Comparative Analysis of Two Soybean Cultivars Revealed Tolerance Mechanisms Underlying Soybean Adaptation to Flooding
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Flooding Treatment
2.2. Determination of Chlorophyll Content
2.3. Chlorophyll Fluorescence Imaging
2.4. Biochemical Indicators
2.5. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.6. Statistical Analysis
3. Results
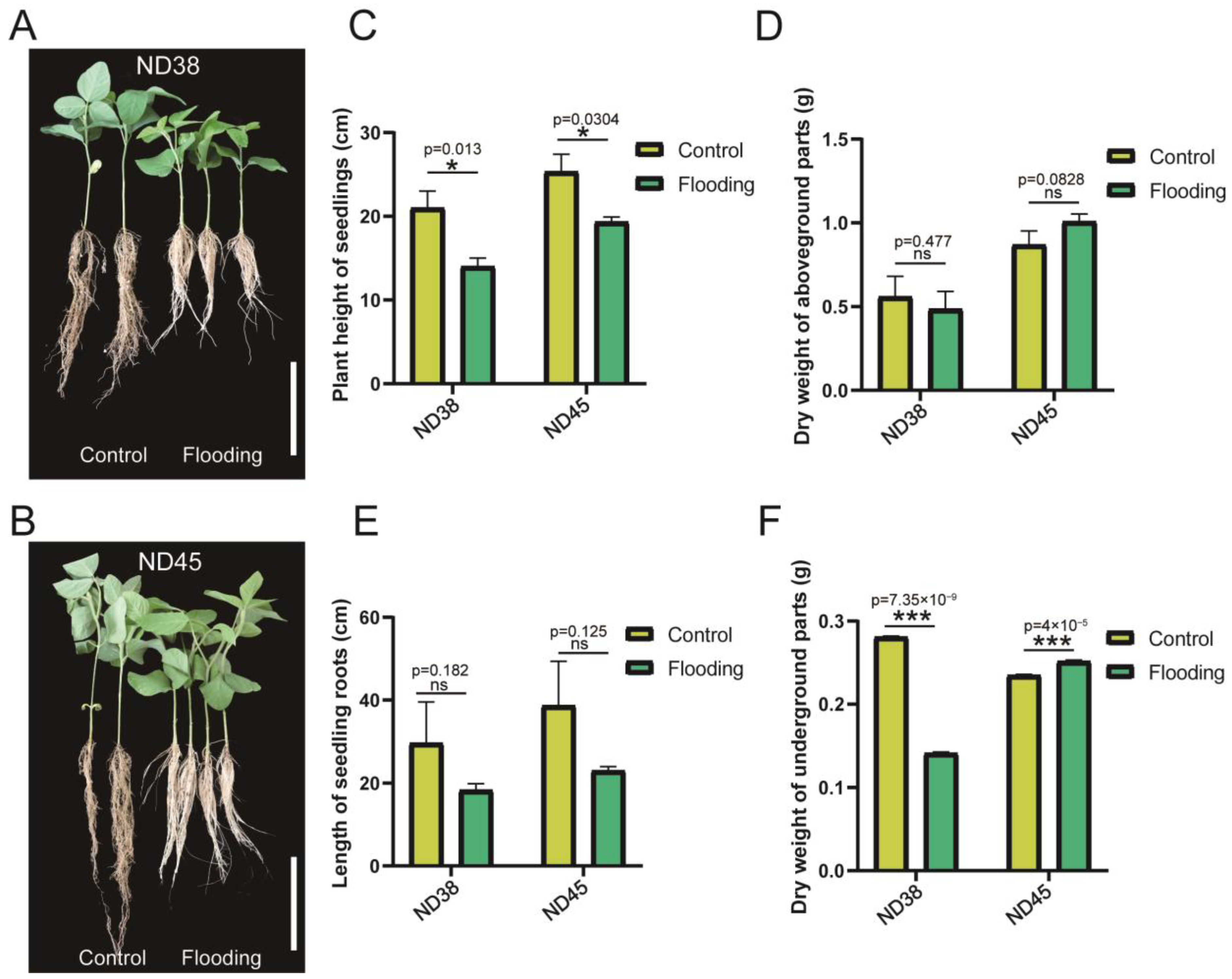
3.1. Flooding Treatment Influenced the Normal Growth of Soybean Plants from Both Cultivars
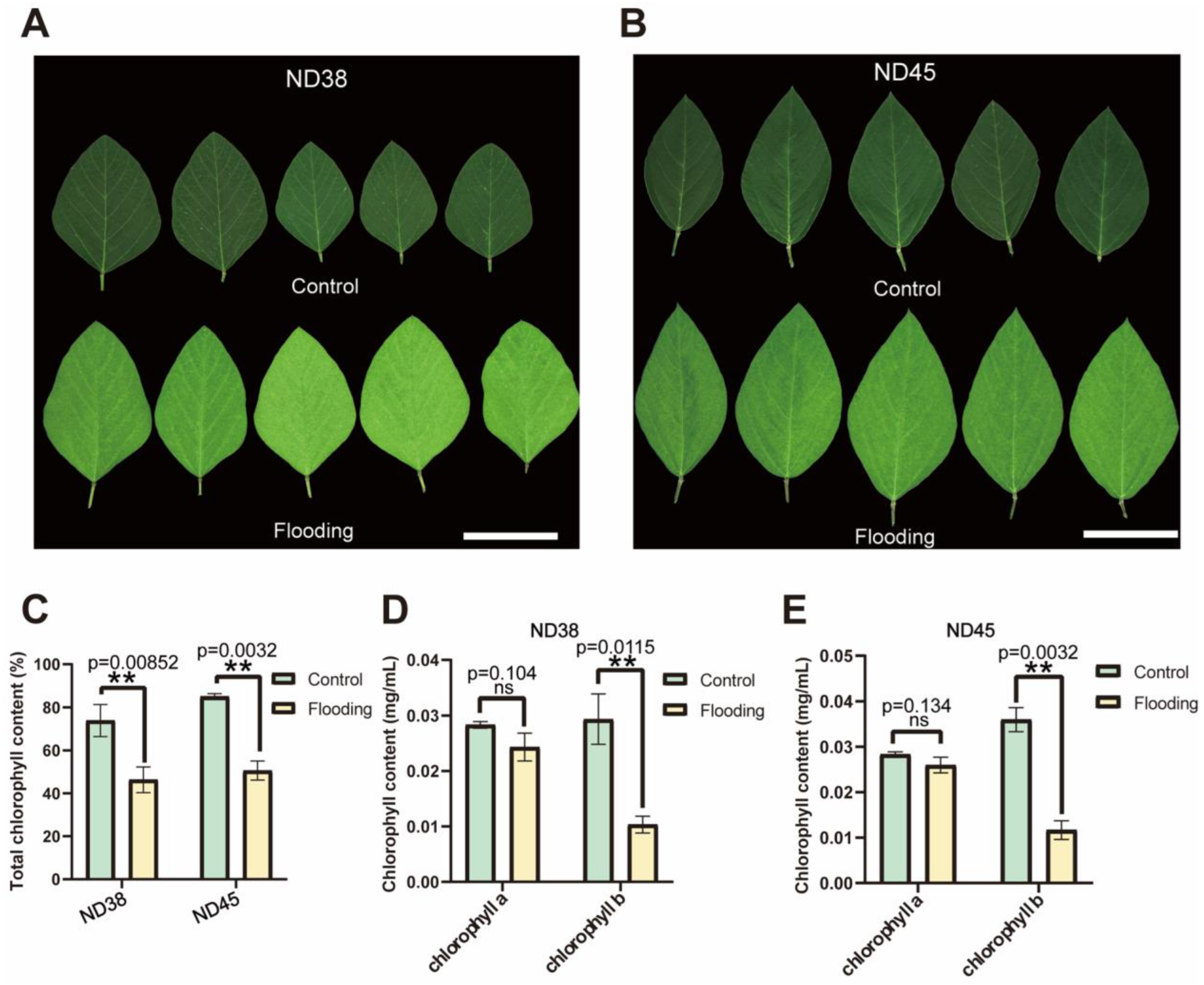
3.2. Flooding Treatment Affected Chlorophyll Metabolism
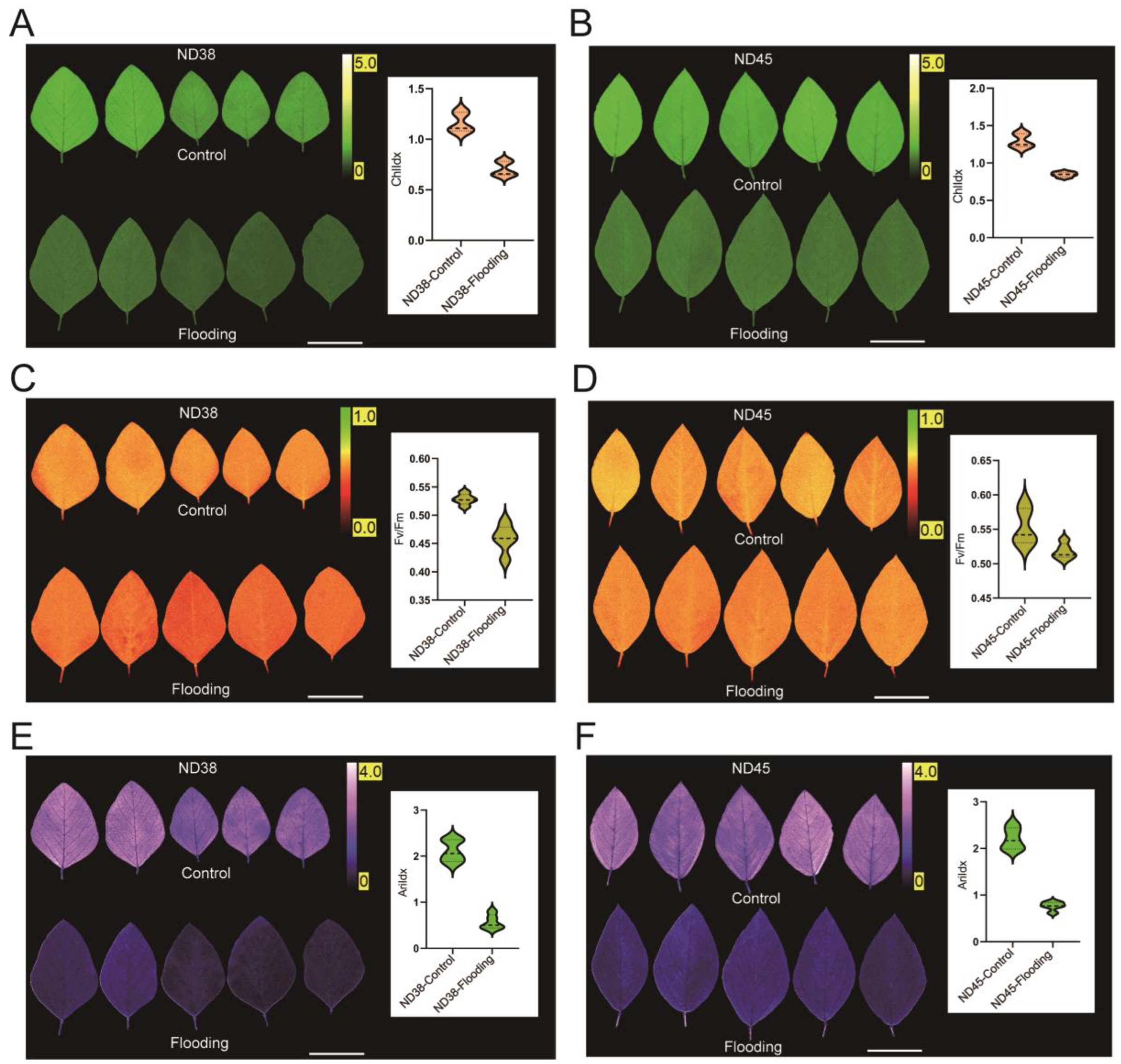
3.3. Flooding Treatment Impacted Photosynthetic Capacity in Soybean Leaves
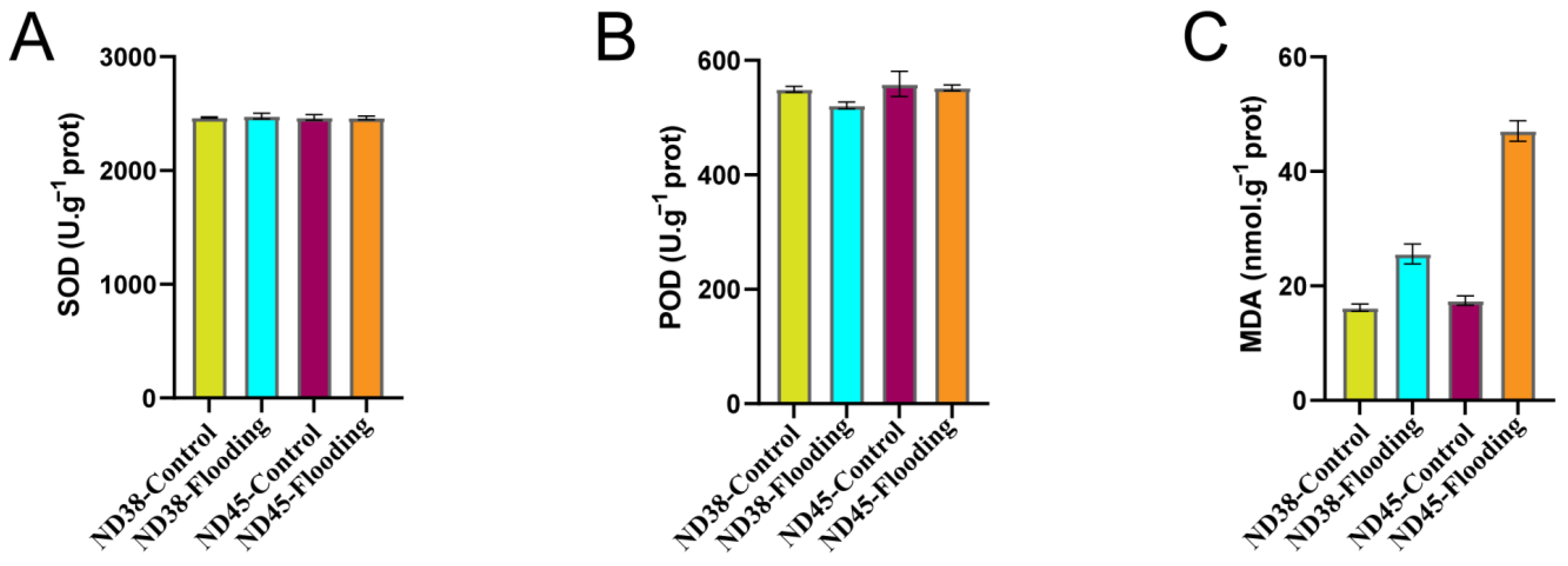
3.4. Flooding Treatment Induced Changes in Key Oxidative Indicators
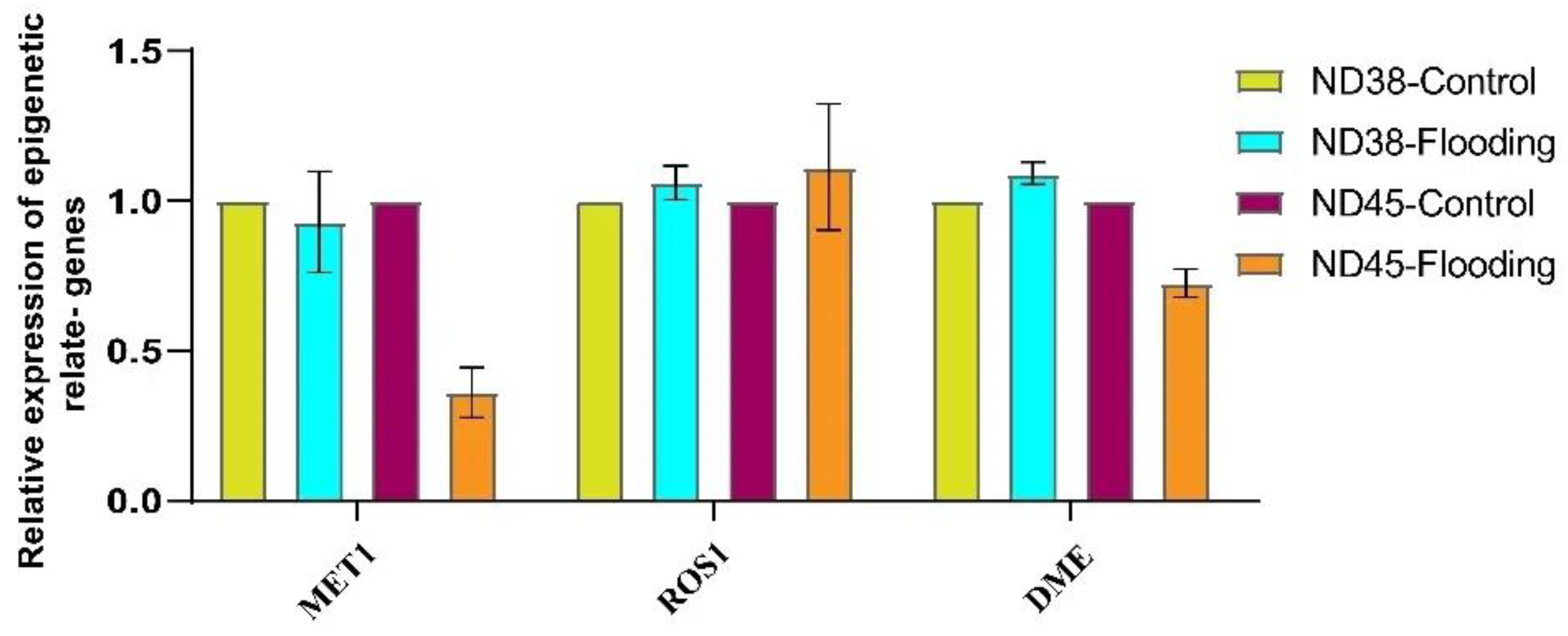
3.5. Flooding Treatment Altered Epigenetic Modifications in Soybean Plants
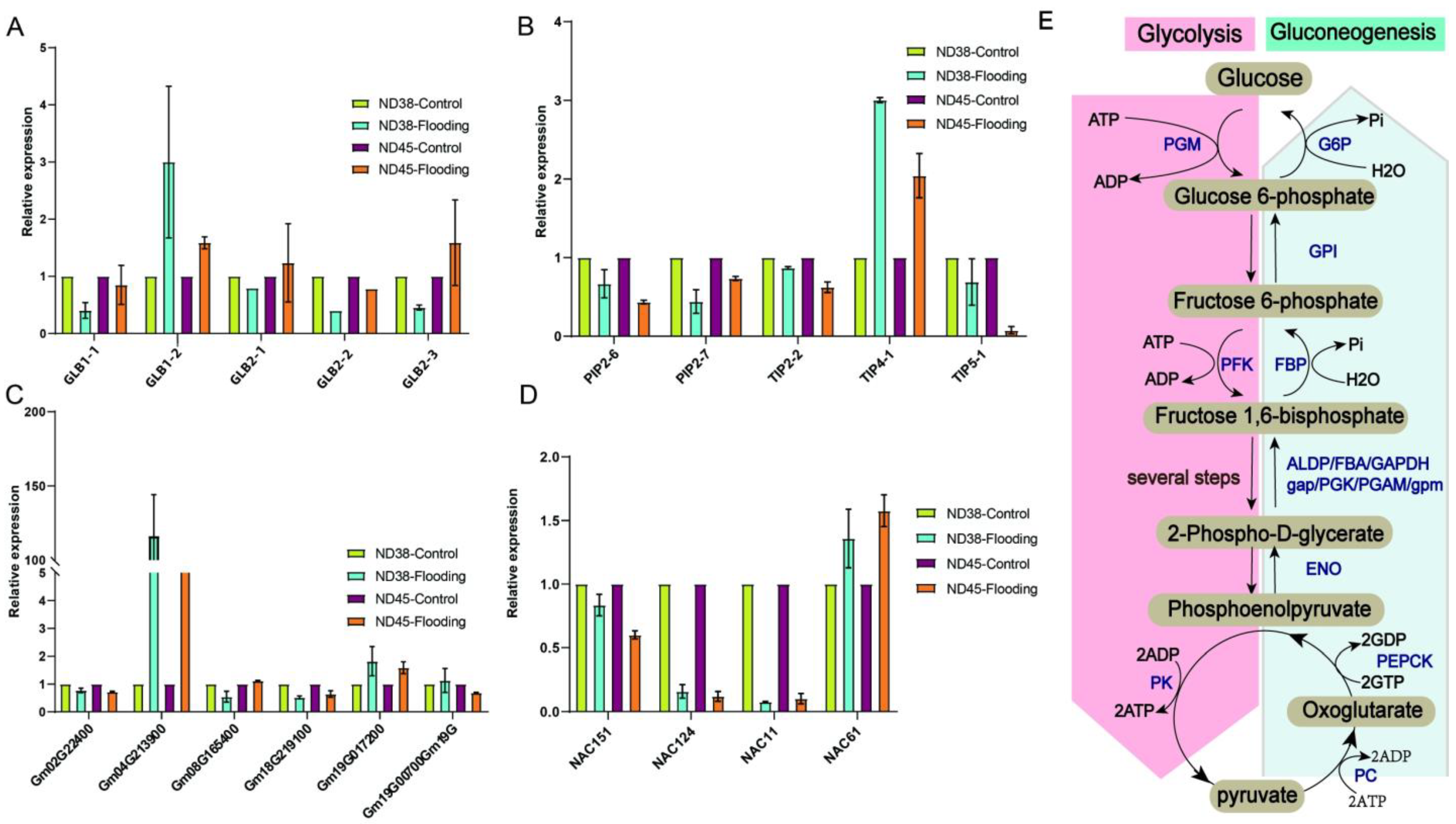
3.6. Flooding Treatment Affected the Expression of Environmental Adaptation Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pazdernik, D.L.; Killam, A.S.; Orf, J.H. Analysis of amino and fatty acid composition in soybean seed, using near infrared reflectance spectroscopy. Agron. J. 1997, 89, 679–685. [Google Scholar] [CrossRef]
- Yin, Y.; Fatufe, A.A.; Blachier, F.; Blachier, F. Soya bean meal and its extensive use in livestock feeding and nutrition. In Soybean and Nutrition; InTech: Houston, TX, USA, 2011; pp. 369–384. [Google Scholar]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for Soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef]
- Nguyen, C.X.; Paddock, K.J.; Zhang, Z.; Stacey, M.G. GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol. 2021, 229, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Bisht, A.; Saini, D.K.; Kaur, B.; Batra, R.; Kaur, S.; Kaur, I.; Jindal, S.; Malik, P.; Sandhu, P.K.; Kaur, A.; et al. Multi-omics assisted breeding for biotic stress resistance in soybean. Mol. Biol. Rep. 2023, 50, 3787–3814. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Lu, S.; Fang, C.; Liu, H.; Dong, L.; Li, H.; Su, T.; Li, S.; Wang, L.; Cheng, Q. Diverse flowering responses subjecting to ambient high temperature in soybean under short-day conditions. Plant Biotechnol. J. 2023, 21, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Nie, T.; Tang, Y.; Lu, D.; Wang, T.; Zhang, Z.; Chen, P.; Li, T.; Meng, L.; Jiao, Y. Responses of Soybean Water Supply and Requirement to Future Climate Conditions in Heilongjiang Province. Agriculture 2022, 12, 1035. [Google Scholar] [CrossRef]
