pp. 3–10. [Google Scholar]
- Lopez, M.E.; Olutoye, O.O. Breast embryology, anatomy, and physiology. In Endocrine Surgery in Children; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1, pp. 365–376. [Google Scholar]
- Youssef, H.M.K.; Radi, D.A.; Abd El-Azeem, M.A. Expression of TSP50, SERCA2 and IL-8 in Colorectal Adenoma and Carcinoma: Correlation to Clinicopathological Factors. Pathol. Oncol. Res. 2021, 27, 1609990. [Google Scholar] [CrossRef]
- Song, Z.B.; Wu, P.; Ni, J.S.; Liu, T.; Fan, C.; Bao, Y.L.; Wu, Y.; Sun, L.G.; Yu, C.L.; Huang, Y.X.; et al. Testes-specific protease 50 promotes cell proliferation via inhibiting activin signaling. Oncogene 2017, 36, 5948–5957. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, S.; Wang, S.; Song, Z.; Zheng, L.; Wang, G.; Sun, Y.; Bao, Y. miR-4709-3p Inhibits Cell Proliferation by Downregulating TSP50 Expression in Breast Cancer Cells. DNA Cell Biol. 2021, 40, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.X.; Li, J.W.; Li, X.L.; Zhang, L.L.; Lang, Y.; Song, Z.B.; Yu, C.L.; Yang, X.G.; Zhao, H.F.; Sun, J.L.; et al. PRSS50-mediated inhibition of MKP3/ERK signaling is crucial for meiotic progression and sperm quality. Zool. Res. 2024, 45, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Frey, M.K.; Sandler, G.; Sobolev, R.; Kim, S.H.; Chambers, R.; Bassett, R.Y.; Martineau, J.; Sapra, K.J.; Boyd, L.; Curtin, J.P.; et al. Multigene panels in Ashkenazi Jewish patients yield high rates of actionable mutations in multiple non-BRCA cancer-associated genes. Gynecol. Oncol. 2017, 146, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.R.; Doyle, M.A.; Ryland, G.L.; Rowley, S.M.; Choong, D.Y.H.; Tothill, R.W.; Thorne, H.; Barnes, D.R.; Li, J.; Ellul, J.; et al. Exome Sequencing Identifies Rare Deleterious Mutations in DNA Repair Genes FANCC and BLM as Potential Breast Cancer Susceptibility Alleles. PLoS Genet. 2012, 8, e1002894. [Google Scholar] [CrossRef]
- Pang, Q.; Christianson, T.A.; Keeble, W.; Diaz, J.; Faulkner, G.R.; Reifsteck, C.; Olson, S.; Bagby, G.C. The Fanconi anemia complementation group C gene product: Structural evidence of multifunctionality. Blood 2001, 98, 1392–1401. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef]
- Aubé, M.; Lafrance, M.; Brodeur, I.; Delisle, M.-C.; Carreau, M. Fanconi anemia genes are highly expressed in primitive CD34 + hematopoietic cells’. BMC Blood Disord. 2003, 3, 1. [Google Scholar] [CrossRef][Green Version]
- Matthews, L.; Gopinath, G.; Gillespie, M.; Caudy, M.; Croft, D.; de Bono, B.; Garapati, P.; Hemish, J.; Hermjakob, H.; Jassal, B.; et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009, 37 (Suppl. 1), D619–D622. [Google Scholar] [CrossRef]
- Pan, Z.W.; Wang, X.J.; Chen, T.; Ding, X.W.; Jiang, X.; Gao, Y.; Mo, W.J.; Huang, Y.; Lou, C.J.; Cao, W.M. Deleterious mutations in DNA repair gene FANCC exist in BRCA1/2-negative Chinese familial breast and/or ovarian cancer patients. Front. Oncol. 2019, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Dörk, T.; Peterlongo, P.; Mannermaa, A.; Bolla, M.K.; Wang, Q.; Dennis, J.; Ahearn, T.; Andrulis, I.L.; Anton-Culver, H.; Arndt, V.; et al. Two truncating variants in FANCC and breast cancer risk. Sci. Rep. 2019, 9, 12524. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.H.; Ke, H.C.; Lin, W.C.; Nian, F.S.; Huang, C.W.; Cheng, H.Y.; Hsu, C.S.; Granata, T.; Chang, C.H.; Castellotti, B.; et al. Novel lissencephaly-associated NDEL1 variant reveals distinct roles of NDE1 and NDEL1 in nucleokinesis and human cortical malformations. Acta Neuropathol. 2024, 147, 13. [Google Scholar] [CrossRef]
