Genome-Wide Identification and Expression Analysis of the CaM Gene Family in Tree Peony (Paeonia ostii) During the Pistil Pollination Process
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Methods
2.2.1. Identification of CaM Gene Family Members and Protein Characterization in Tree Peony
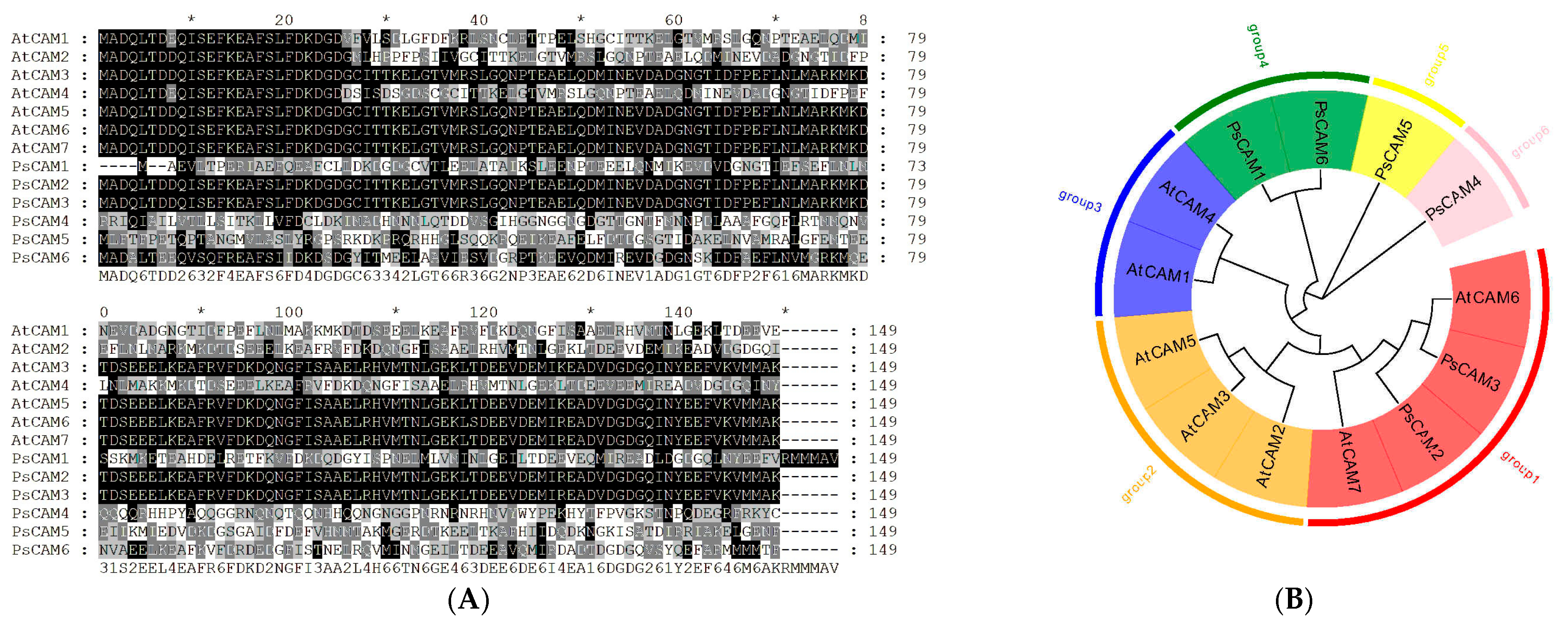
2.2.2. Phylogenetic Analysis of CaM Gene Family Members
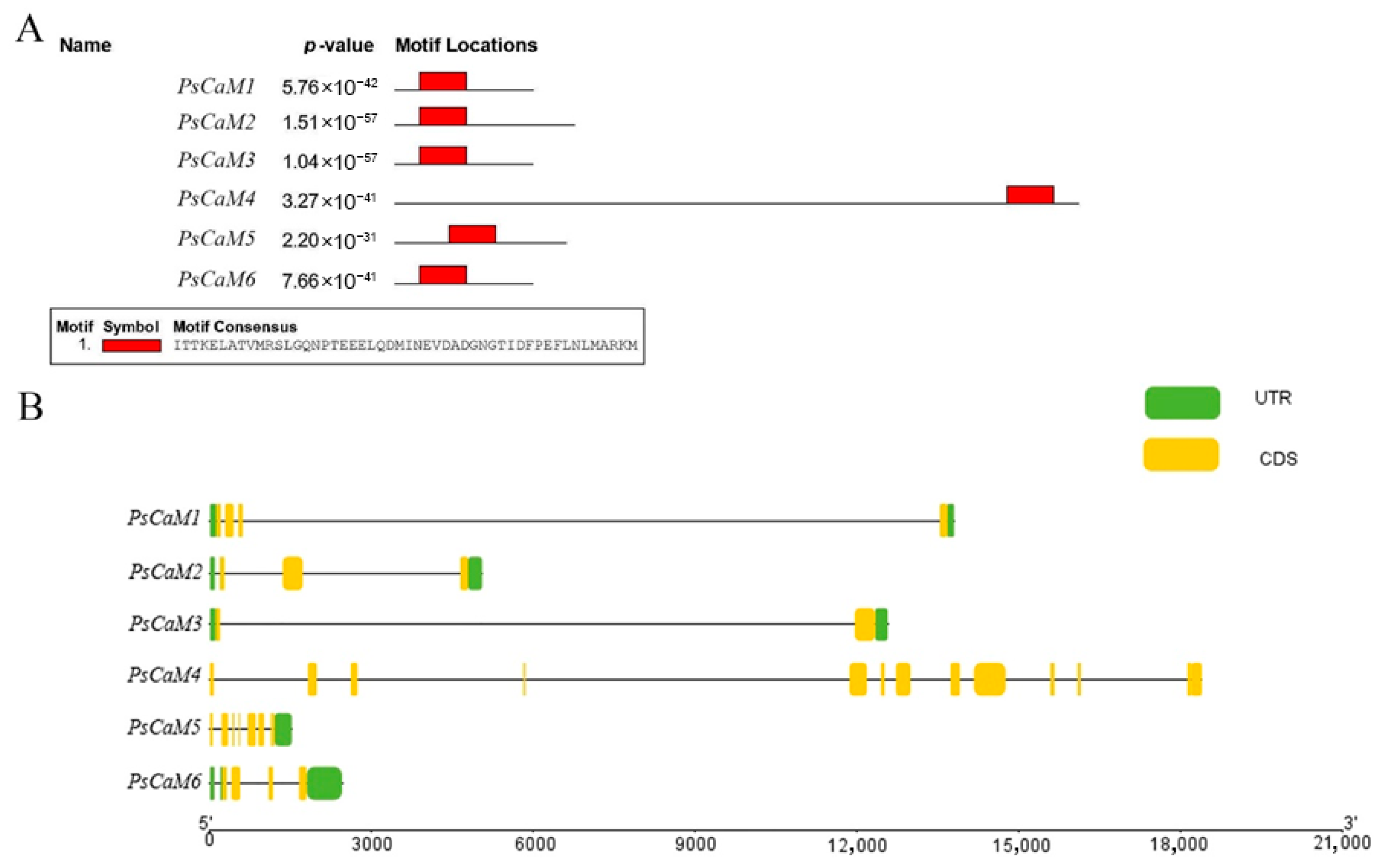
2.2.3. Conserved Motif and Gene Structure Analysis of CaM Genes
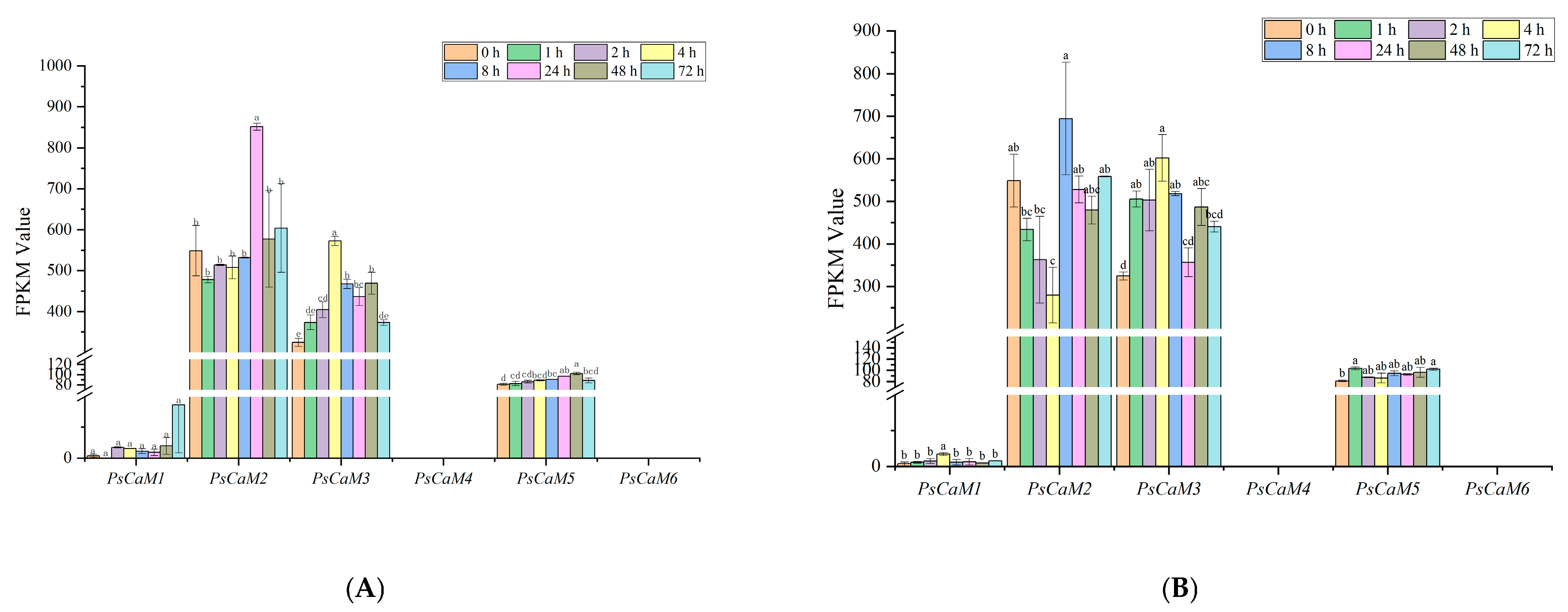
2.2.4. Expression Analysis of PsCaM Genes Based on RNA-Seq Data
2.2.5. qRT-PCR Validation of PsCaM Gene Expression
3. Results
3.1. Identification and Physicochemical Properties of CaM Gene Family Members in Paeonia suffruticosa
3.2. Chromosomal Localization of PsCaM Genes
3.3. Phylogenetic Analysis of CaM Family Members
3.4. Conserved Motif and Gene Structure Analysis of PsCaMs
3.5. Cis-Regulatory Elements in Promoters of PsCaMs
3.6. Expression Patterns of PsCaM Family Genes During Pistil Development Following Pollination in P. ostii ‘Fengdan’
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.Y.; Guo, L.L.; Wang, Z.Y.; Zhao, D.H.; Guo, D.L.; Carlson, J.E.; Yin, W.L.; Hou, X.G. Genome-wide association study of twenty-three flowering phenology traits and four floral agronomic traits in tree peony (Paeonia section Moutan DC.) reveals five genes known to regulate flowering time. Hortic. Res. 2023, 10, uhac263. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Guo, Q.; Zhang, K.Y.; Wang, H.; Jia, C.S.; Guo, D.L.; Guo, L.L.; Hou, X.G. Dormancy-release, germination and seedling growth of Paeonia ostii ‘Fengdan’ seeds under measures of physical and chemical treatment and sowing. PLoS ONE 2022, 17, e0270767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.; Wang, X.; Bao, J.Y.; He, X.N.; Leri, Y.; He, C.L.; Hou, X.G. Bumblebee pollination ensures the stability of both yield and quality of the woody oil crop Paeonia ostii ‘Fengdan’. Basic Appl. Ecol. 2024, 79, 38–45. [Google Scholar] [CrossRef]