- Wang, Q.; Ning, Z.; Awan, S.A.; Gao, J.; Chen, J.; Lei, Y.; Tan, X.; Wu, X.; Wu, Y.; Liu, C. Far-Red light mediates light energy capture and distribution in soybeans (Glycine max L.) under the shade. Plant Physiol. Biochem. 2023, 204, 108130. [Google Scholar] [CrossRef]
- Liu, M.; Linna, C.; Ma, S.; Ma, Q.; Guo, J.; Wang, F.; Wang, L. Effects of biochar with inorganic and organic fertilizers on agronomic traits and nutrient absorption of soybean and fertility and microbes in purple soil. Front. Plant Sci. 2022, 13, 871021. [Google Scholar] [CrossRef]
- Jumrani, K.; Bhatia, V.S. Impact of combined stress of high temperature and water deficit on growth and seed yield of soybean. Physiol. Mol. Biol. Plants 2018, 24, 37–50. [Google Scholar] [CrossRef]
- Staniak, M.; Szpunar-Krok, E.; Kocira, A. Responses of Soybean to Selected Abiotic Stresses—Photoperiod, Temperature and Water. Agriculture 2023, 13, 146. [Google Scholar] [CrossRef]
- Bagale, S.; Abdelhamid, M. Nutrient Management for Soybean Crops. Int. J. Agron. 2021, 2021, 3304634. [Google Scholar] [CrossRef]
- Eulenstein, F.; Lana, M.; Schlindwein, S.; Sheudzhen, A.; Tauschke, M.; Behrend, A.; Guevara, E.; Meira, S. Trends of soybean yields under climate change scenarios. Horticulturae 2016, 3, 10. [Google Scholar] [CrossRef]
- Zong, C.; Zhao, J.; Wang, Y.; Wang, L.; Chen, Z.; Qi, Y.; Bai, Y.; Li, W.; Wang, W.; Ren, H. Identification of Gene–Allele System Conferring Alkali-Tolerance at Seedling Stage in Northeast China Soybean Germplasm. Int. J. Mol. Sci. 2024, 25, 2963. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wei, W.; Tao, J.J.; Lu, X.; Bian, X.H.; Hu, Y.; Cheng, T.; Yin, C.C.; Zhang, W.K.; Chen, S.Y. Nuclear factor Y subunit GmNFYA competes with GmHDA13 for interaction with GmFVE to positively regulate salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 2362–2379. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Mahmood, A.; Maqbool, R.; Albaqami, M.; Sher, A.; Sattar, A.; Bakhsh, G.; Nawaz, M.; Hassan, M.U.; Al-Yahyai, R. Key insights to develop drought-resilient soybean: A review. J. King Saud. Univ.-Sci. 2022, 34, 102089. [Google Scholar] [CrossRef]
- Wang, X.; Komatsu, S. Proteomic techniques for the development of flood-tolerant soybean. Int. J. Mol. Sci. 2020, 21, 7497. [Google Scholar] [CrossRef]
- Jia, W.; Ma, M.; Chen, J.; Wu, S. Plant morphological, physiological and anatomical adaption to flooding stress and the underlying molecular mechanisms. Int. J. Mol. Sci. 2021, 22, 1088. [Google Scholar] [CrossRef]
- Liu, J.; Li, X.; Li, Y.; Sirisrisakulchai, J.; Kang, X.; Cui, J. Decomposition and Driving Factors of Total Factor Productivity of Food Crops in the Yellow River Basin, China. Agriculture 2024, 14, 547. [Google Scholar] [CrossRef]
