Seasonal Stability Assessment of Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Normalization in Bombus terrestris
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects and Sampling Procedure
2.2. Genes and Primer Design
2.3. RNA Extraction and Reverse Transcription
2.4. Quantitative Real-Time PCR
2.5. Selection of Reference Gene Expression Stability
2.6. Validation of the Selected Reference Genes
2.7. Data Analysis
3. Results
3.1. Performance of qRT-PCR Primer
3.2. Cycle Threshold (Ct) Range of Reference Genes
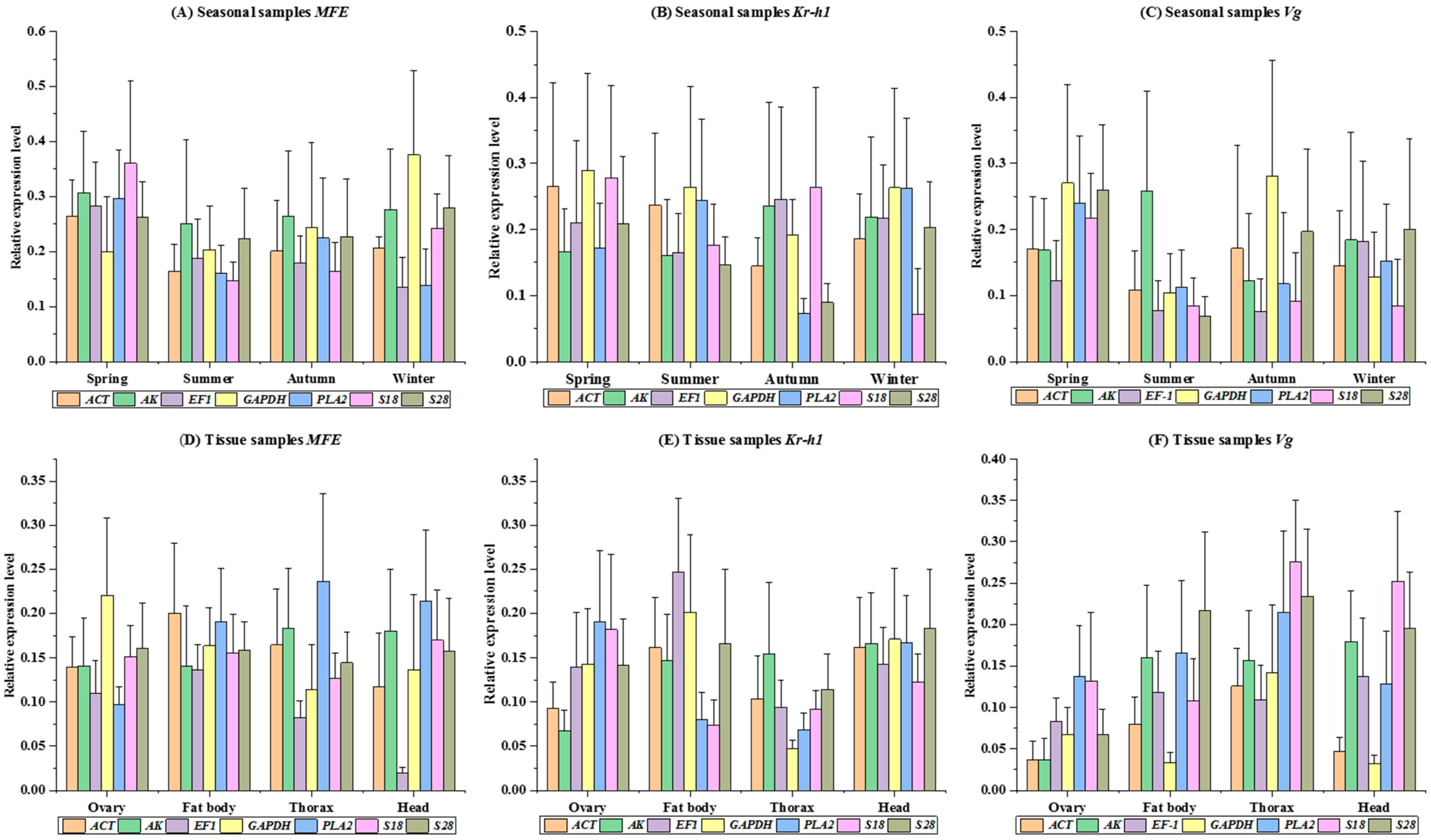
3.3. Stability of Candidate Reference Genes for Seasoning
3.4. Stability of Candidate Reference Genes for Tissue
3.5. RefFinder Ranking of Reference Genes
3.6. Endogenous Control Gene Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Velthuis, H.H.; Van Doorn, A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 2006, 37, 421–451. [Google Scholar] [CrossRef]
- Thorne, B.L.; Breisch, N.L.; Muscedere, M.L. Evolution of eusociality and the soldier caste in termites: Influence of intraspecific competition and accelerated inheritance. Proc. Natl. Acad. Sci. USA 2003, 100, 12808–12813. [Google Scholar] [CrossRef]
- Asbury, K.; McBride, T.; Bawn, R. Can genomic research make a useful contribution to social policy? R. Soc. Open Sci. 2022, 9, 220873. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Chandler, D.; Cooper, E.; Prince, G. Are there risks to wild European bumblebees from using commercial stocks of domesticated Bombus terrestris for crop pollination? J. Apic. Res. 2019, 58, 665–681. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Jones, B.M.; Pan, H.; Li, C.; Harpur, B.A.; Kent, C.F.; Zayed, A.; Ioannidis, P.; Waterhouse, R.M.; Kingwell, C.; et al. Developmental plasticity shapes social traits and selection in a facultatively eusocial bee. Proc. Natl. Acad. Sci. USA 2020, 117, 13615–13625. [Google Scholar] [CrossRef] [PubMed]
