A Theoretical Study of the Interaction of PARP-1 with Natural and Synthetic Inhibitors: Advances in the Therapy of Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Computational Details
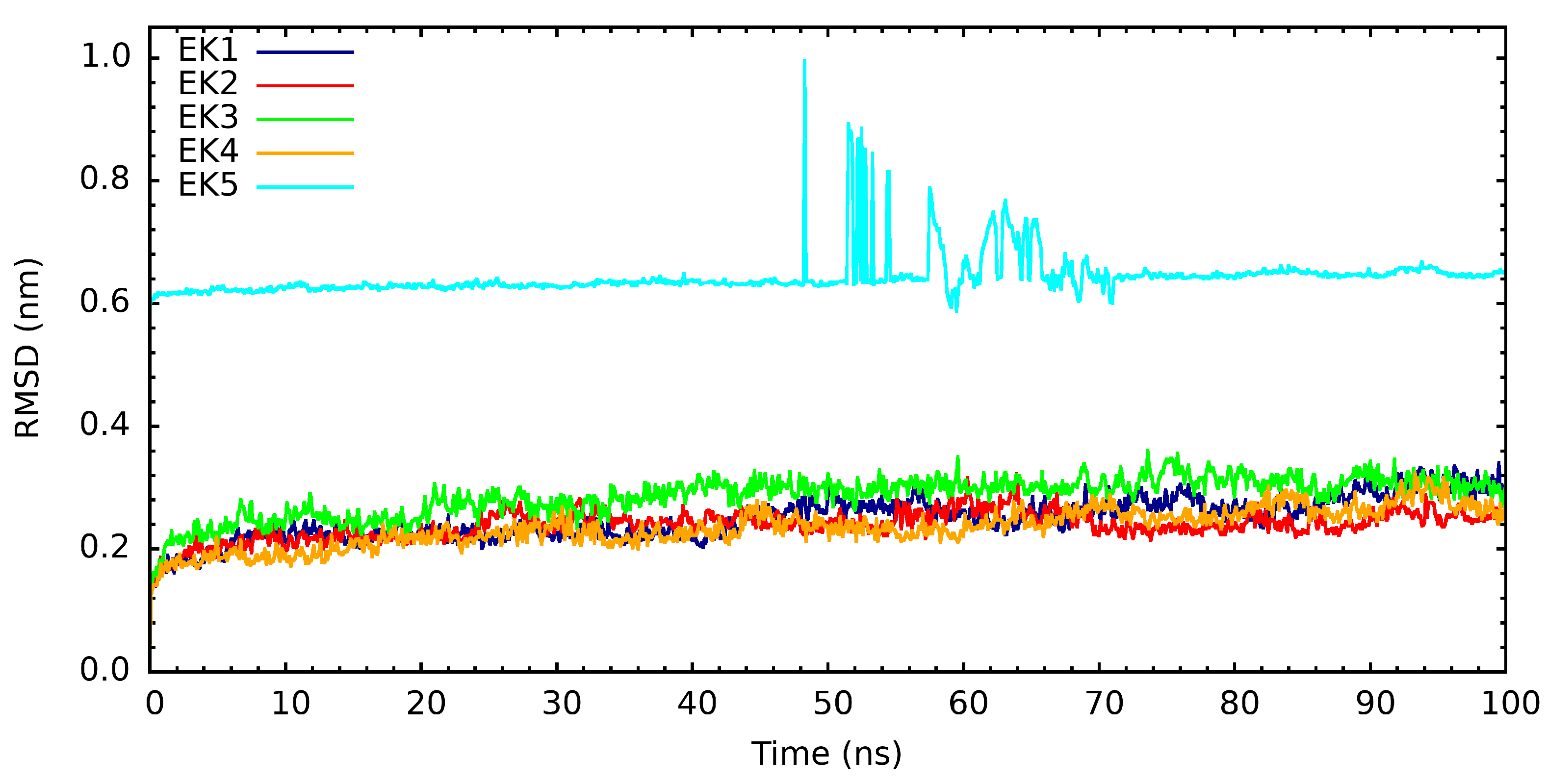
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramos Águila, Y.d.l.C.; Marimón Torres, E.R.; Crespo González, C.; Junco Sena, B.; Valiente Morejón, W. Cáncer de mama, su caracterización epidemiológica. Rev. De Cienc. Médicas De Pinar Del Río 2015, 19, 619–629. [Google Scholar]
- Vallejos-Sologuren, C.S. Situación del Cáncer en el Perú. Diagnóstico 2020, 59, 77–85. [Google Scholar] [CrossRef]
- Richard, I.A.; Burgess, J.T.; O’Byrne, K.J.; Bolderson, E. Beyond PARP1: The Potential of Other Members of the Poly(ADP-Ribose) Polymerase Family in DNA Repair and Cancer Therapeutics. Front. Cell Dev. Biol. 2022, 9, 801200. [Google Scholar] [CrossRef]
- Quiñonero Muñoz, F.J. Estudio de PARP-1 en Células Tumorales y Células Madre de Cáncer de Páncreas. Implicación en la Resistencia Tumoral y uso Como Diana Terapéutica. Ph.D. Thesis, Universidad de Granada, Granada, Spain, 2023. [Google Scholar]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomarkers Amp; Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, B.; Nowak, A.Z.; Romanowicz, H. Breast Cancer—Epidemiology, Classification, Pathogenesis and Treatment (Review of Literature). Cancers 2022, 14, 2569. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, M.; Zeinomar, N.; Tehranifar, P.; Terry, M.B. Are Global Breast Cancer Incidence and Mortality Patterns Related to Country-Specific Economic Development and Prevention Strategies? J. Glob. Oncol. 2018, 4, 1–16. [Google Scholar] [CrossRef]
- Borri, F.; Granaglia, A. Pathology of triple negative breast cancer. Semin. Cancer Biol. 2021, 72, 136–145. [Google Scholar] [CrossRef]
- Gong, R.; Ma, Z.; He, L.; Jiang, S.; Cao, D.; Cheng, Y. Identification and evaluation of a novel PARP1 inhibitor for the treatment of triple-negative breast cancer. Chem. Biol. Interact. 2023, 382, 110567. [Google Scholar] [CrossRef]
- Rakha, E.A.; Ellis, I.O. Triple-negative/basal-like breast cancer: Review. Pathology 2009, 41, 40–47. [Google Scholar] [CrossRef]
- Mustacchi, G.; De Laurentiis, M. The role of taxanes in triple-negative breast cancer: Literature review. Drug Des. Devel. Ther. 2015, 9, 4303–4318. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, H.; Song, X.; Yang, Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020, 60, 14–27. [Google Scholar] [CrossRef]
- Chopra, N.; Tovey, H.; Pearson, A.; Cutts, R.; Toms, C.; Proszek, P.; Hubank, M.; Dowsett, M.; Dodson, A.; Daley, F.; et al. Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat. Commun. 2020, 11, 2662. [Google Scholar] [CrossRef]