- Li, T.; Li, J.; Zhang, D.D. Yellow River flooding during the past two millennia from historical documents. Prog. Phys. Geogr. Earth Environ. 2020, 44, 661–678. [Google Scholar] [CrossRef]
- Wei, K.; Ouyang, C.; Duan, H.; Li, Y.; Chen, M.; Ma, J.; An, H.; Zhou, S. Reflections on the catastrophic 2020 Yangtze River Basin flooding in southern China. Innovation 2020, 1, 100038. [Google Scholar] [CrossRef]
- Khan, M.N.; Ahmed, I.; Ud Din, I.; Noureldeen, A.; Darwish, H.; Khan, M. Proteomic insight into soybean response to flooding stress reveals changes in energy metabolism and cell wall modifications. PLoS ONE 2022, 17, e0264453. [Google Scholar] [CrossRef] [PubMed]
- Kigel, J. Seed germination in arid and semiarid regions. In Seed Development and Germination; Routledge: Oxfordshire, UK, 2017; pp. 645–699. [Google Scholar]
- Kabrick, J.M.; Dey, D.C.; Van Sambeek, J.; Coggeshall, M.V.; Jacobs, D.F. Quantifying flooding effects on hardwood seedling survival and growth for bottomland restoration. New For. 2012, 43, 695–710. [Google Scholar] [CrossRef]
- Purcell, L.C.; Salmeron, M.; Ashlock, L. Soybean growth and development. In Arkansas Soybean Production Handbook; K-State Research and Extension: Manhattan, NY, USA, 2014; Volume 197, pp. 1–8. [Google Scholar]
- Mishra, N.; Jiang, C.; Chen, L.; Paul, A.; Chatterjee, A.; Shen, G. Achieving abiotic stress tolerance in plants through antioxidative defense mechanisms. Front. Plant Sci. 2023, 14, 1110622. [Google Scholar] [CrossRef]
- Du, H.; Shen, X.; Huang, Y.; Huang, M.; Zhang, Z. Overexpression of Vitreoscilla hemoglobin increases waterlogging tolerance in Arabidopsis and maize. BMC Plant Biol. 2016, 16, 35. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wang, X.; Peng, F.; Zhao, Y. Molecular cloning and characterization of a nonsymbiotic hemoglobin gene (GLB1) from Malus hupehensis Rehd. with heterologous expression in tomato. Mol. Biol. Rep. 2012, 39, 8075–8082. [Google Scholar] [CrossRef] [PubMed]
- Koltun, A.; Fuhrmann-Aoyagi, M.B.; Cardoso Moraes, L.A.; Lima Nepomuceno, A.; Simoes Azeredo Goncalves, L.; Mertz-Henning, L.M. Uncovering the roles of hemoglobins in soybean facing water stress. Gene 2022, 810, 146055. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.; Zhu, W.; Zhang, B.; Huang, Q.; Su, X. Waterlogging tolerance and wood properties of transgenic Populus alba × glandulosa expressing Vitreoscilla hemoglobin gene (Vgb). J. For. Res. 2020, 32, 831–839. [Google Scholar] [CrossRef]
- Reddy, P.S.; Rao, T.S.R.B.; Sharma, K.K.; Vadez, V. Genome-wide identification and characterization of the aquaporin gene family in Sorghum bicolor (L.). Plant Gene 2015, 1, 18–28. [Google Scholar] [CrossRef]