- Jedlička, P.; Ernst, U.R.; Votavová, A.; Hanus, R.; Valterová, I. Gene expression dynamics in major endocrine regulatory pathways along the transition from solitary to social life in a bumblebee, Bombus terrestris. Front. Physiol. 2016, 7, 574. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D.; Lepais, O.; O’Connor, S.; Osborne, J.L.; Sanderson, R.A.; Cussans, J.; Goffe, L.; Darvill, B. Effects of land use at a landscape scale on bumblebee nest density and survival. J. Appl. Ecol. 2010, 47, 1207–1215. [Google Scholar] [CrossRef]
- Arrese, E.L.; Soulages, J.L. Insect fat body: Energy, metabolism, and regulation. Annu. Rev. Entomol. 2010, 55, 207–225. [Google Scholar] [CrossRef] [PubMed]
- Hartfelder, K.; Cnaani, J.; Hefetz, A. Caste-specific differences in ecdysteroid titers in early larval stages of the bumblebee Bombus terrestris. J. Insect Physiol. 2000, 46, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Nijhout, H.F.; Emlen, D.J. Competition among body parts in the development and evolution of insect morphology. Proc. Natl. Acad. Sci. USA 1998, 95, 3685–3689. [Google Scholar] [CrossRef]
- Lourenço, A.P.; Mackert, A.; dos Santos Cristino, A.; Simões, Z.L.P. Validation of reference genes for gene expression studies in the honeybee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 2008, 39, 372–385. [Google Scholar] [CrossRef]
- Bomtorin, A.D.; Mackert, A.; Rosa, G.C.C.; Moda, L.M.; Martins, J.R.; Bitondi, M.M.G.; Hartfelder, K.; Simões, Z.L.P. Juvenile hormone biosynthesis gene expression in the corpora allata of honeybee (Apis mellifera L.) female castes. PLoS ONE 2014, 9, e86923. [Google Scholar] [CrossRef]
- Shpigler, H.; Amsalem, E.; Huang, Z.Y.; Cohen, M.; Siegel, A.J.; Hefetz, A.; Bloch, G. Gonadotropic and physiological functions of juvenile hormone in bumblebee (Bombus terrestris) workers. PLoS ONE 2014, 9, e100650. [Google Scholar] [CrossRef]
- He, Q.; Zhang, Y. Kr-h1, a cornerstone gene in insect life history. Front. Physiol. 2022, 13, 905441. [Google Scholar] [CrossRef] [PubMed]