- Soto-Perez, J.; Baumgartner, M.; Kanadia, R.N. Role of NDE1 in the Development and Evolution of the Gyrified Cortex. Front. Neurosci. 2020, 14, 617513. [Google Scholar] [CrossRef]
- Yao, M.; Pan, Y.; Ren, T.; Yang, C.; Lei, Y.; Xing, X.; Zhang, L.; Cui, X.; Zheng, Y.; Xing, L.; et al. Loss of Dip2b leads to abnormal neural differentiation from mESCs. Stem Cell Res. Ther. 2023, 14, 248. [Google Scholar] [CrossRef]
- Musa, G.; Cazorla-Vázquez, S.; van Amerongen, M.J.; Stemmler, M.P.; Eckstein, M.; Hartmann, A.; Braun, T.; Brabletz, T.; Engel, F.B. Gpr126 (Adgrg6) is expressed in cell types known to be exposed to mechanical stimuli. Ann. N. Y. Acad. Sci. 2019, 1456, 96–108. [Google Scholar] [CrossRef]
- Gowhari Shabgah, A.; Jadidi-Niaragh, F.; Mohammadi, H.; Ebrahimzadeh, F.; Oveisee, M.; Jahanara, A.; Gholizadeh Navashenaq, J. The Role of Atypical Chemokine Receptor D6 (ACKR2) in Physiological and Pathological Conditions; Friend, Foe, or Both? Front. Immunol. 2022, 13, 861931. [Google Scholar] [CrossRef] [PubMed]
- Dalaie, K.; Yassaee, V.R.; Behnaz, M.; Yazdanian, M.; Jafari, F.; Farimani, R.M. Relationship of the rs10850110 and rs11611277 polymorphisms of the MYO1H gene with non-syndromic mandibular prognathism in the Iranian population. Dent. Med. Probl. 2020, 57, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Spielmann, M.; Hernandez-Miranda, L.R.; Ceccherini, I.; Weese-Mayer, D.E.; Kragesteen, B.K.; Harabula, I.; Krawitz, P.; Birchmeier, C.; Leonard, N.; Mundlos, S. Mutations in MYO1H cause a recessive form of central hypoventilation with autonomic dysfunction. J. Med. Genet. 2017, 54, 754–761. [Google Scholar] [CrossRef]
- Alerasool, N.; Leng, H.; Lin, Z.Y.; Gingras, A.C.; Taipale, M. Identification and functional characterization of transcriptional activators in human cells. Mol. Cell 2022, 82, 677–695. [Google Scholar] [CrossRef]
- Yuan, Y.; Qi, P.; Xiang, W.; Yanhui, L.; Yu, L.; Qing, M. Multi-Omics Analysis Reveals Novel Subtypes and Driver Genes in Glioblastoma. Front. Genet. 2020, 11, 565341. [Google Scholar] [CrossRef] [PubMed]
- Roci, I.; Watrous, J.D.; Lagerborg, K.A.; Jain, M.; Nilsson, R. Mapping choline metabolites in normal and transformed cells. Metabolomics Off. J. Metabolomic Soc. 2020, 16, 125. [Google Scholar] [CrossRef]
- Li Yifei Shen, X.; Yang, X.; Lian, F.; Li Yanping Li, J.; Huang, Y.; Shen, W.; Liu, H. CHDH, a key mitochondrial enzyme, plays a diagnostic role in metabolic disorders diseases and tumor progression. Front. Genet. 2023, 14, 1240650. [Google Scholar] [CrossRef]
- Martinelli, D.; Schiff, M.; Semeraro, M.; Agolini, E.; Novelli, A.; Dionisi-Vici, C. CUGC for lysinuric protein intolerance (LPI). Eur. J. Hum. Genet. 2020, 28, 1129–1134. [Google Scholar] [CrossRef]
- Carpenter, D.; Robinson, R.L.; Quinnell, R.J.; Ringrose, C.; Hogg, M.; Casson, F.; Booms, P.; Iles, D.E.; Halsall, P.J.; Steele, D.S.; et al. Genetic variation in RYR1 and malignant hyperthermia phenotypes. Br. J. Anaesth. 2009, 103, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Spaccavento, A.; Rodríguez, M.d.R.; Meretta, A.; Elissamburu, P.; Carvelli, V.; Gobbo, M.; Rosa, D.; Masoli, O.; Conde, D.; Costabel, J.P. Prevalence of transthyretin amyloid cardiomyopathy in patients admitted for acute heart failure. Curr. Probl. Cardiol. 2024, 49, 102385. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, D.R.; Alexander, A.A.; Tagoe, C.; Garvey, W.T.; Williams, S.M.; Tishkoff, S.; Modiano, D.; Sirima, S.B.; Kalidi, I.; Toure, A.; et al. The prevalence and distribution of the amyloidogenic transthyretin (TTR) V122I allele in Africa. Mol. Genet. Genom. Med. 2016, 4, 548–556. [Google Scholar] [CrossRef] [PubMed]