- Cheng, X.-F.; Wu, H.-H.; Zou, Y.-N.; Wu, Q.-S.; Kuča, K. Mycorrhizal response strategies of trifoliate orange under well-watered, salt stress, and waterlogging stress by regulating leaf aquaporin expression. Plant Physiol. Biochem. 2021, 162, 27–35. [Google Scholar] [CrossRef]
- Castonguay, Y.; Nadeau, P.; Simard, R. Effects of flooding on carbohydrate and ABA levels in roots and shoots of alfalfa. Plant Cell Environ. 1993, 16, 695–702. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, W.; Hashiguchi, A.; Nishimura, M.; Tian, J.; Komatsu, S. Metabolic profiles of flooding-tolerant mechanism in early-stage soybean responding to initial stress. Plant Mol. Biol. 2017, 94, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Chen, T.; Wang, X.; Cao, J.; Li, X.; Xu, X.; Chen, L.; Xia, Q.; Dong, Y.; Huang, L.; et al. Physiological and Expressional Regulation on Photosynthesis, Starch and Sucrose Metabolism Response to Waterlogging Stress in Peanut. Front. Plant Sci. 2021, 12, 601771. [Google Scholar] [CrossRef]
- Borrego-Benjumea, A.; Carter, A.; Tucker, J.R.; Yao, Z.; Xu, W.; Badea, A. Genome-Wide Analysis of Gene Expression Provides New Insights into Waterlogging Responses in Barley (Hordeum vulgare L.). Plants 2020, 9, 240. [Google Scholar] [CrossRef]
- Rauf, M.; Arif, M.; Fisahn, J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 2013, 25, 4941–4955. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Y.; Li, D.; Wang, Y.; Zhou, R.; Wang, L.; Zhang, Y.; Yu, J.; Gong, H.; You, J.; et al. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum. PLoS ONE 2018, 13, e0199262. [Google Scholar] [CrossRef]
- Venkategowda, R.; Cao, M.; Zheng, L.; Li, J.; Mao, Y.; Zhang, R.; Niu, X.; Geng, M.; Zhang, X.; Huang, W.; et al. Transcriptomic profiling suggests candidate molecular responses to waterlogging in cassava. PLoS ONE 2022, 17, e0261086. [Google Scholar] [CrossRef]
- Bhanu, B.D.; Alluri, A.; Shanker, A.K.; Ulaganathan, K. DNA methylation in plants and its role in abiotic stress tolerance. In Climate Change and Crop Stress Molecules to Ecosystems; Academic Press: Cambridge, MA, USA, 2022; pp. 539–564. [Google Scholar]
- Pan, R.; Xu, Y.H.; Xu, L.; Zhou, M.X.; Jiang, W.; Wang, Q.; Zhang, W.Y. Methylation Changes in Response to Hypoxic Stress in Wheat Regulated by Methyltransferases. Russ. J. Plant Physiol. 2020, 67, 323–333. [Google Scholar] [CrossRef]
- Dossa, K.; Mmadi, M.A.; Zhou, R.; Zhou, Q.; Yang, M.; Cisse, N.; Diouf, D.; Wang, L.H.; Zhang, X.R. The contrasting response to drought and waterlogging is underpinned by divergent DNA methylation programs associated with transcript accumulation in sesame. Plant Sci. 2018, 277, 207–217. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, T.; Liu, C.; Liu, C.; Jiang, Z.; Zhang, Z.; Ali, S.; Li, Z.; Wang, J.; Sun, S. A DNA demethylase reduces seed size by decreasing the DNA methylation of AT-rich transposable elements in soybean. Commun. Biol. 2024, 7, 613. [Google Scholar] [CrossRef]