- Cardoso-Júnior, C.A.M.; Oldroyd, B.; Ronai, I. Vitellogenin expression in the ovaries of adult honeybee workers provides insights into the evolution of reproductive and social traits. Insect Mol. Biol. 2021, 30, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Wynants, E.; Van Dun, C.; Lenaerts, N.; Princen, S.A.; Tuyttens, E.; Shpigler, H.Y.; Wenseleers, T.; Van Oystaeyen, A. Uncovering the role of juvenile hormone in ovary development and egg laying in bumblebees. J. Insect Physiol. 2023, 150, 104557. [Google Scholar] [CrossRef] [PubMed]
- Shpigler, H.Y.; Cohen, T.M.; Ben-Shimol, E.; Ben-Betzalel, R.; Levin, E. Juvenile hormone functions as a metabolic rate accelerator in bumblebees (Bombus terrestris). Horm. Behav. 2021, 136, 105073. [Google Scholar] [CrossRef] [PubMed]
- De Kort, C.; Granger, N. Regulation of JH titers: The relevance of degradative enzymes and binding proteins. In Archives of Insect Biochemistry and Physiology: Published in Collaboration with the Entomological Society of America; Agricultural Lhriz~ersity: Wageningen, The Netherlands; University of North Carolina at Chapel Hill: Chapel Hill, NC, USA, 1996; Volume 33, pp. 1–26. [Google Scholar]
- Santos, C.G.; Humann, F.C.; Hartfelder, K. Juvenile hormone signaling in insect oogenesis. Curr. Opin. Insect Sci. 2019, 31, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.J.; Lee, K.Y.; Lee, Y.B.; Lee, M.Y.; Sankar, K.; Park, J.D. Current status of insect pollinators use for horticultural crops in Korea, 2020. J. Apic. 2021, 36, 111–123. [Google Scholar]
- Sankar, K.; Yoon, H.J.; Lee, Y.B.; Lee, K.Y. Evaluation of Reference Genes for Real-Time Quantitative PCR Analysis in Tissues from Bumblebees (Bombus terrestris) of Different Lines. Int. J. Mol. Sci. 2022, 23, 14371. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef]