| Genomic Coordinate | ETI | Gene Name | Ref# | Alt# | Genotype of Affected Individuals | HGVSc | HGVSp | NTP |
|---|---|---|---|---|---|---|---|---|
| Chr3:46714301 (rs145256818) | ENST00000460241 | PRSS50 | G | A | G/A | c.671C>T | p.Pro224Leu | 7 |
| Chr9:95172132 (rs766909460) | ENST00000289081 | FANCC | TA | T | TA/T | c.360del | p.His120GlnfsTer24 | 1 |
| Chr9:95172134 (rs750003253) | ENST00000289081 | FANCC | TGAGA | T | TGAGA/T | c.355_358del | p.Ser119IlefsTer24 | 1 |
| Genomic Coordinate | ETI | Gene Name | Ref# | Alt# | Genotype of Affected Individuals | HGVSc | HGVSp | NTP |
|---|---|---|---|---|---|---|---|---|
| Chr14:22813019 (rs764284986) | ENST00000397532 | SLC7A7 | A | G | A/G | c.380T>C | p.Ile127Thr | 9 |
| Chr16:15687426 (Novel) | ENST00000396355 | NDE1 | C | G | C/G | c.438C>G | p.Ile146Met | 9 |
| Chr12:50731373 (rs147225936) | ENST00000301180 | DIP2B | T | C | T/C | c.3646T>C | p.Tyr1216His | 8 |
| Chr6:142415804 (rs1204304460) | ENST00000367609 | ADGRG6 | G | A | G/A | c.2678G>A | p.Arg893Gln | 6 |
| Chr3:53823630 (rs765283936) | ENST00000315251 | CHDH | C | T | C/T | c.379G>A | p.Val127Ile | 6 |
| Chr3:42865498 (rs577087357) | ENST00000422265 | ACKR2 | ATGGCACC | A | ATGGCACC/A | c.1010_1016del | p.Pro337LeufsTer21 | 1 |
| Chr12:109447180 (rs200225794) | ENST00000310903 | MYO1H | T | C | T/C | c.3115T>C | p.Ter1039ArgextTer1 | 1 |
| Chr3:49276455 (rs868287641) | ENST00000343010 | C3orf62 | CCT | C | CCT/C | c.416_417del | p.Glu139GlyfsTer20 | 1 |
| Chr14:19747835 (Novel) | ENST00000642117 | OR4Q3 | CATGAACCCCCAGCT | C | CATGAACCCCCAGCT/C | c.436_449del | p.Asn146ProfsTer32 | 1 |
| Chr6:29044316 (rs149813138) | ENST00000377175 | OR2W1 | G | A | G/A | c.860C>T | p.Pro287Leu | 7 |
| Genomic Coordinate | ETI | Gene Name | Ref Allele | Alt Allele | Genotype | HGVSc | HGVSp | NTP |
|---|---|---|---|---|---|---|---|---|
| Chr19:38512366 (rs61739911) | ENST00000359596 | RYR1 | C | T | C/T | c.9355C>T | p.Arg3119Cys | 7 |
| Chr18:31595139 (rs1555631393) | ENST00000237014 | TTR | G | A | G/A | c.220G>A | p.Glu74Lys | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danbaki, A.S.; Asamoah, C.O.; Mensah, G.O.; Tsri, B.; Busch, T.D.; Arthur, F.K.N.; Kyei, I.; Blay, L.K.; Mensah, S.; Adeyemo, A.A.; et al. Pathogenic FANCC Variants Are Associated with Accessory Breasts in a Sub-Saharan African Multiplex Family. Curr. Issues Mol. Biol. 2025, 47, 875. https://doi.org/10.3390/cimb47110875
Danbaki AS, Asamoah CO, Mensah GO, Tsri B, Busch TD, Arthur FKN, Kyei I, Blay LK, Mensah S, Adeyemo AA, et al. Pathogenic FANCC Variants Are Associated with Accessory Breasts in a Sub-Saharan African Multiplex Family. Current Issues in Molecular Biology. 2025; 47(11):875. https://doi.org/10.3390/cimb47110875
Chicago/Turabian StyleDanbaki, Abass Shaibu, Christian Opoku Asamoah, Gideon Okyere Mensah, Bruce Tsri, Tamara D. Busch, Fareed Kow Nanse Arthur, Ishmael Kyei, Lawrence Kobina Blay, Samuel Mensah, Adebowale A. Adeyemo, and et al. 2025. "Pathogenic FANCC Variants Are Associated with Accessory Breasts in a Sub-Saharan African Multiplex Family" Current Issues in Molecular Biology 47, no. 11: 875. https://doi.org/10.3390/cimb47110875
APA StyleDanbaki, A. S., Asamoah, C. O., Mensah, G. O., Tsri, B., Busch, T. D., Arthur, F. K. N., Kyei, I., Blay, L. K., Mensah, S., Adeyemo, A. A., Butali, A., Donkor, P., & Gowans, L. J. J. (2025). Pathogenic FANCC Variants Are Associated with Accessory Breasts in a Sub-Saharan African Multiplex Family. Current Issues in Molecular Biology, 47(11), 875. https://doi.org/10.3390/cimb47110875