- Coelho, F.S.; Miranda, S.S.; Moraes, J.L.; Hemerly, A.S.; Ballesteros, H.G.F.; Santa-Catarina, C.; Dos Santos, R.C.; de Almeida, F.A.; Silveira, V.; Macedo, A. DNA methylation impacts soybean early development by modulating hormones and metabolic pathways. Physiol. Plant. 2024, 176, e14492. [Google Scholar] [CrossRef]
- Pinheiro, G.L.; Marques, C.S.; Costa, M.D.; Reis, P.A.; Alves, M.S.; Carvalho, C.M.; Fietto, L.G.; Fontes, E.P. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 2009, 444, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Fleurat-Lessard, P.; Michonneau, P.; Maeshima, M.; Drevon, J.-J.; Serraj, R. The distribution of aquaporin subtypes (PIP1, PIP2 and γ-TIP) is tissue dependent in soybean (Glycine max) root nodules. Ann. Bot. 2005, 96, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Hou, X.; Li, J.; Han, Y.; Zhang, Y.; Feng, N.; Du, J.; Zhang, W.; Zheng, D.; Fang, S. High-resolution DNA methylome reveals that demethylation enhances adaptability to continuous cropping comprehensive stress in soybean. BMC Plant Biol. 2019, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.-J.; Liu, N.; Zhang, G.-W.; Niu, F.-G.; Xu, S.-C.; Gong, Y.-M. Investigation of the AQP family in soybean and the promoter activity of TIP2; 6 in heat stress and hormone responses. Int. J. Mol. Sci. 2019, 20, 262. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Li, W.; Zhang, Y.; Xia, C.; Liu, Y.; Wang, C.; Xu, R.; Zhang, L. Identification of genes/proteins related to submergence tolerance by transcriptome and proteome analyses in soybean. Sci. Rep. 2019, 9, 14688. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.-X.; Zhang, Y.-C.; Chen, P.-L.; Zhang, F.-F.; Li, J.; Yan, F.; Dong, Y.; Feng, B.-L. How Does the Waterlogging Regime Affect Crop Yield? A Global Meta-Analysis. Front. Plant Sci. 2021, 12, 634898. [Google Scholar] [CrossRef]
- Ploschuk, R.A.; Miralles, D.J.; Colmer, T.D.; Ploschuk, E.L.; Striker, G.G. Waterlogging of Winter Crops at Early and Late Stages: Impacts on Leaf Physiology, Growth and Yield. Front. Plant Sci. 2018, 9, 1863. [Google Scholar] [CrossRef]
- de San Celedonio, R.P.; Abeledo, L.G.; Miralles, D.J. Identifying the critical period for waterlogging on yield and its components in wheat and barley. Plant Soil. 2014, 378, 265–277. [Google Scholar] [CrossRef]