- Obayashi, T.; Kinoshita, K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 2009, 16, 249–260. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Inoue, M.N.; Yokoyama, J.; Washitani, I. Displacement of Japanese native bumblebees by the recently introduced Bombus terrestris (L.) (Hymenoptera: Apidae). J. Insect Conserv. 2008, 12, 135–146. [Google Scholar] [CrossRef]
- Namin, S.M.; Huang, J.; An, J.; Jung, C. Genetic variation and phylogenetic relationships of commercial populations of Bombus ignitus (Hymenoptera, Apidae) with wild populations in Eastern Asia. J. Hymenopt. Res. 2023, 96, 495–506. [Google Scholar] [CrossRef]
- Rojas-Hernandez, N.; Véliz, D.; Vega-Retter, C. Selection of suitable reference genes for gene expression analysis in gills and liver of fish under field pollution conditions. Sci. Rep. 2019, 9, 3459. [Google Scholar] [CrossRef] [PubMed]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- García-Reina, A.; Rodríguez-García, M.J.; Galián, J. Validation of reference genes for quantitative real-time PCR in tiger beetles across sexes, body parts, sexual maturity and immune challenge. Sci. Rep. 2018, 8, 10743. [Google Scholar] [CrossRef] [PubMed]
- Robledo, D.; Hernández-Urcera, J.; Cal, R.M.; Pardo, B.G.; Sánchez, L.; Martínez, P.; Viñas, A. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genom. 2014, 15, 648. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Cassan-Wang, H.; Soler, M.; Yu, H.; Camargo, E.L.O.; Carocha, V.; Ladouce, N.; Savelli, B.; Paiva, J.A.; Leplé, J.-C.; Grima-Pettenati, J. Reference genes for high-throughput quantitative reverse transcription–PCR analysis of gene expression in organs and tissues of Eucalyptus grown in various environmental conditions. Plant Cell Physiol. 2012, 53, 2101–2116. [Google Scholar] [CrossRef]
- Herath, S.; Dai, H.; Erlich, J.; Au, A.Y.; Taylor, K.; Succar, L.; Endre, Z.H. Selection and validation of reference genes for normalisation of gene expression in ischaemic and toxicological studies in kidney disease. PLoS ONE 2020, 15, e0233109. [Google Scholar] [CrossRef]
- Barnes, M.; Freudenberg, J.; Thompson, S.; Aronow, B.; Pavlidis, P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005, 33, 5914–5923. [Google Scholar] [CrossRef]