- Zhang, K.L.; Yao, L.J.; Zhang, Y.; Baskin, J.M.; Baskin, C.C.; Xiong, Z.M.; Tao, J. A review of the seed biology of Paeonia species (Paeoniaceae), with particular reference to dormancy and germination. Planta 2019, 249, 291–303. [Google Scholar] [CrossRef]
- Wang, H.; Wei, S.; He, Y.L.; Wang, X.H.; Li, Y.Y.; Wei, D.F.; Wang, Z.Y.; Guo, L.L.; Shaaban, M.; Hou, X.G. Characterization of agronomic and seed oil features for different cultivars of tree peony. Plants 2023, 12, 3112. [Google Scholar] [CrossRef]
- Zhang, K.Y.; He, C.L.; Hou, X.G. Influence of pollination methods on fruit development, fruit yield and oil quality in oil tree peony. Sci. Hortic. 2022, 295, 110877. [Google Scholar] [CrossRef]
- Shi, J.; Wang, X.Y.; Zhang, S.L.; Huo, Z.P.; Liu, S.D. The influence of culture medium components on the germination of peony pollen and the growth of pollen tubes. J. Henan Univ. Sci. Technol. (Nat. Sci. Ed.) 2013, 34, 75–80+1. [Google Scholar]
- Wang, H.; Zhang, C.H.; Huang, Y.L. Research progress on factors affecting plant pollen and preservation methods. Chin. Agric. Sci. Bull. 2023, 39, 49–54. [Google Scholar]
- Luo, J.Y.; Tao, Q.; Wang, Y.; Pei, S.Y.; Zou, X.X.; Yuan, F. The role of calcium signaling in pollen germination and pollen tube growth. Trop. Crops J. 2025, 1–15. [Google Scholar]
- Zhou, Z.G.; Zheng, S.; UI Haq, S.I.; Zhang, D.F.; Qiu, Q.S. Regulation of pollen tube growth by cellular pH and ions. J. Plant Physiol. 2022, 277, 153792. [Google Scholar] [CrossRef]
- Zuo, N.; Chen, J.; Lv, Y.G. Advance progress in plant calmodulin and calmodulin-binding proteins structure biology. Cereals Oils 2016, 29, 1–5. [Google Scholar]
- Gong, M.; Cao, Z.X. Regulation of calcium and calmodulin on pollen germination and growth of pollen tube. Plant Physiol. Commun. 1995, 31, 321–328. [Google Scholar]
