Integrative Taxonomy Reveals New Insights into the Species Validity of the Neocaridina davidi-N. denticulata-N. heteropoda Complex and Mitogenomic Phylogeny of Caridean Shrimps
Abstract
1. Introduction
2. Materials and Methods
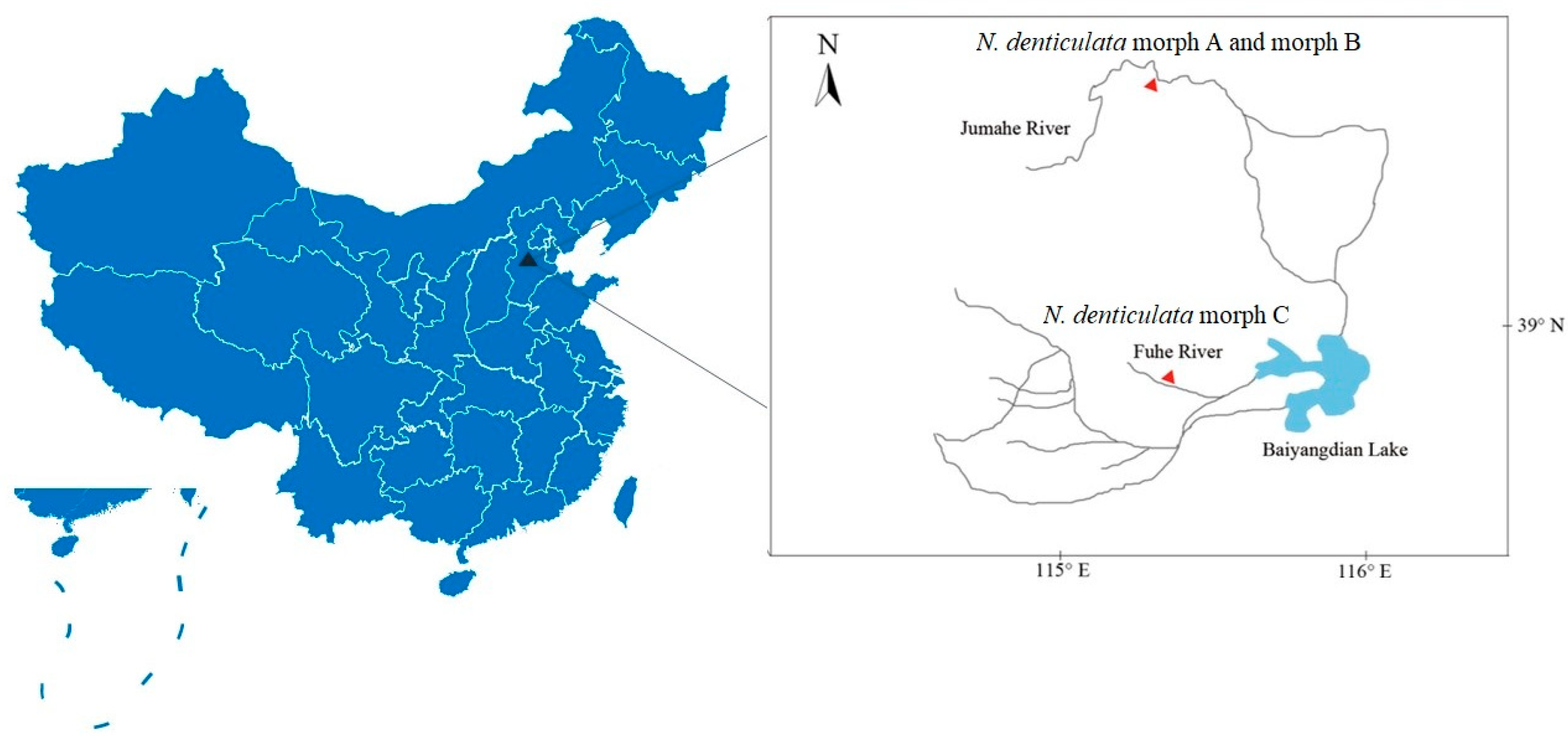