- Ma, S.; Hou, J.; Wang, Y.; Wang, M.; Zhang, W.; Fan, Y.; Huang, Z. Post-flowering Soil Waterlogging Curtails Grain Yield Formation by Restricting Assimilates Supplies to Developing Grains. Front. Plant Sci. 2022, 13, 944308. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, C.; Meng, Y.; Liu, X.; Gao, Y.; Liu, Z.; Ma, S. Physiological Mechanism of Waterlogging Stress on Yield of Waxy Maize at the Jointing Stage. Plants 2023, 12, 3034. [Google Scholar] [CrossRef]
- Yin, D.; Sun, D.; Han, Z.; Ni, D.; Norris, A.; Jiang, C.-Z. PhERF2, an ethylene-responsive element binding factor, plays an essential role in waterlogging tolerance of petunia. Hortic. Res. 2019, 6, 83. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef]
- Komatsu, S.; Thibaut, D.; Hiraga, S.; Kato, M.; Chiba, M.; Hashiguchi, A.; Tougou, M.; Shimamura, S.; Yasue, H. Characterization of a novel flooding stress-responsive alcohol dehydrogenase expressed in soybean roots. Plant Mol. Biol. 2011, 77, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Valliyodan, B.; Prince, S.; Wan, J.; Nguyen, H. Characterization of the XTH Gene Family: New Insight to the Roles in Soybean Flooding Tolerance. Int. J. Mol. Sci. 2018, 19, 2705. [Google Scholar] [CrossRef]
- Oliveira, F.K.d.; Da-Silva, C.J.; Garcia, N.; Agualongo, D.A.P.; de Oliveira, A.C.B.; Kanamori, N.; Takasaki, H.; Urano, K.; Shinozaki, K.; Nakashima, K.; et al. The overexpression of NCED results in waterlogging sensitivity in soybean. Plant Stress. 2022, 3, 100047. [Google Scholar] [CrossRef]
- Pan, J.; Sharif, R.; Xu, X.; Chen, X. Mechanisms of Waterlogging Tolerance in Plants: Research Progress and Prospects. Front. Plant Sci. 2020, 11, 627331. [Google Scholar] [CrossRef]
- Yijun, G.; Zhiming, X.; Jianing, G.; Qian, Z.; Rasheed, A.; Hussain, M.I.; Ali, I.; Shuheng, Z.; Hassan, M.U.; Hashem, M.; et al. The intervention of classical and molecular breeding approaches to enhance flooding stress tolerance in soybean—An review. Front. Plant Sci. 2022, 13, 1085368. [Google Scholar] [CrossRef]
- Sathi, K.S.; Masud, A.A.C.; Falguni, M.R.; Ahmed, N.; Rahman, K.; Hasanuzzaman, M.; Nikalje, G. Screening of Soybean Genotypes for Waterlogging Stress Tolerance and Understanding the Physiological Mechanisms. Adv. Agric. 2022, 2022, 5544665. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Epigenetic regulation during salinity and drought stress in plants: Histone modifications and DNA methylation. Plant Gene 2017, 11, 199–204. [Google Scholar] [CrossRef]
- Singroha, G.; Kumar, S.; Gupta, O.P.; Singh, G.P.; Sharma, P. Uncovering the epigenetic marks involved in mediating salt stress tolerance in plants. Front. Genet. 2022, 13, 811732. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Feng, L.; Li, J.; He, Z. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 2015, 6, 267. [Google Scholar] [CrossRef]