- Zhang, W.; Zhou, R.G.; Gao, Y.J.; Zheng, S.Z.; Xu, P.; Zhang, S.Q.; Sun, D.Y. Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 2009, 149, 1773–1784. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, L.; Cheng, M.; Sun, J.F.; Ji, X.M.; Ullah, A.; Xie, G.S. Calmodulins and calmodulin-like proteins-mediated plant organellar calcium signaling networks under abiotic stress. Crop J. 2024, 12, 1321–1332. [Google Scholar] [CrossRef]
- Chu, M.X.; Li, J.J.; Zhang, J.Y.; Shen, S.F.; Li, C.N.; Gao, Y.J.; Zhang, S.Q. AtCaM4 interacts with a Sec14-like protein, PATL1, to regulate freezing tolerance in Arabidopsis in a CBF-independent manner. J. Exp. Bot. 2018, 69, 5241–5253. [Google Scholar] [CrossRef]
- Yu, X.J.; Cao, S.Y.; Dong, Y.M.; Bi, B.L.; Zhang, Y.H.; Xu, J.Q. Research progress on the regulation of pollen growth and development by calcium-binding proteins. Northwest Bot. J. 2016, 36, 2121–2127. [Google Scholar] [CrossRef]
- Duan, H.S.; Wang, C.T.; Wei, F.J. Identification and characterization of the CaM gene family in maize. Cereal Sci. 2023, 31, 30–38. [Google Scholar]
- Yu, B.W.; Yan, S.S.; Zhou, H.Y.; Dong, R.Y.; Lei, J.J.; Chen, C.M.; Cao., B.H. Overexpression of CsCaM3 improves high temperature tolerance in cucumber. Front. Plant Sci. 2018, 9, 797. [Google Scholar] [CrossRef]
- Li, T.; Liu, Z.; Lv, T.X.; Xu, Y.X.; Wei, Y.; Liu, W.T.; Wei, Y.J.; Liu, L.; Wang, A.D. Phosphorylation of MdCYTOKININ RESPONSE FACTOR4 suppresses ethylene biosynthesis during apple fruit ripening. Plant Physiol. 2023, 191, 694–714. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, J.; Wang, Y.H.; Wang, J.C.; Yu, Y.; Long, Y.; Wang, Y.L.; Zhang, H.; Ren, Y.L.; Chen, J.; et al. OsCNGC13 promotes seed-setting rate by facilitating pollen tube growth in stylar tissues. PLos Genet. 2017, 13, e1006906. [Google Scholar] [CrossRef]
- Yuan, J.H.; Jiang, S.J.; Jian, J.B.; Liu, M.Y.; Yue, Z.; Xu, J.B.; Li, J.; Xu, C.Y.; Lin, L.H.; Jing, Y.; et al. Genomic basis of the giga-chromosomes and giga-genome of tree peony Paeonia ostii. Nat. Commun. 2022, 13, 7328. [Google Scholar] [CrossRef]