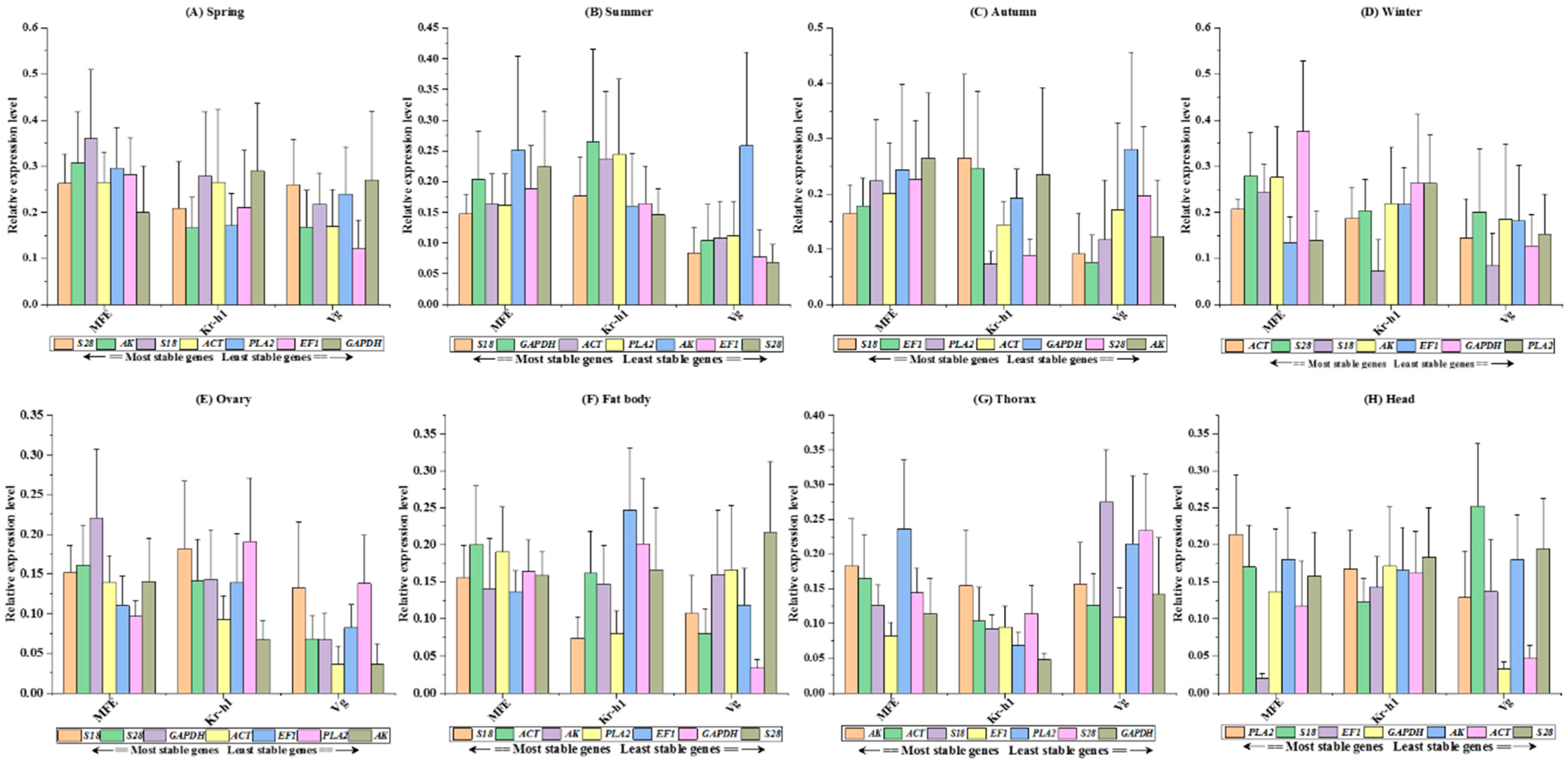
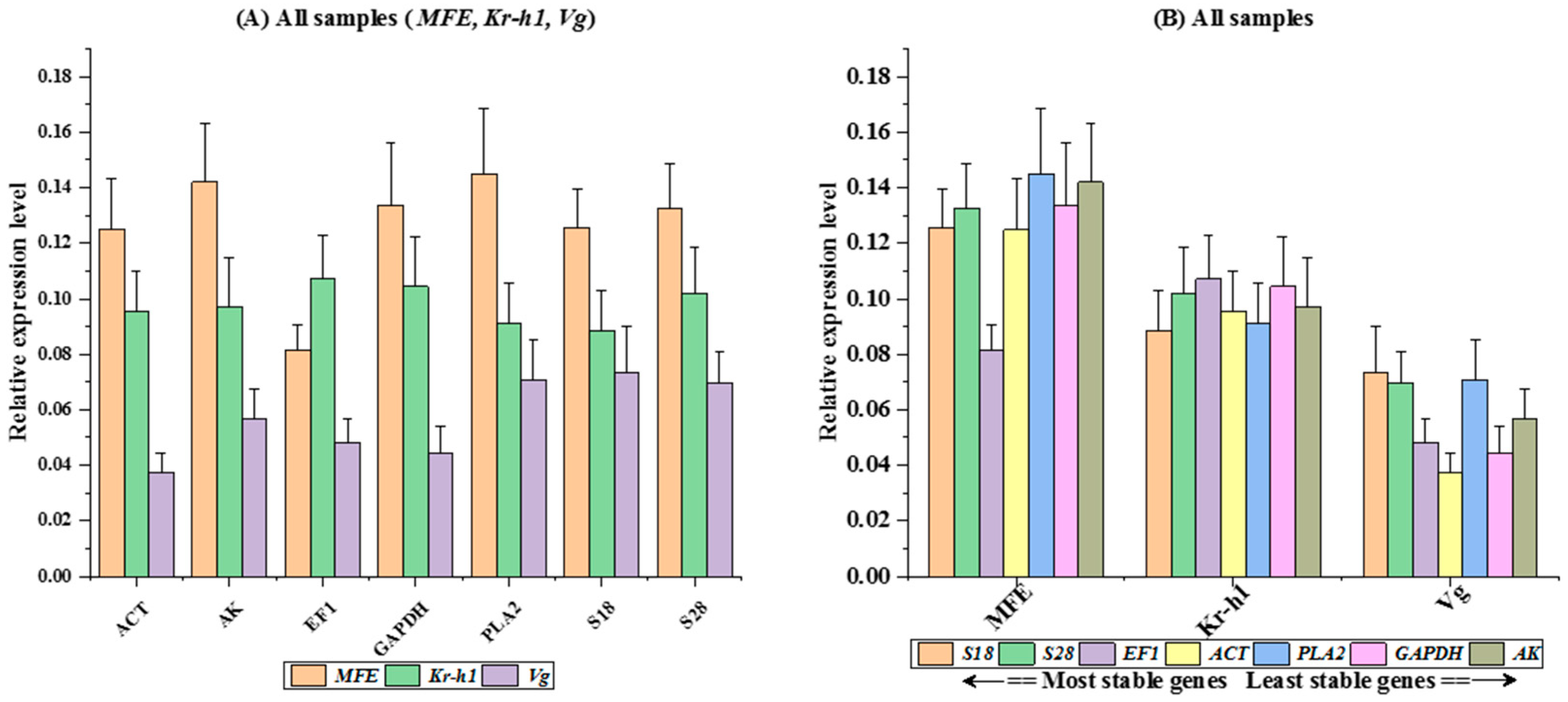
| Rank | geNorm | Norm Finder | Best Keeper | delta-Ct | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | (MV) | Gene | (SV) | Gene | (SD) | Gene | (SD) | Gene | Geomean Ranking Value | |
| Spring | ||||||||||
| 1 | AK | 0.79 | S28 | 0.33 | S28 | 0.29 | S28 | 0.98 | S28 | 1.31 |
| 2 | ACT | 0.79 | S18 | 0.42 | PLA2 | 0.63 | S18 | 1.1 | AK | 2.71 |
| 3 | S28 | 0.86 | AK | 0.78 | ACT | 0.67 | AK | 1.22 | S18 | 2.82 |
| 4 | S18 | 0.94 | PLA2 | 0.9 | S18 | 0.73 | EF1 | 1.26 | ACT | 3.22 |
| 5 | EF1 | 0.99 | EF1 | 0.93 | EF1 | 0.81 | PLA2 | 1.29 | PLA2 | 3.93 |
| 6 | PLA2 | 1.05 | ACT | 1.01 | AK | 0.85 | ACT | 1.3 | EF1 | 4.72 |
| 7 | GAPDH | 1.3 | GAPDH | 1.8 | GAPDH | 1.78 | GAPDH | 1.93 | GAPDH | 7 |
| Summer | ||||||||||
| 1 | ACT | 0.77 | S18 | 0.47 | S18 | 0.32 | S18 | 1.11 | S18 | 1.31 |
| 2 | GAPDH | 0.77 | GAPDH | 0.76 | AK | 0.73 | GAPDH | 1.26 | GAPDH | 1.86 |
| 3 | S18 | 0.91 | ACT | 0.94 | GAPDH | 0.80 | ACT | 1.35 | ACT | 1.81 |
| 4 | PLA2 | 1.00 | PLA2 | 1.00 | PLA2 | 0.95 | PLA2 | 1.41 | PLA2 | 4.00 |
| 5 | AK | 1.10 | EF1 | 1.05 | S28 | 1.03 | EF1 | 1.48 | AK | 4.35 |
| 6 | EF1 | 1.22 | AK | 1.09 | EF1 | 1.07 | AK | 1.49 | EF1 | 5.47 |
| 7 | S28 | 1.45 | S28 | 1.84 | ACT | 0.14 | S28 | 2.02 | S28 | 6.43 |
| Autumn | ||||||||||
| 1 | EF1 | 0.66 | S18 | 0.25 | S18 | 0.46 | S18 | 1.29 | S18 | 1.00 |
| 2 | S18 | 0.66 | PLA2 | 0.75 | EF1 | 0.71 | EF1 | 1.47 | EF1 | 1.86 |
| 3 | PLA2 | 1.02 | EF1 | 0.80 | PLA2 | 0.93 | PLA2 | 1.48 | PLA2 | 2.71 |
| 4 | GAPDH | 1.22 | ACT | 1.13 | ACT | 0.93 | ACT | 1.64 | ACT | 4.42 |
| 5 | S28 | 1.33 | GAPDH | 1.46 | S28 | 0.96 | GAPDH | 1.81 | GAPDH | 4.94 |
| 6 | ACT | 1.47 | S28 | 1.50 | GAPDH | 1.09 | S28 | 1.84 | S28 | 5.47 |
| 7 | AK | 1.67 | AK | 1.99 | AK | 1.56 | AK | 2.18 | AK | 7.00 |
| Winter | ||||||||||