- Feng, F.Y.; Speers, C.; Liu, M.; Jackson, W.C.; Moon, D.; Rinkinen, J.; Wilder-Romans, K.; Jagsi, R.; Pierce, L.J. Targeted radiosensitization with PARP1 inhibition: Optimization of therapy and identification of biomarkers of response in breast cancer. Breast Cancer Res. Treat. 2014, 147, 81–94. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- King, M.C.; Marks, J.H.; Mandell, J.B. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 2003, 302, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Zatreanu, D.; Robinson, H.M.R.; Alkhatib, O.; Boursier, M.; Finch, H.; Geo, L.; Grande, D.; Grinkevich, V.; Heald, R.A.; Langdon, S.; et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 2021, 12, 3636. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H.; Yang, D.; Yang, Q.; Tang, H.; Fu, Y.X.; Wan, Y. Differential regulation of breast cancer bone metastasis by PARP1 and PARP2. Nat. Commun. 2020, 11, 1578. [Google Scholar] [CrossRef]
- O’Neil, N.J.; Bailey, M.L.; Hieter, P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017, 18, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Lundine, D.; Annor, G.K.; Canar, J.; Ellison, V.; Polotskaia, A.; Donabedian, P.L.; Reiner, T.; Khramtsova, G.F.; Olopade, O.I.; et al. Gain-of-Function Mutant p53 R273H Interacts with Replicating DNA and PARP1 in Breast Cancer. Cancer Res. 2020, 80, 394–405. [Google Scholar] [CrossRef]
- Ohmoto, A.; Yachida, S. Current status of poly(ADP-ribose) polymerase inhibitors and future directions. Onco Targets Ther. 2017, 10, 5195–5208. [Google Scholar] [CrossRef] [PubMed]