- Satyakam; Zinta, G.; Singh, R.K.; Kumar, R. Cold adaptation strategies in plants—An emerging role of epigenetics and antifreeze proteins to engineer cold resilient plants. Front. Genet. 2022, 13, 909007. [Google Scholar] [CrossRef]
- Molinier, J. Genome and epigenome surveillance processes underlying UV exposure in plants. Genes 2017, 8, 316. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kumar, S.; Qian, W. Active DNA demethylation: Mechanism and role in plant development. Plant Cell Rep. 2018, 37, 77–85. [Google Scholar] [CrossRef]
- Bhattarai, K.; Poudel, M.R. DNA methylation and plants response to biotic and abiotic stress. Trends Sci. 2022, 19, 5102. [Google Scholar] [CrossRef]
- Roldán-Arjona, T.; Ariza, R.R. DNA demethylation. In DNA and RNA Modification Enzymes: Structure, Mechanisms, Functions and Evolution; CRC Press: Boca Raton, FL, USA, 2009; pp. 149–161. [Google Scholar]
- De Oliveira, M.R.; Wu, C.; Harrison, D.; Florez-Palacios, L.; Acuna, A.; Da Silva, M.P.; Ravelombola, S.F.; Winter, J.; Rupe, J.; Shakiba, E.; et al. Response to selection to different breeding methods for soybean flood tolerance. Crop Sci. 2022, 62, 648–660. [Google Scholar] [CrossRef]
- Becana, M.; Yruela, I.; Sarath, G.; Catalán, P.; Hargrove, M.S. Plant hemoglobins: A journey from unicellular green algae to vascular plants. New Phytol. 2020, 227, 1618–1635. [Google Scholar] [CrossRef]
- Song, L.; Nguyen, N.; Deshmukh, R.K.; Patil, G.B.; Prince, S.J.; Valliyodan, B.; Mutava, R.; Pike, S.M.; Gassmann, W.; Nguyen, H.T. Soybean TIP Gene Family Analysis and Characterization of GmTIP1;5 and GmTIP2;5 Water Transport Activity. Front. Plant Sci. 2016, 7, 1564. [Google Scholar] [CrossRef]
- Tan, X.; Xu, H.; Khan, S.; Equiza, M.A.; Lee, S.H.; Vaziriyeganeh, M.; Zwiazek, J.J. Plant water transport and aquaporins in oxygen-deprived environments. J. Plant Physiol. 2018, 227, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.-G.; Kong, C.-C.; Yan, K.; Zhang, H.; Luo, Y.-M.; Xie, Z.-H. Elucidation of the molecular responses to waterlogging in Sesbania cannabina roots by transcriptome profiling. Sci. Rep. 2017, 7, 9256. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.J.; Yu, Z.; Xing, G.; Zhao, T.; Gai, J. Establishment of evaluation procedure for soybean seed-flooding tolerance and its application to screening for tolerant germplasm sources. Legume Res.-Int. J. 2017, 41, 34–40. [Google Scholar] [CrossRef]
- Pokhrel, A.; Shrestha, R.; Dangi, S.R. Screening of Soybean Genotypes to Short Period of Flooding. Agron. J. Nepal. 2021, 5, 97–104. [Google Scholar] [CrossRef]
| Gene ID | Forward Primer Sequence | Reverse Primer Sequence | Reference |
|---|---|---|---|
| GmDMEs | AATCCAACTGGGCATCCAGG | TGAACTTGGGGCTTGTGTGT | [43] |
| GmMET1 | ACGCTGAGAAGACAACCACA | CACTTGCAGTTGCATGGGTC | [44] |