- Li, Y.Y.; Wang, C.; Guo, Q.; Song, C.W.; Wang, X.H.; Guo, L.L.; Hou, X.G. Characteristics of PoVIN3, a key gene of vernalization pathway, affects flowering time. Int. J. Mol. Sci. 2022, 23, 140033. [Google Scholar] [CrossRef]
- Wang, H.B.; Feng, M.C.; Zhong, X.Q.; Yu, Q.; Que, Y.X.; Xu, L.P.; Guo, J.L. Identification of saccharum CaM gene family and function characterization of ScCaM1 during cold and oxidant exposure in Pichia pastoris. Genes Genom. 2022, 45, 103–122. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, R.E. Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 697–725. [Google Scholar] [CrossRef] [PubMed]
- Minnes, L.; Shaw, D.J.; Cossins, B.P.; Donaldson, P.M.; Greetham, G.M.; Towrie, N.; Parker, A.W.; Baker, M.G.; Henry, A.J.; Taylor, R.J.; et al. Quantifying Secondary Structure Changes in Calmodulin Using 2D-IR Spectroscopy. Anal. Chem. 2017, 89, 10898–10906. [Google Scholar] [CrossRef] [PubMed]
- Gao, L. Systematic Evolution of Plant CaM/CML Gene Family and Its Structural and Expression Characteristics in Lotus. Master’s Thesis, Hubei University, Wuhan, China, 2023. [Google Scholar]
- Mccormack, E.; Braam, J. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 2003, 159, 585–598. [Google Scholar] [CrossRef]
- Boonburapong, B.; Buaboocha, T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007, 7, 4. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.W.; Chen, W.Y.; Liu, L.B.; Su, Y.H.; Li, Y.; Jia, W.Z.; Jiao, B.; Wang, J.; Yang, F.; Dong, F.S.; et al. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in wheat (Triticum aestivum L.). Plant Signal. Behav. 2022, 17, 2013646. [Google Scholar] [CrossRef]
- Vandelle, E.; Vannozzi, A.; Wong, D.; Danzi, D.; Digby, A.M.; Santo, S.D.; Astegno, A. Identification, characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine reveal likely roles in stress responses. Plant Physiol. Biochem. 2018, 129, 221–237. [Google Scholar] [CrossRef]