- Lodovichi, S.; Mercatanti, A.; Cervelli, T.; Galli, A. Computational analysis of data from a genome-wide screening identifies new PARP1 functional interactors as potential therapeutic targets. Oncotarget 2019, 10, 2722–2737. [Google Scholar] [CrossRef]
- Murai, J.; Huang, S.Y.; Das, B.B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012, 72, 5588–5599. [Google Scholar] [CrossRef]
- Chiarugi, A. A snapshot of chemoresistance to PARP inhibitors. Trends Pharmacol. Sci. 2012, 33, 42–48. [Google Scholar] [CrossRef]
- Montoni, A.; Robu, M.; Pouliot, E.; Shah, G.M. Resistance to PARP-Inhibitors in Cancer Therapy. Front. Pharmacol. 2013, 4, 18. [Google Scholar] [CrossRef]
- Shen, Y.; Aoyagi-Scharber, M.; Wang, B. Trapping Poly(ADP-Ribose) Polymerase. J. Pharmacol. Exp. Ther. 2015, 353, 446–457. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Wei, Z.L.; Juan, W.; Tong, D.; Juan, L.X.; Sa, L.Y.; Jie, H.F.M.; Xiao, G.; Xiang, L.G.; Jie, H.M.; Xu, C. Curcumol inhibits breast cancer growth via NCL/ERα36 and the PI3K/AKT pathway. Food Funct. 2023, 14, 874–885. [Google Scholar] [CrossRef]
- Xu, H.; Shen, X.; Li, X.; Yang, X.; Chen, C.; Luo, D. The natural product dehydrocurvularin induces apoptosis of gastric cancer cells by activating PARP-1 and caspase-3. Apoptosis 2023, 28, 525–538. [Google Scholar] [CrossRef]