| GmNAC11 | TTGCCACCCGGTTTTAGGTT | CTGCAATGATGGAAACGGGC | [45] |
| GmNAC61 | CCTCAGAAGGTTCCAAGTGC | TCCTGCAGATAGCCCAAGAT | |
| GmNAC124 | TCCATACCCTTGACCGTTTC | CCTTGCACCATTTGGGTACT | |
| GmNAC151 | TCTGTCGAAGCTGAAGACGA | TGGTCTTCCAGTAGCCCCCTA | |
| GmPIP2-6 | CTGGAACCGGCATTAACCCT | GCTCCAACAAACGGTCCAAC | [46] |
| GmPIP2-7 | TTTCTGGCGAGGAAGGTGTC | CCAAACCGGTTCCTTTGCTG | |
| GmROS1 | ACCATCAAAGAACGGGGCAT | TGCAGTGTTAAAAGCCGCAC | [47] |
| GmTIP2-2 | TTTGTAGGTGTCTCCGTCGC | ATGCTCTTGGCGGTGATGAA | [48] |
| GmTIP4-1 | CATCTTTCGTTCCCTGCTCT | AGTTGCCTGTCCTCCAGAGA | |
| GmTIP5-1 | CCTCACGGAAGTTGATGCCT | CCATTGCAAAGGTCACAGCC | |
| Gm02G222400 | TGGGAAGAAGCCATGGTCAC | TCTGAGGCACCATCAGCAAG | [49] |
| Gm04G213900 | AGCTGGAAAGCCATTGGTGA | AAAGGTGTCTGTCCCTTGGC | |
| Gm08G165400 | ACGTGAATGATGCTTTCGGC | GAATCCTGCAACAGAGGGCT | |
| Gm18G219100 | GTGGAGATTGGTCAGGGCTC | AAATATGCGACCAGCCCCAA | |
| Gm19G000700 | AGTCCATTGGAGAGCCTTGC | CTTAGCTGTAGACCCGCCAC | |
| Gm19G017200 | CAACCAGATGCCCTTGCCTA | GAAGGTCGGTTGCCTGAGAA | |
| GmGLB1-1 | AGCAGTGCCTGAAATGTGGT | GATTTGATGGCTTCGGCCAG | [29] |
| GmGLB1-2 | CCATGCCGTGTCTGTCTTTG | ACGCCGGTTCTAAAATGGGT | |
| GmGLB2-1 | CCGCACTTGGTTCTATCCAT | GCTGCTGCCAATTCATCATA | |
| GmGLB2-2 | TGATGCCACACTTGGTCCTA | GCCATTGCCTTCTTAATTGC | |
| GmGLB2-3 | ACCTGCAGCAAAGGACTTGT | GCTTTTTGGGCATGGATAGA | |
| GmACT11 | GGTGGTTCTATCTTGGCATC | CTTTCGCTTCAATAACCCTA | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; An, J.; Liang, J.; Yang, W.; Zeng, Z.; Zhang, M.; Wu, H.; Liu, S.; Cao, X. Comparative Analysis of Two Soybean Cultivars Revealed Tolerance Mechanisms Underlying Soybean Adaptation to Flooding. Curr. Issues Mol. Biol. 2024, 46, 12442-12456. https://doi.org/10.3390/cimb46110739
Yu X, An J, Liang J, Yang W, Zeng Z, Zhang M, Wu H, Liu S, Cao X. Comparative Analysis of Two Soybean Cultivars Revealed Tolerance Mechanisms Underlying Soybean Adaptation to Flooding. Current Issues in Molecular Biology. 2024; 46(11):12442-12456. https://doi.org/10.3390/cimb46110739
Chicago/Turabian StyleYu, Xiaobo, Jiangang An, Jianqiu Liang, Wenying Yang, Zhaoqiong Zeng, Mingrong Zhang, Haiying Wu, Sichen Liu, and Xiaoning Cao. 2024. "Comparative Analysis of Two Soybean Cultivars Revealed Tolerance Mechanisms Underlying Soybean Adaptation to Flooding" Current Issues in Molecular Biology 46, no. 11: 12442-12456. https://doi.org/10.3390/cimb46110739
APA StyleYu, X., An, J., Liang, J., Yang, W., Zeng, Z., Zhang, M., Wu, H., Liu, S., & Cao, X. (2024). Comparative Analysis of Two Soybean Cultivars Revealed Tolerance Mechanisms Underlying Soybean Adaptation to Flooding. Current Issues in Molecular Biology, 46(11), 12442-12456. https://doi.org/10.3390/cimb46110739