- Crego, C.G.; Hess, J.; Yardeni, G.; Harpe, M.A.; Priemer, C.; Beclin, F.; Saadain, S.; Cauz-Santos, L.A.; Temsch, E.M.; Weiss-Schneeweiss, H.; et al. CAM evolution is associated with gene family expansion in an explosive bromeliad radiation. Plant Cell 2024, 10, 4109–4131. [Google Scholar] [CrossRef]
- Li, Q.H.; Gao, L.; Yu, F.; Lv, S.Y.; Yang, P.F. Evolution and diversification of CaM/CML gene family in green plants. Plant Physiol. Biochem. 2023, 2021, 107922. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, W.; Xu, Y.P.; Cao, J.Y.; Braam, J.; Cai, X.Z. Genome-wide identification and functional analyses of calmodulin genes in solanaceous species. BMC Plant Biol. 2013, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.D.; Ge, W.Y.; Yan, H.B.; Yang, K.; Guan, J.F. Cloning and expression analysis of calmodulin gene from “Yali” pears. J. Agric. Univ. Hebei 2016, 39, 58–63. [Google Scholar] [CrossRef]
- Nie, F.; Fang, H.L.; Liu, H.P.; Qi, X.W.; Yu, F.; Li, L.; Liang, C.Y. Cloning and expression analysis on FcCaM gene from Ficus carica. J. Plant Resour. Environ. 2024, 33, 1–10. [Google Scholar]
- Zhou, L.; Fu, Y.; Yang, Z. A genome-wide functional characterization of Arabidopsis regulatory calcium sensors in pollen tubes. J. Integr. Plant Biol. 2009, 51, 751–761. [Google Scholar] [CrossRef]
- Gao, Q.; Wang, C.; Xi, Y.; Shao, Q.L.; Li, L.G.; Luan, S. A receptor-channel trio conducts Ca2+ signalling for pollen tube reception. Nature 2022, 607, 534–539. [Google Scholar] [CrossRef]
- El Mahi, H.; Pérez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef]


| Primer Name | Forward Primer | Reverse Primer |
|---|---|---|
| PsCaM1 | 5′-GATGGAGATGGCTGCGTTAC-3′ | 5′-CAACTCGTCGTGTGCTTCAG-3′ |
| PsCaM2 | 5′-GGTTGATGCTGATGGGAATG-3′ | 5′-TGAAGCCGTTTTGGTCCT-3′ |
| PsCaM3 | 5′-CGGTTGTATCACCACGAAAG-3′ | 5′-ATTCTTCCTCAGAGTCGGTGTC-3′ |
| PsCaM4 | 5′-GTTTCTGGGATTCACGGAGG-3′ | 5′-GGACCACCATTACCGTTCTG-3′ |
| PsCaM5 | 5′-AACACAGCCTACAGCCAATG-3′ | 5′-TGTCATCTCAAAGCCCAGTG-3′ |
| PsCaM6 | 5′-GAAGAACTGGCAGCGGTCAT-3′ | 5′-CCTCTGCGACATTTTCCTGC-3′ |
| EF-1α | 5′-CCGCCAGAGAGGCTGCTAAT-3′ | 5′-GCAATGTGGGAAGTGTGGCA-3′ |