- Meng, J.; Yuan, Y.; Li, Y.; Yuan, B. Effects of hirsuteine on MDA-MB-453 breast cancer cell proliferation. Oncol. Lett. 2022, 25, 4. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.S.; Feng, P.P.; Zhang, Y.Y.; Wang, F.Z.; Wang, G.L.; Fei, H.R. Scutebarbatine A induces ROS-mediated DNA damage and apoptosis in breast cancer cells by modulating MAPK and EGFR/Akt signaling pathway. Chem. Biol. Interact. 2023, 378, 110487. [Google Scholar] [CrossRef]
- Lim, J.S.; Kyung, S.Y.; Jeon, Y.; Kim, I.S.; Kwak, J.H.; Kim, H.S. Anticancer effects of the HDAC inhibitor, 3β,6β-dihydroxyurs-12-en-27-oic acid, in MCF-7 breast cancer cells via the inhibition of Akt/mTOR pathways. Oncol. Rep. 2023, 49, 43. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.M.; Abd El-Karim, S.S.; Mahmoud, A.H.; Amr, A.E.E.; Al-Omar, M.A. A Comparative Study of the Anticancer Activity and PARP-1 Inhibiting Effect of Benzofuran-Pyrazole Scaffold and Its Nano-Sized Particles in Human Breast Cancer Cells. Molecules 2019, 24, 2413. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, D.; Girimanchanaika, S.S.; Dukanya, D.; Rangappa, S.; Yang, J.R.; Pandey, V.; Lobie, P.E.; Basappa, B. Design and Activity of Novel Oxadiazole Based Compounds That Target Poly(ADP-ribose) Polymerase. Molecules 2022, 27, 703. [Google Scholar] [CrossRef] [PubMed]
- Syam, Y.M.; Anwar, M.M.; Abd El-Karim, S.S.; Elokely, K.M.; Abdelwahed, S.H. New Quinoxaline-Based Derivatives as PARP-1 Inhibitors: Design, Synthesis, Antiproliferative, and Computational Studies. Molecules 2022, 27, 4924. [Google Scholar] [CrossRef]