| 1 | S28 | 0.78 | ACT | 0.82 | ACT | 0.45 | ACT | 1.31 | ACT | 1.00 |
| 2 | ACT | 0.78 | S28 | 0.84 | S18 | 0.55 | S28 | 1.31 | S28 | 1.86 |
| 3 | S18 | 0.87 | S18 | 0.92 | S28 | 0.56 | S18 | 1.36 | S18 | 2.71 |
| 4 | AK | 1.00 | EF1 | 1.03 | AK | 0.67 | EF1 | 1.43 | AK | 4.47 |
| 5 | GAPDH | 1.25 | AK | 1.10 | EF1 | 0.86 | AK | 1.47 | EF1 | 4.68 |
| 6 | EF1 | 1.36 | GAPDH | 1.16 | GAPDH | 1.09 | GAPDH | 1.55 | GAPDH | 5.73 |
| 7 | PLA2 | 1.44 | PLA2 | 1.38 | PLA2 | 1.20 | PLA2 | 1.64 | PLA2 | 7.00 |
| Rank | geNorm | Norm Finder | Best Keeper | delta-Ct | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | (MV) | Gene | (SV) | Gene | (SD) | Gene | (SD) | Gene | Geomean Ranking Value | |
| Ovary | ||||||||||
| 1 | S18 | 0.58 | S28 | 0.28 | S18 | 0.27 | S18 | 1.04 | S18 | 1.18 |
| 2 | S28 | 0.58 | S18 | 0.29 | GAPDH | 0.41 | S28 | 1.10 | S28 | 1.56 |
| 3 | GAPDH | 0.68 | GAPDH | 0.30 | S28 | 0.46 | GAPDH | 1.12 | GAPDH | 2.71 |
| 4 | EF1 | 0.96 | ACT | 1.33 | ACT | 0.91 | ACT | 1.62 | ACT | 4.22 |
| 5 | ACT | 1.13 | EF1 | 1.37 | AK | 1.08 | EF1 | 1.62 | EF1 | 5.14 |
| 6 | PLA2 | 1.29 | PLA2 | 1.4 | PLA2 | 1.14 | PLA2 | 1.65 | PLA2 | 6.00 |
| 7 | AK | 1.40 | AK | 1.43 | EF1 | 1.21 | AK | 1.67 | AK | 6.43 |
| Fat body | ||||||||||
| 1 | S18 | 0.29 | ACT | 0.14 | S18 | 0.2 | S18 | 0.77 | S18 | 1.18 |
| 2 | ACT | 0.29 | S18 | 0.14 | ACT | 0.22 | ACT | 0.78 | ACT | 1.41 |
| 3 | AK | 0.53 | AK | 0.58 | AK | 0.35 | AK | 0.89 | AK | 3.00 |
| 4 | PLA2 | 0.56 | PLA2 | 0.76 | PLA2 | 0.45 | PLA2 | 0.97 | PLA2 | 4.00 |
| 5 | GAPDH | 0.67 | EF1 | 0.76 | GAPDH | 0.61 | EF1 | 1.07 | EF1 | 5.47 |
| 6 | EF1 | 0.81 | GAPDH | 0.95 | EF1 | 0.7 | GAPDH | 1.12 | GAPDH | 5.47 |
| 7 | S28 | 1.02 | S28 | 1.47 | S28 | 1.23 | S28 | 1.54 | S28 | 7.00 |
| Thorax | ||||||||||
| 1 | ACT | 0.45 | ACT | 0.22 | S18 | 0.23 | AK | 1.27 | AK | 1.41 |
| 2 | AK | 0.45 | AK | 0.22 | AK | 0.35 | ACT | 1.292 | ACT | 1.56 |
| 3 | S18 | 0.53 | S18 | 0.26 | ACT | 0.39 | S18 | 1.315 | S18 | 2.28 |
| 4 | EF1 | 0.83 | PLA2 | 1.39 | EF1 | 1.14 | EF1 | 1.675 | EF1 | 4.22 |