| Gene ID | Number of Amino Acids | Molecular Weight/kD | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| PsCaM1 | 453 | 35,954.28 | 5.30 | 31.79 | 36.20 | 0.72 | Cytomembrane, cytoplasm |
| PsCaM2 | 585 | 46,161.92 | 5.21 | 40.72 | 31.11 | 0.75 | Cytomembrane, cytoplasm |
| PsCaM3 | 450 | 36,013.58 | 5.26 | 34.73 | 29.11 | 0.67 | Cytomembrane, cytoplasm |
| PsCaM4 | 2217 | 184,707.70 | 4.95 | 41.17 | 32.48 | 0.73 | Cytomembrane |
| PsCaM5 | 558 | 43,948.14 | 5.25 | 35.04 | 35.30 | 0.74 | Cytomembrane |
| PsCaM6 | 450 | 35,528.73 | 5.30 | 19.16 | 33.78 | 0.68 | Vacuole |
| Genes | ABRE | MBS | LTR | ARE | G-Box |
|---|---|---|---|---|---|
| PsCaM1 | 7 | 0 | 0 | 2 | 10 |
| PsCaM2 | 2 | 0 | 0 | 2 | 11 |
| PsCaM3 | 2 | 1 | 0 | 2 | 10 |
| PsCaM4 | 2 | 0 | 1 | 1 | 7 |
| PsCaM3 | 2 | 1 | 0 | 6 | 2 |
| PsCaM6 | 2 | 0 | 1 | 1 | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, G.; Lv, S.; Zhao, Y.; Li, Y.; Hou, X. Genome-Wide Identification and Expression Analysis of the CaM Gene Family in Tree Peony (Paeonia ostii) During the Pistil Pollination Process. Curr. Issues Mol. Biol. 2025, 47, 816. https://doi.org/10.3390/cimb47100816
Zhao G, Lv S, Zhao Y, Li Y, Hou X. Genome-Wide Identification and Expression Analysis of the CaM Gene Family in Tree Peony (Paeonia ostii) During the Pistil Pollination Process. Current Issues in Molecular Biology. 2025; 47(10):816. https://doi.org/10.3390/cimb47100816
Chicago/Turabian StyleZhao, Guodong, Shuran Lv, Yuxin Zhao, Yuying Li, and Xiaogai Hou. 2025. "Genome-Wide Identification and Expression Analysis of the CaM Gene Family in Tree Peony (Paeonia ostii) During the Pistil Pollination Process" Current Issues in Molecular Biology 47, no. 10: 816. https://doi.org/10.3390/cimb47100816
APA StyleZhao, G., Lv, S., Zhao, Y., Li, Y., & Hou, X. (2025). Genome-Wide Identification and Expression Analysis of the CaM Gene Family in Tree Peony (Paeonia ostii) During the Pistil Pollination Process. Current Issues in Molecular Biology, 47(10), 816. https://doi.org/10.3390/cimb47100816