- Sadeghian, Z.; Bayat, M.; Safari, F. Synthesis and in vitro evaluation of antitumor activity of spiro[indolo[2,1-b]quinazoline-pyrano[2,3-d]pyrimidine] and spiro[indolo[2,1-b]quinazoline-pyrido[2,3-d]pyrimidine] derivatives by using 2D and 3D cell culture models. Mol. Divers. 2022, 26, 3173–3184. [Google Scholar] [CrossRef]
- Evans, D.A. History of the Harvard ChemDraw Project. Angew. Chem. Int. Ed. 2014, 53, 11140–11145. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Dodda, L.S.; Cabeza de Vaca, I.; Tirado-Rives, J.; Jorgensen, W.L. LigParGen web server: An automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017, 45, W331–W336. [Google Scholar] [CrossRef]
- Hirshfeld, F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange–correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef]
- Schäfer, A.; Huber, C.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J. Chem. Phys. 1994, 100, 5829–5835. [Google Scholar] [CrossRef]
- Schäfer, A.; Horn, H.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J. Chem. Phys. 1992, 97, 2571–2577. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Kutzner, C.; Páll, S.; Fechner, M.; Esztermann, A.; De Groot, B.L.; Grubmüller, H. More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 2019, 40, 2418–2431. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Strain fluctuations and elastic constants. J. Chem. Phys. 1982, 76, 2662–2666. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Crystal Structure and Pair Potentials: A Molecular-Dynamics Study. Phys. Rev. Lett. 1980, 45, 1196–1199. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Williams, T.; Kelley, C. Gnuplot 5.4: An Interactive Plotting Program. 2010. Available online: http://www.gnuplot.info/ (accessed on 20 August 2024).
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson-Boltzmann Surface Area Method. Mol. Inf. 2012, 31, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. g mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
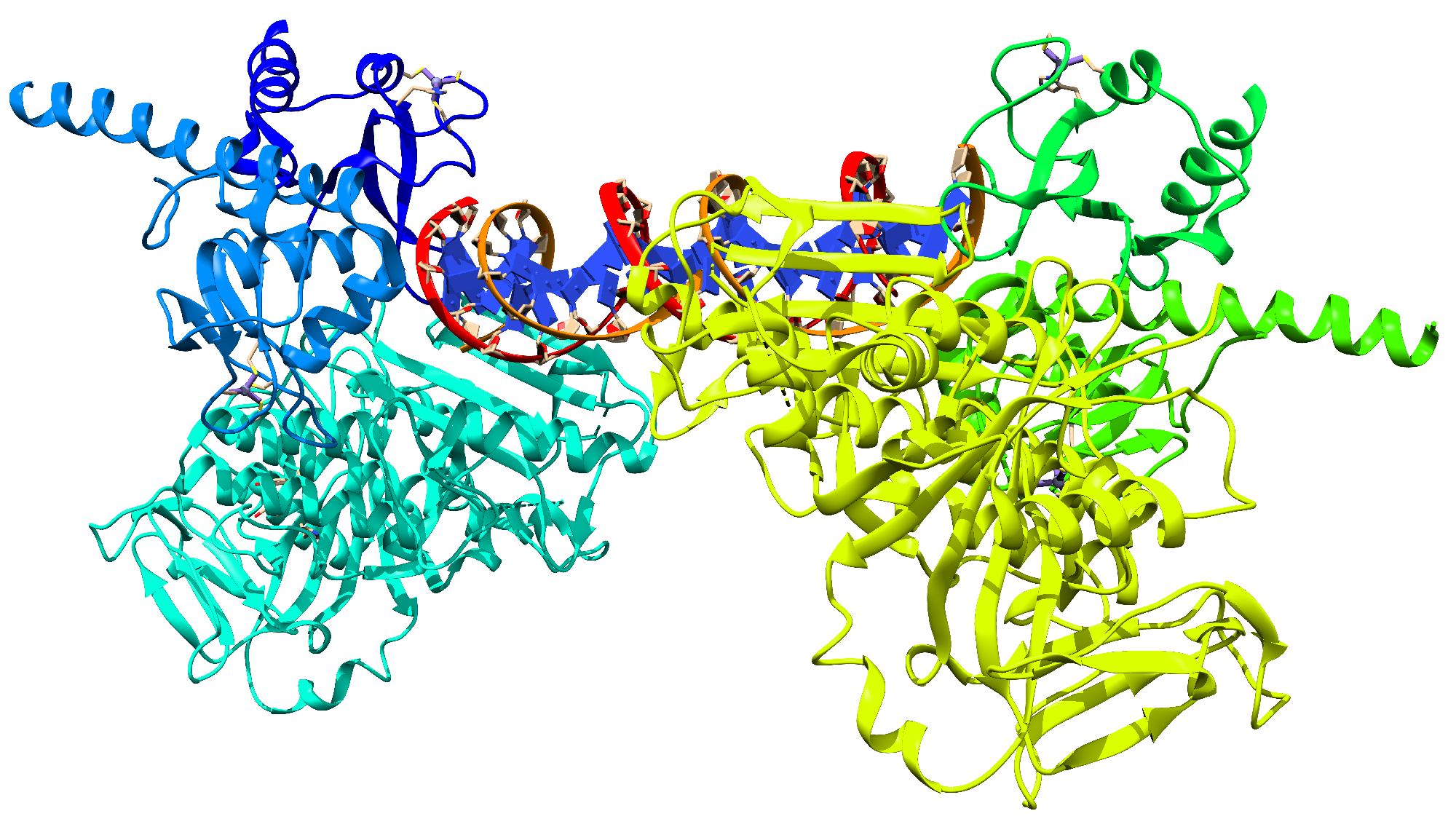
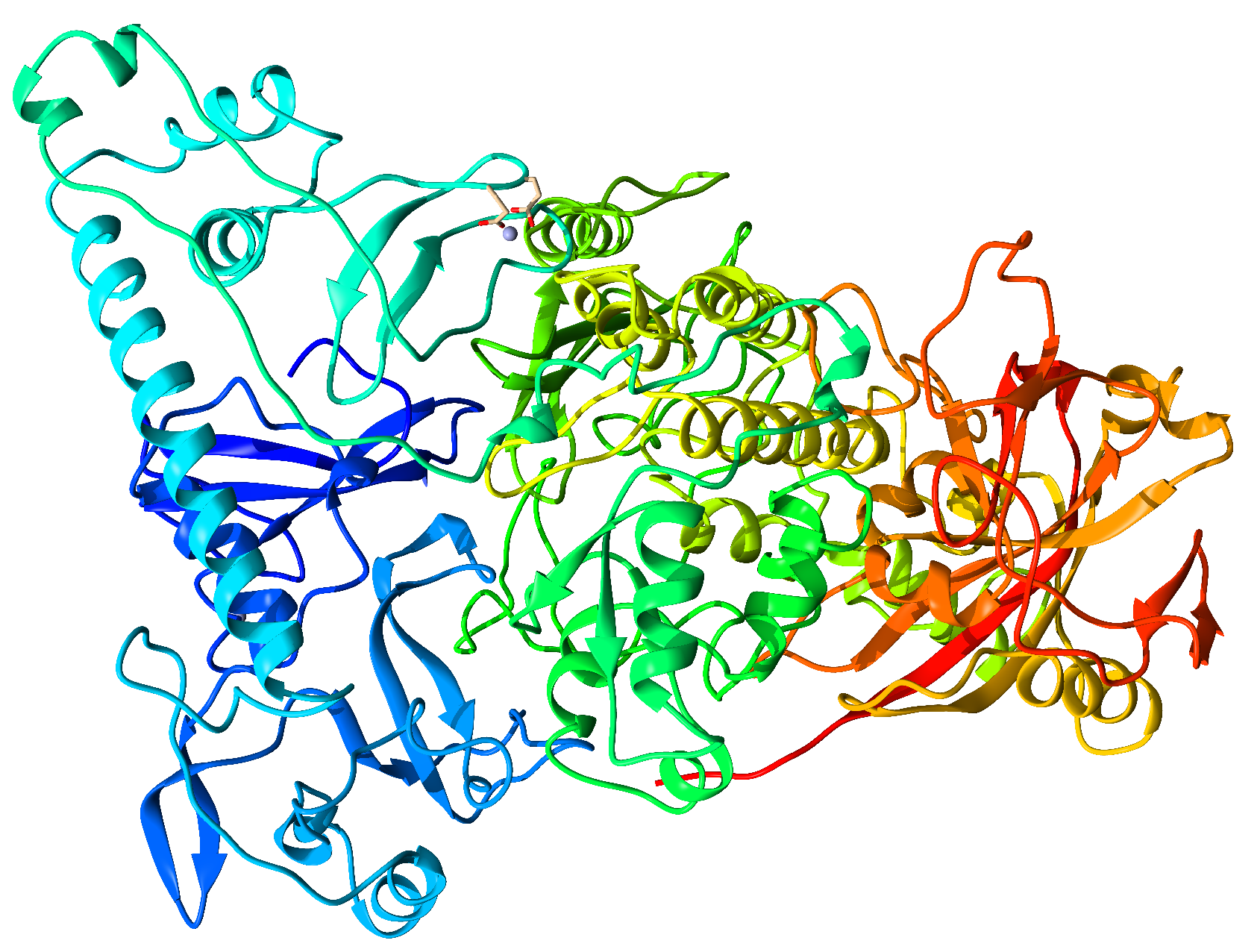
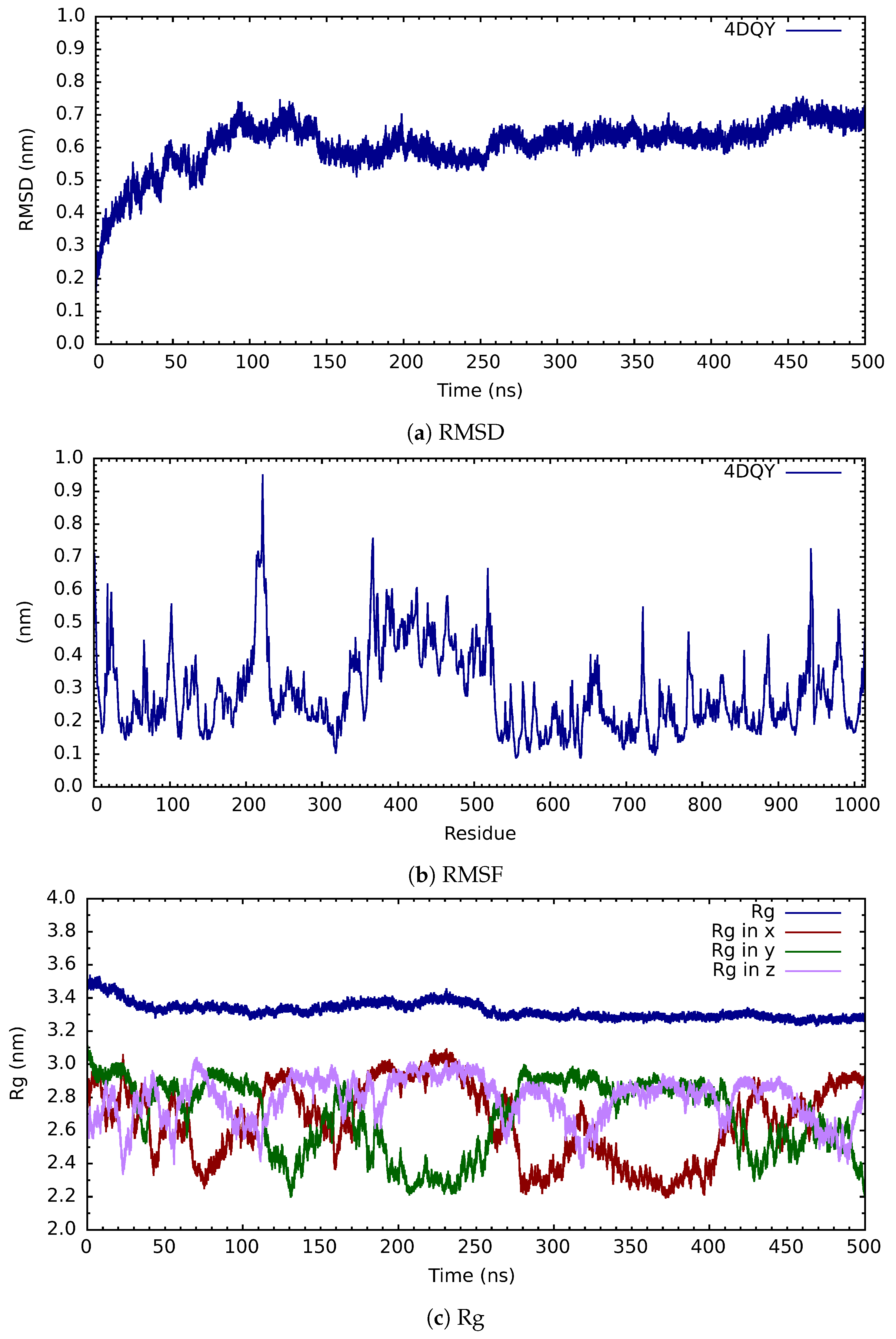
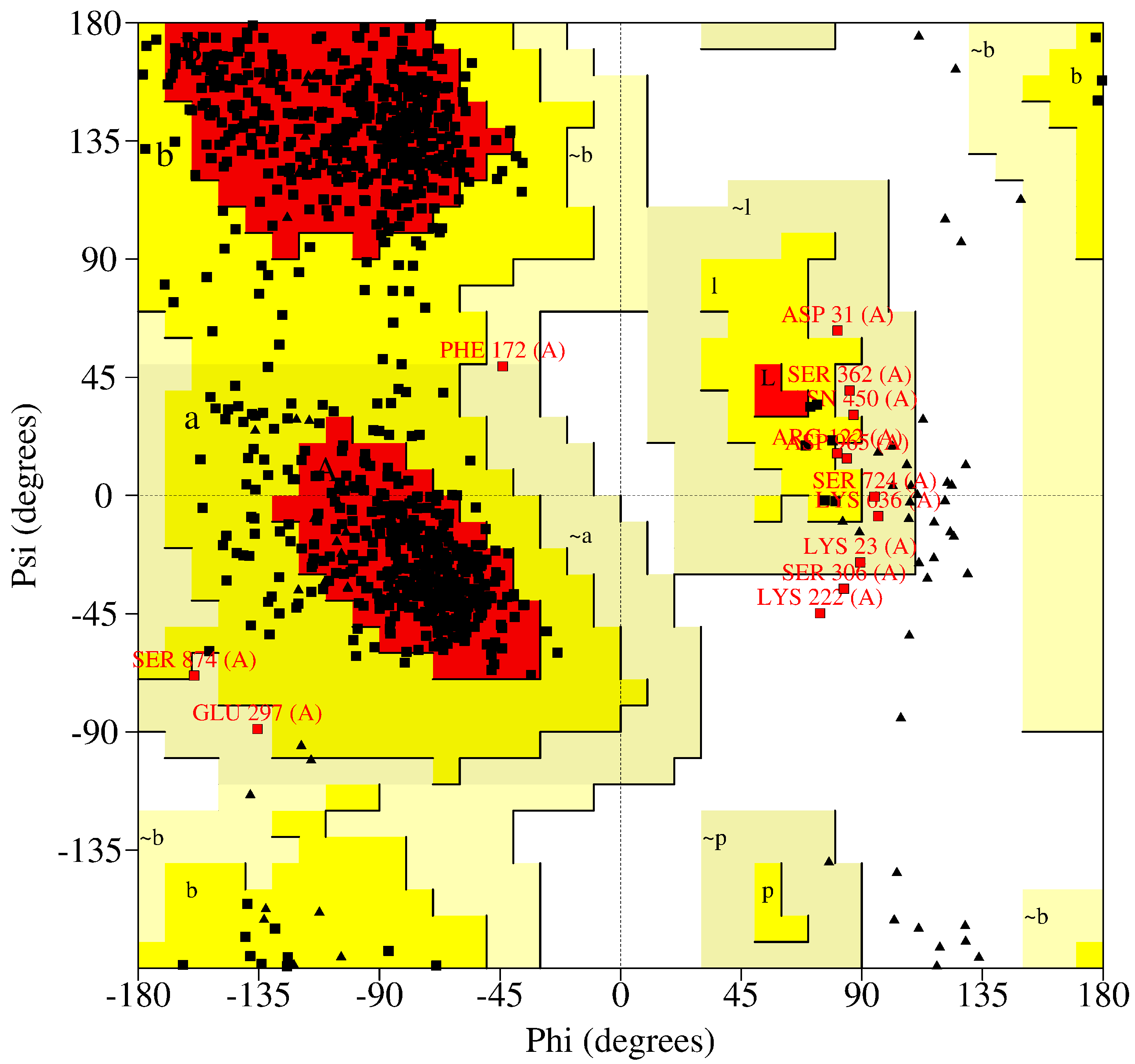
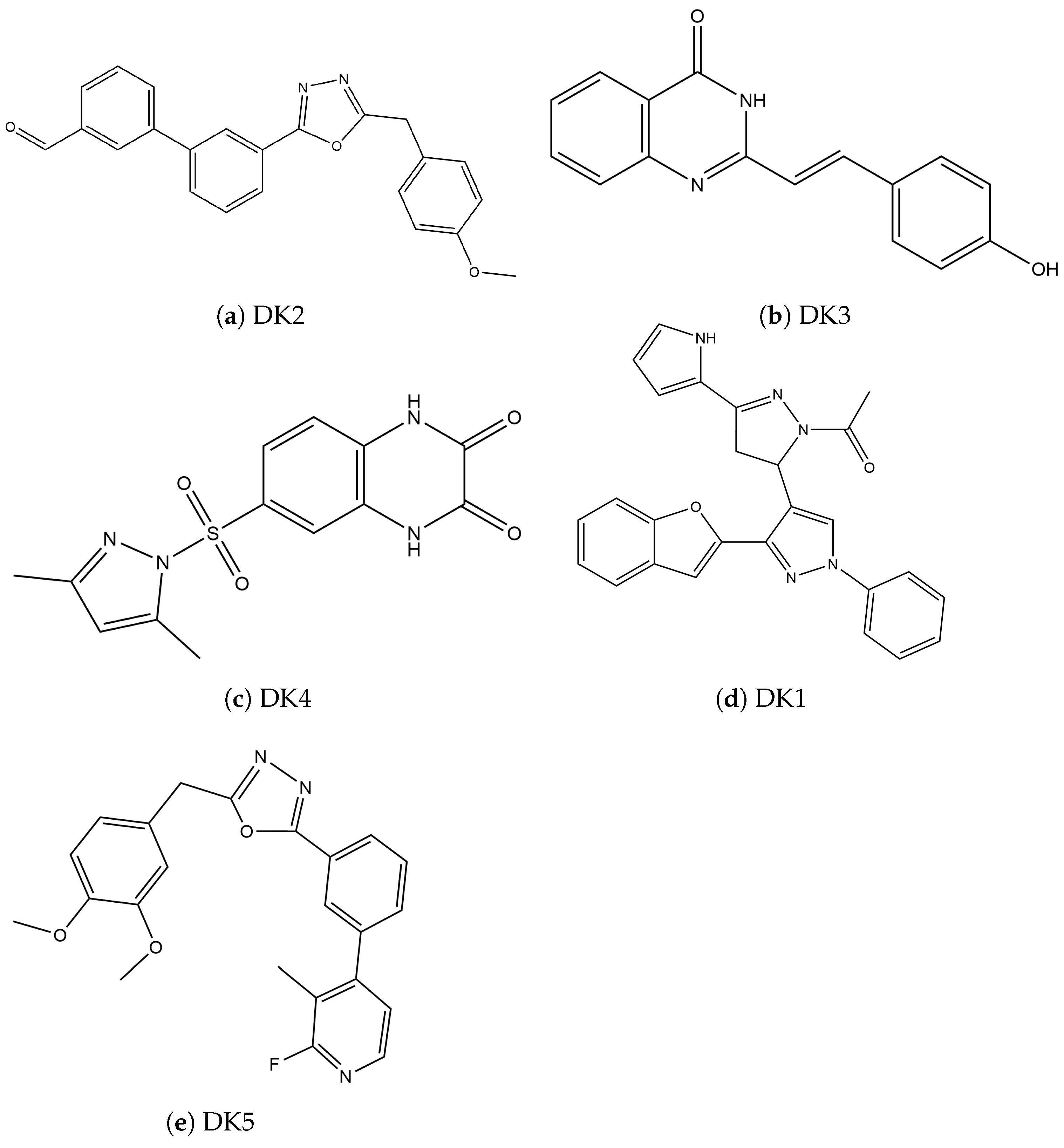

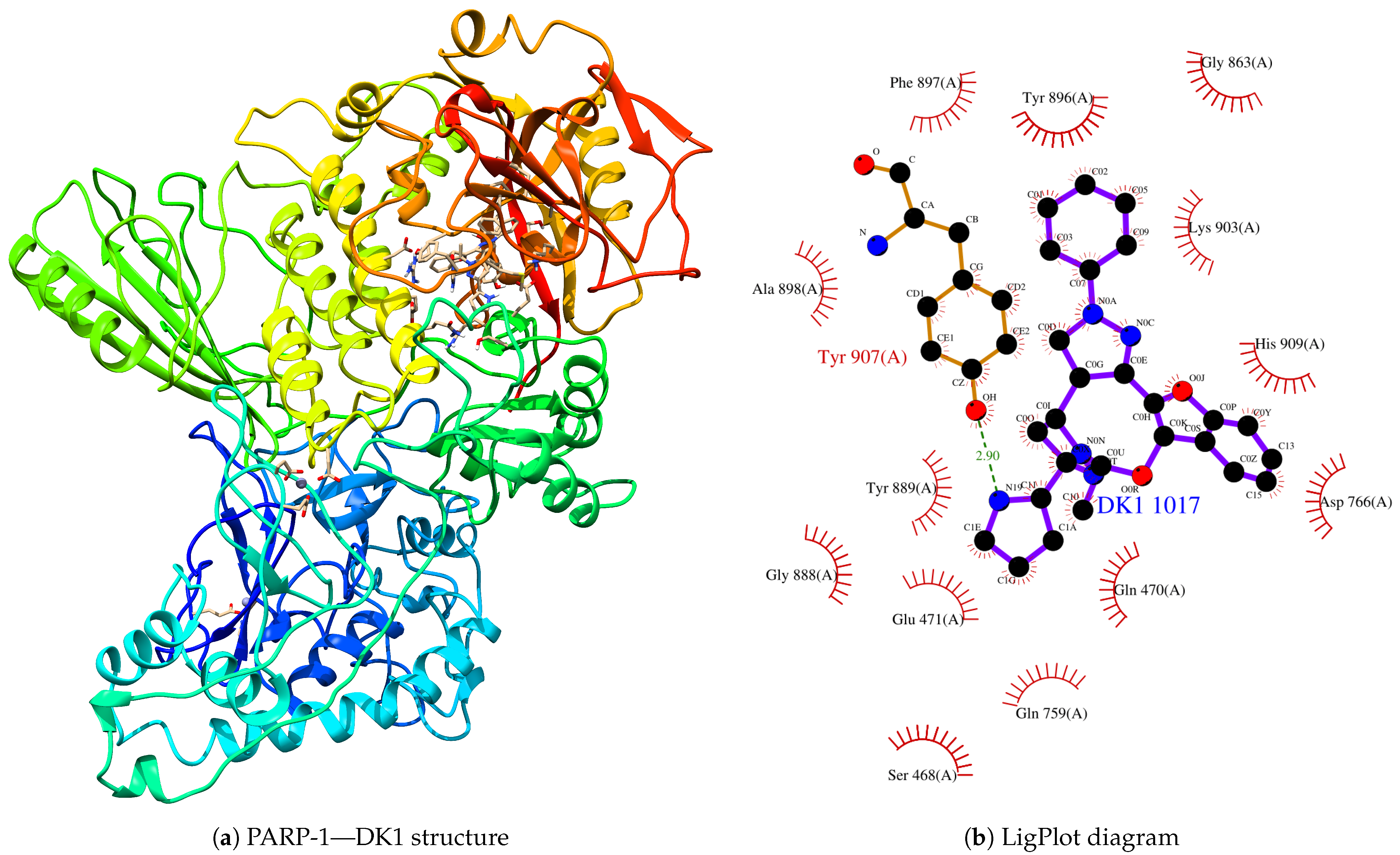
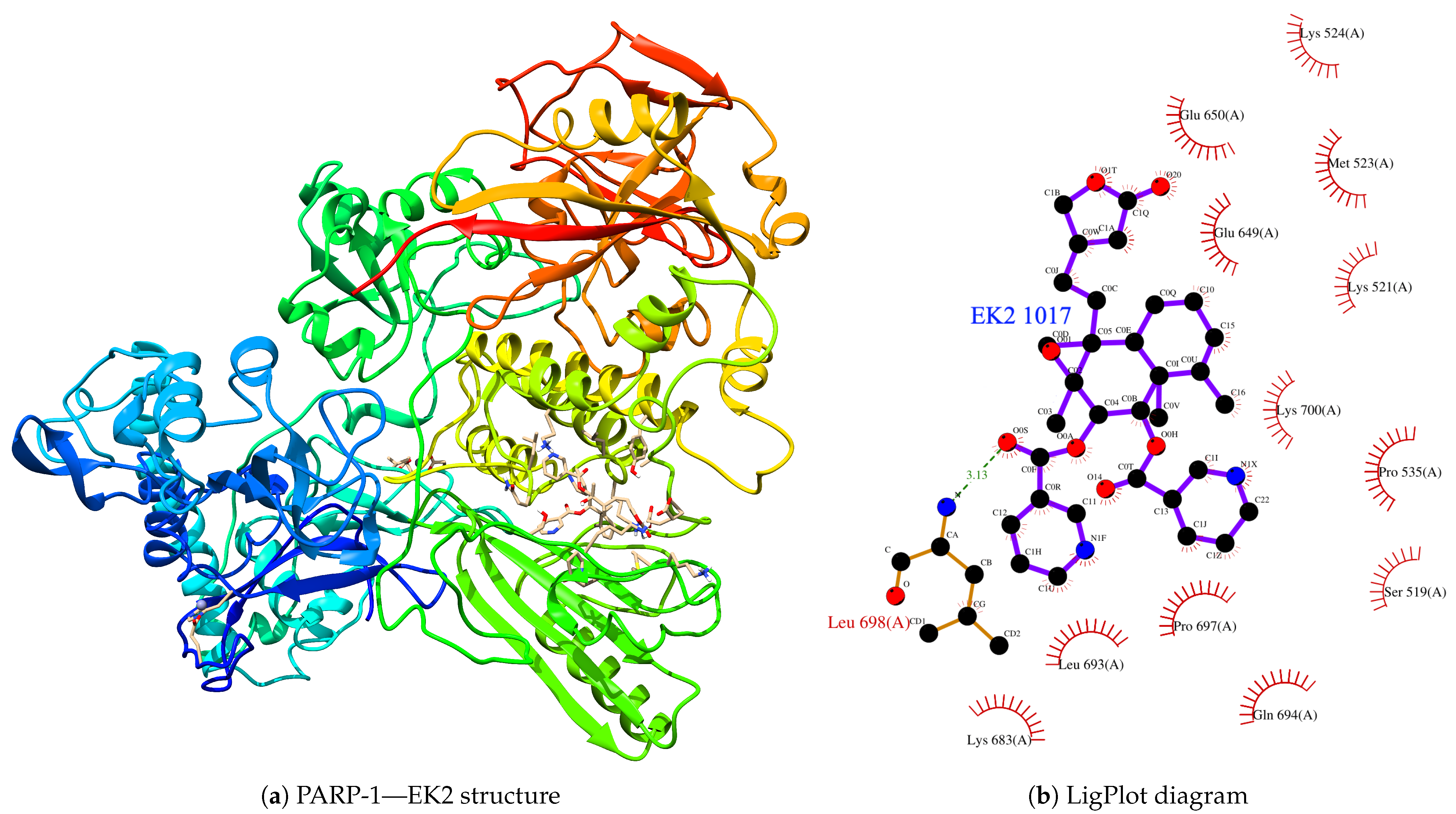
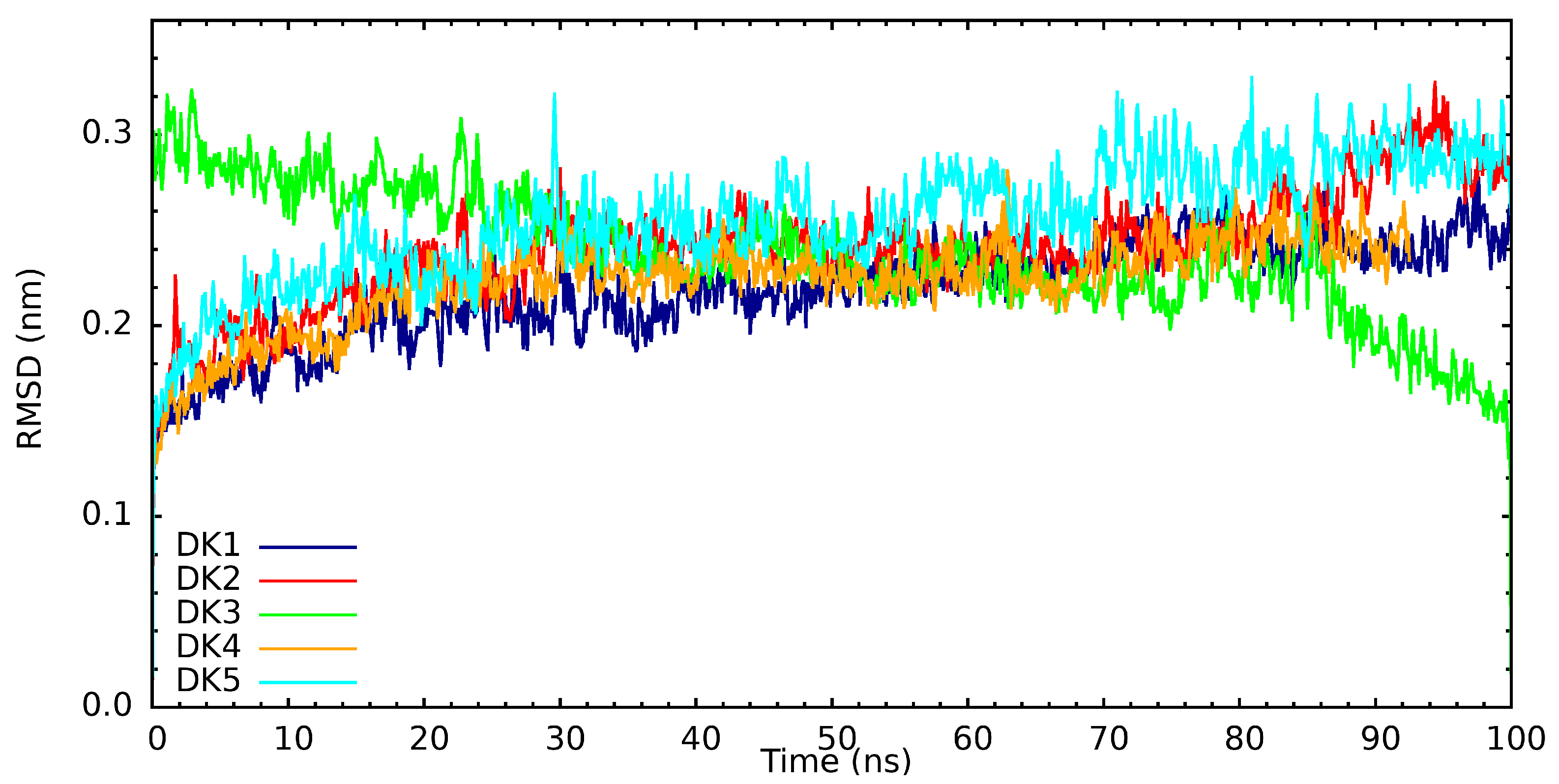

| Protein | Inhibitor | Energy |
|---|---|---|
| PARP-1 | DK1 | −9.999 |
| DK2 | −9.573 | |
| DK3 | −9.294 | |
| DK4 | −7.916 | |
| DK5 | −10.260 | |
| PARP-1 | EK1 | −9.997 |
| EK2 | −9.924 | |
| EK3 | −9.182 | |
| EK5 | −9.520 | |
| EK5 | −8.209 |
| Molecule | Binding Energy 1 |
|---|---|
| DK1 | −99.927 ± 6.615 |
| DK2 | −65.858 ± 9.097 |
| DK3 | −63.680 ± 20.180 |
| DK4 | −82.934 ± 6.662 |
| DK5 | −67.150 ± 6.825 |
| EK1 | −65.414 ± 8.264 |
| EK2 | −88.301 ± 6.897 |