| 5 | S28 | 1.00 | EF1 | 1.39 | PLA2 | 1.14 | PLA2 | 1.858 | PLA2 | 4.94 |
| 6 | PLA2 | 1.34 | S28 | 1.90 | S28 | 1.49 | S28 | 2.00 | S28 | 5.73 |
| 7 | GAPDH | 1.72 | GAPDH | 2.64 | GAPDH | 2.03 | GAPDH | 2.695 | GAPDH | 7.00 |
| Head | ||||||||||
| 1 | PLA2 | 0.36 | S18 | 0.14 | PLA2 | 0.17 | PLA2 | 1.16 | PLA2 | 1.18 |
| 2 | S18 | 0.36 | PLA2 | 0.18 | S18 | 0.32 | S18 | 1.25 | S18 | 1.41 |
| 3 | GAPDH | 1.06 | EF1 | 0.91 | EF1 | 0.52 | EF1 | 1.45 | EF1 | 3.40 |
| 4 | ACT | 1.10 | AK | 1.35 | AK | 1.21 | GAPDH | 1.65 | GAPDH | 4.35 |
| 5 | EF1 | 1.26 | GAPDH | 1.37 | S28 | 1.27 | ACT | 1.73 | AK | 4.89 |
| 6 | AK | 1.38 | ACT | 1.49 | GAPDH | 1.34 | AK | 1.77 | ACT | 5.38 |
| 7 | S28 | 1.59 | S28 | 2.02 | ACT | 1.43 | S28 | 2.13 | S28 | 6.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sankar, K.; Lee, K.-Y.; Kwak, K.-W.; Lee, S.-J.; Lee, Y.-B. Seasonal Stability Assessment of Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Normalization in Bombus terrestris. Curr. Issues Mol. Biol. 2024, 46, 1335-1347. https://doi.org/10.3390/cimb46020085
Sankar K, Lee K-Y, Kwak K-W, Lee S-J, Lee Y-B. Seasonal Stability Assessment of Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Normalization in Bombus terrestris. Current Issues in Molecular Biology. 2024; 46(2):1335-1347. https://doi.org/10.3390/cimb46020085
Chicago/Turabian StyleSankar, Kathannan, Kyeong-Yong Lee, Kyu-Won Kwak, Su-Jin Lee, and Young-Bo Lee. 2024. "Seasonal Stability Assessment of Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Normalization in Bombus terrestris" Current Issues in Molecular Biology 46, no. 2: 1335-1347. https://doi.org/10.3390/cimb46020085
APA StyleSankar, K., Lee, K.-Y., Kwak, K.-W., Lee, S.-J., & Lee, Y.-B. (2024). Seasonal Stability Assessment of Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Normalization in Bombus terrestris. Current Issues in Molecular Biology, 46(2), 1335-1347. https://doi.org/10.3390/cimb46020085