2.1. Specimen Collection and Morphological Examination
2.2. Mitogenome Assembly and Annotation
2.3. Phylogenetic Analysis
2.4. Gene Order Analysis
3. Results
3.1. Morphological Classification
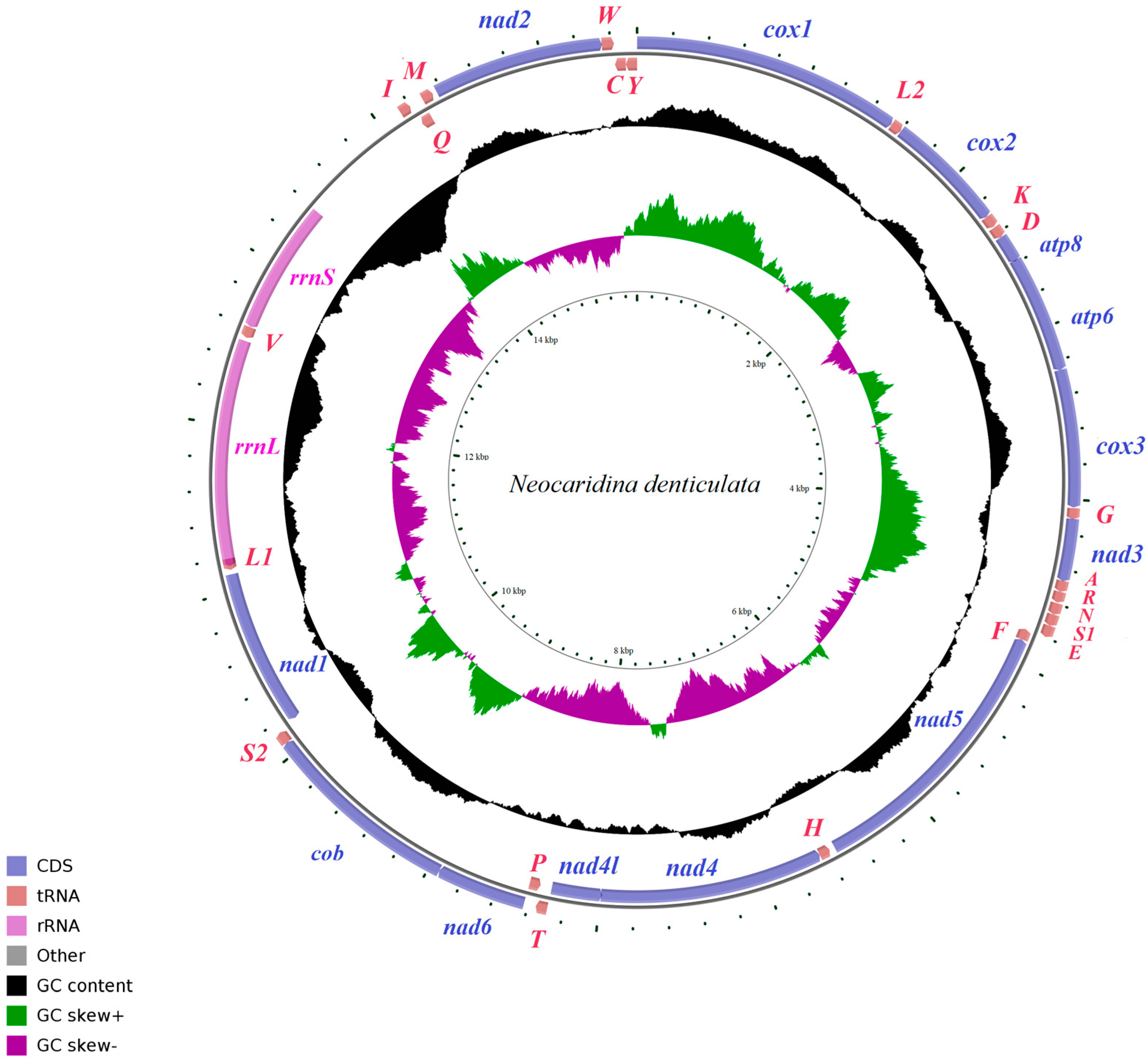