| EK3 | −77.123 ± 6.562 |
| EK4 | −40.123 ± 8.044 |
| EK5 | −55.813 ± 5.506 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turpo-Peqqueña, A.G.; Leiva-Flores, E.K.; Luna-Prado, S.; Gómez, B. A Theoretical Study of the Interaction of PARP-1 with Natural and Synthetic Inhibitors: Advances in the Therapy of Triple-Negative Breast Cancer. Curr. Issues Mol. Biol. 2024, 46, 9415-9429. https://doi.org/10.3390/cimb46090558
Turpo-Peqqueña AG, Leiva-Flores EK, Luna-Prado S, Gómez B. A Theoretical Study of the Interaction of PARP-1 with Natural and Synthetic Inhibitors: Advances in the Therapy of Triple-Negative Breast Cancer. Current Issues in Molecular Biology. 2024; 46(9):9415-9429. https://doi.org/10.3390/cimb46090558
Chicago/Turabian StyleTurpo-Peqqueña, Albert Gabriel, Emily Katherine Leiva-Flores, Sebastián Luna-Prado, and Badhin Gómez. 2024. "A Theoretical Study of the Interaction of PARP-1 with Natural and Synthetic Inhibitors: Advances in the Therapy of Triple-Negative Breast Cancer" Current Issues in Molecular Biology 46, no. 9: 9415-9429. https://doi.org/10.3390/cimb46090558
APA StyleTurpo-Peqqueña, A. G., Leiva-Flores, E. K., Luna-Prado, S., & Gómez, B. (2024). A Theoretical Study of the Interaction of PARP-1 with Natural and Synthetic Inhibitors: Advances in the Therapy of Triple-Negative Breast Cancer. Current Issues in Molecular Biology, 46(9), 9415-9429. https://doi.org/10.3390/cimb46090558


_Kim.png)