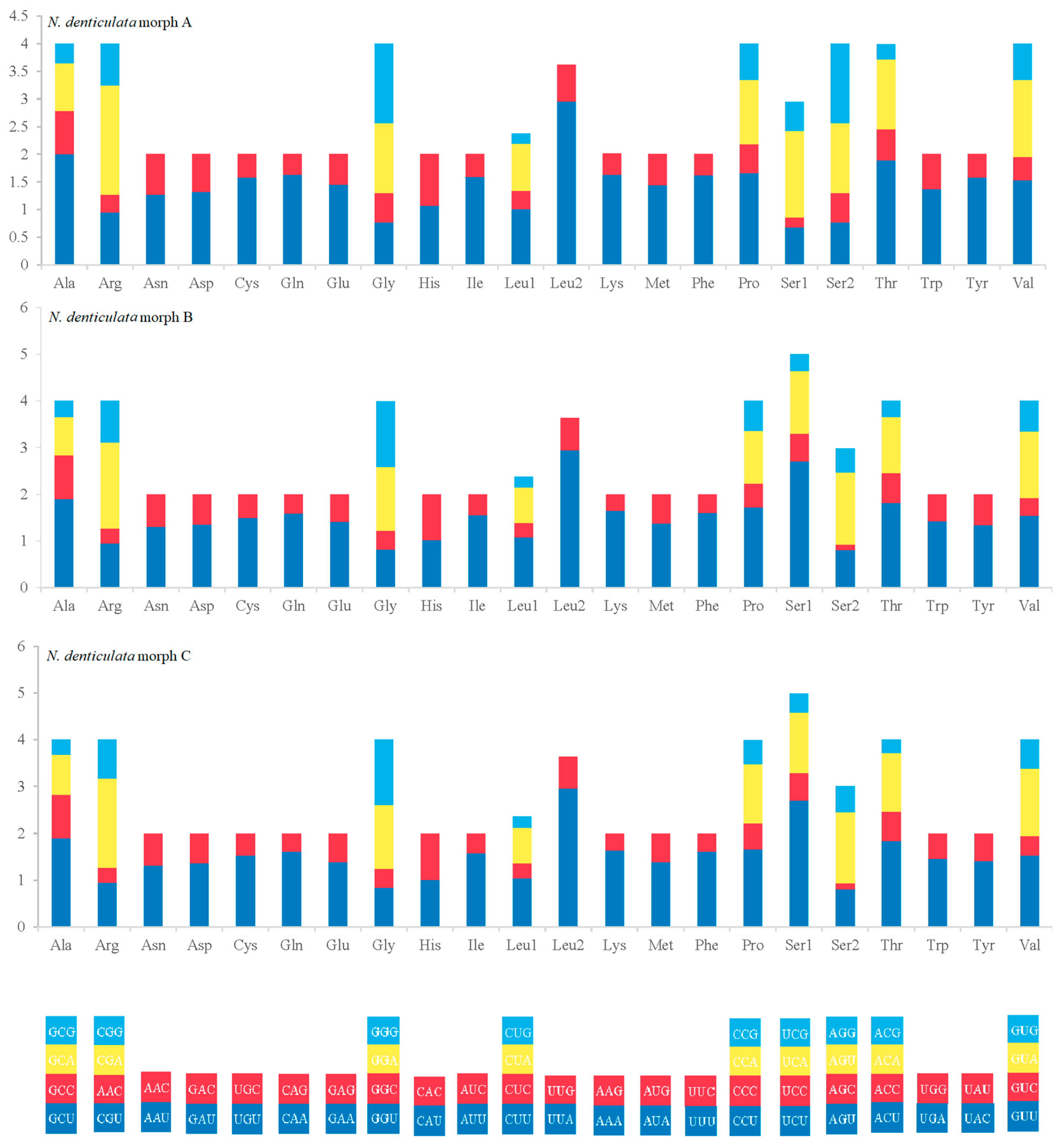
3.2. Organization and Characterization of Mitogenomes
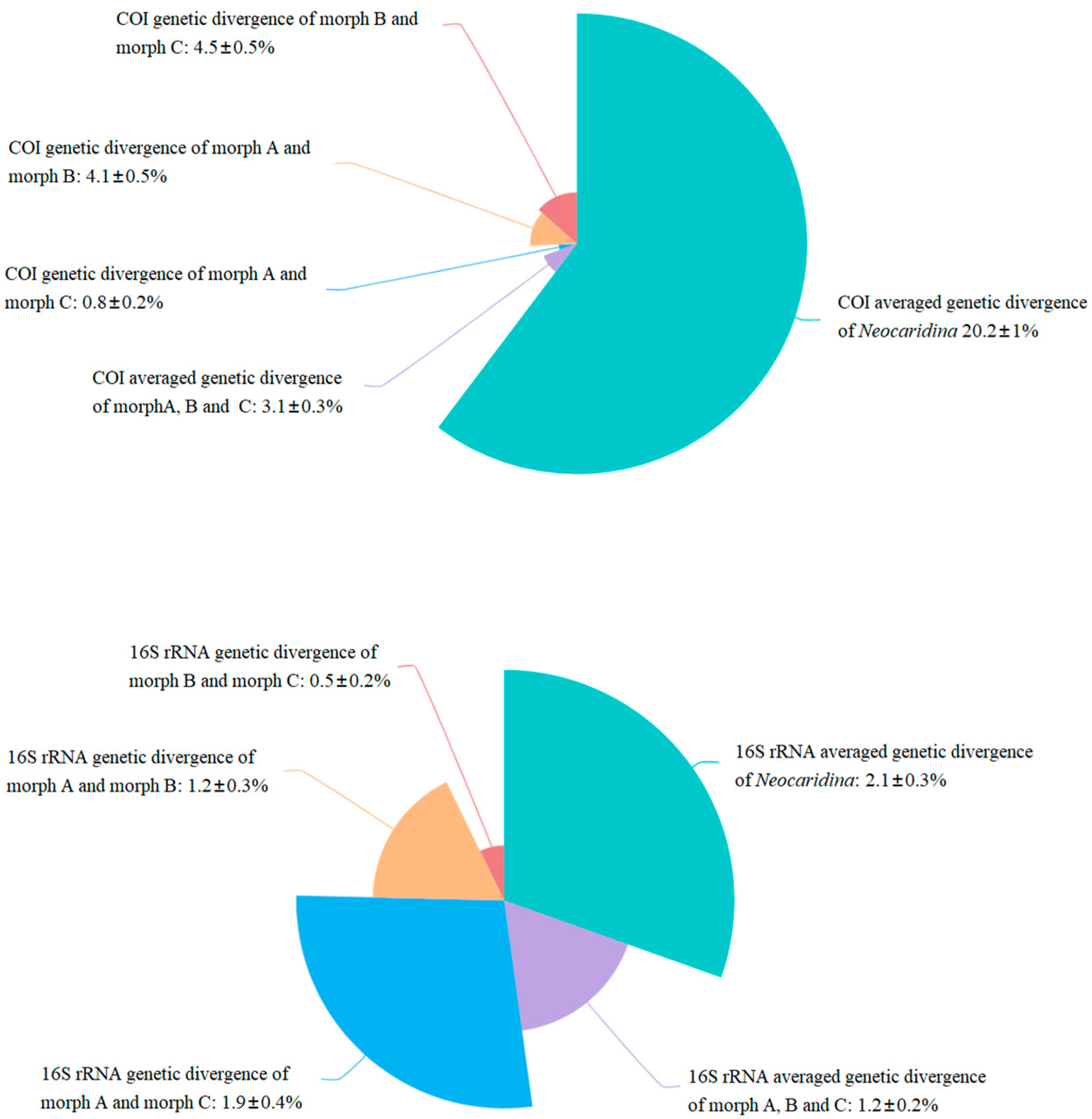
3.3. Genetic Distances
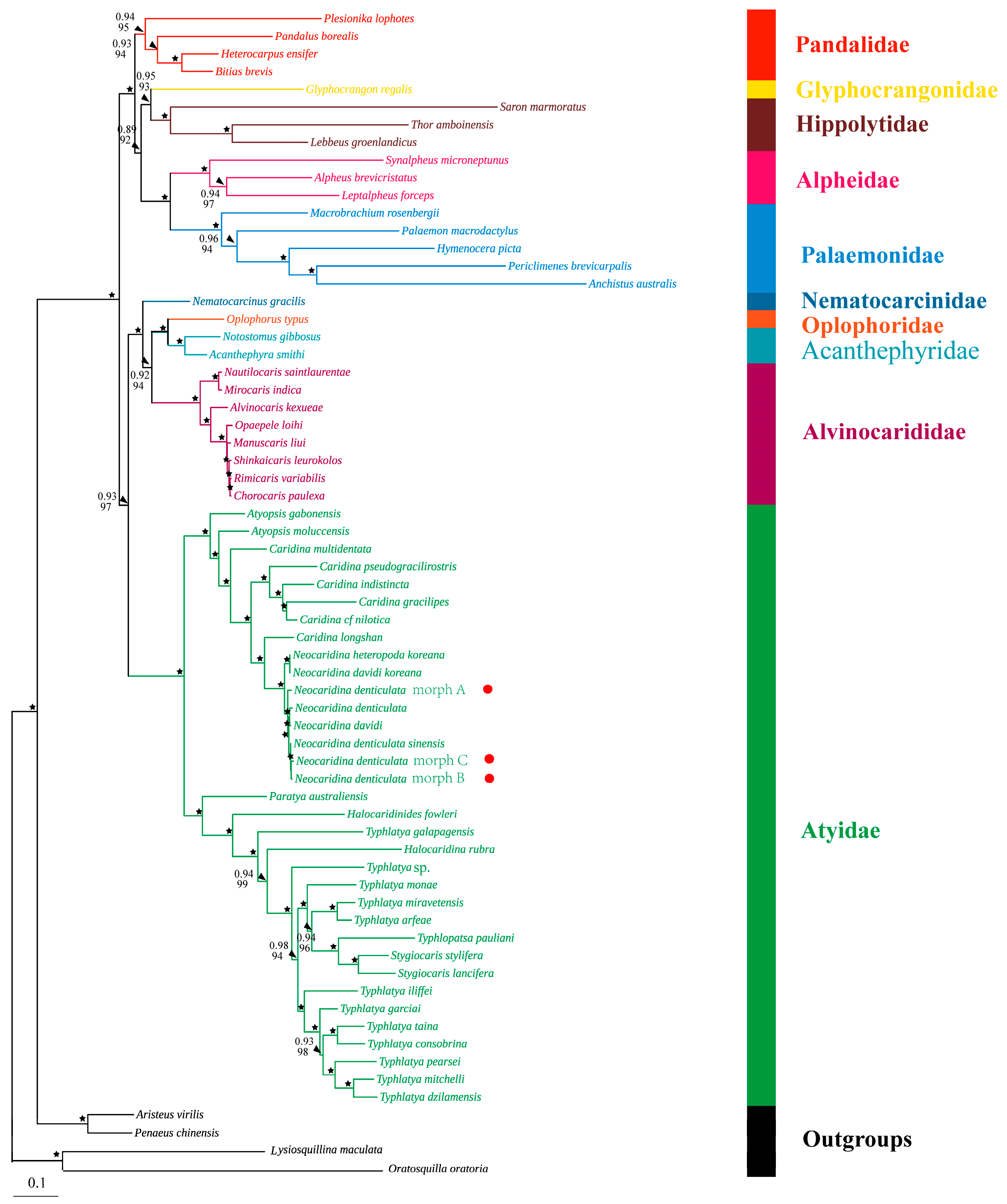
3.4. Phylogenetic Analysis
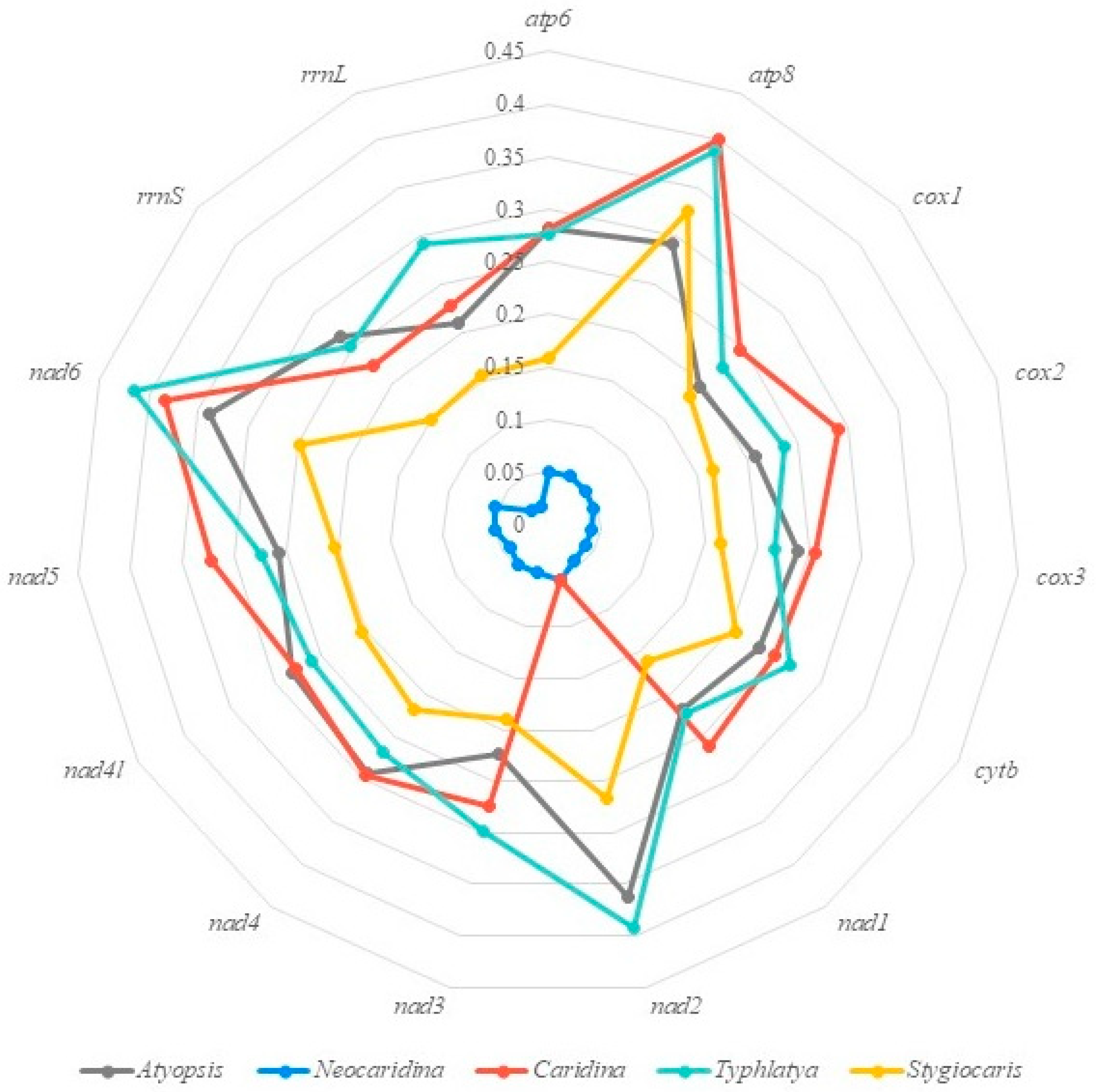
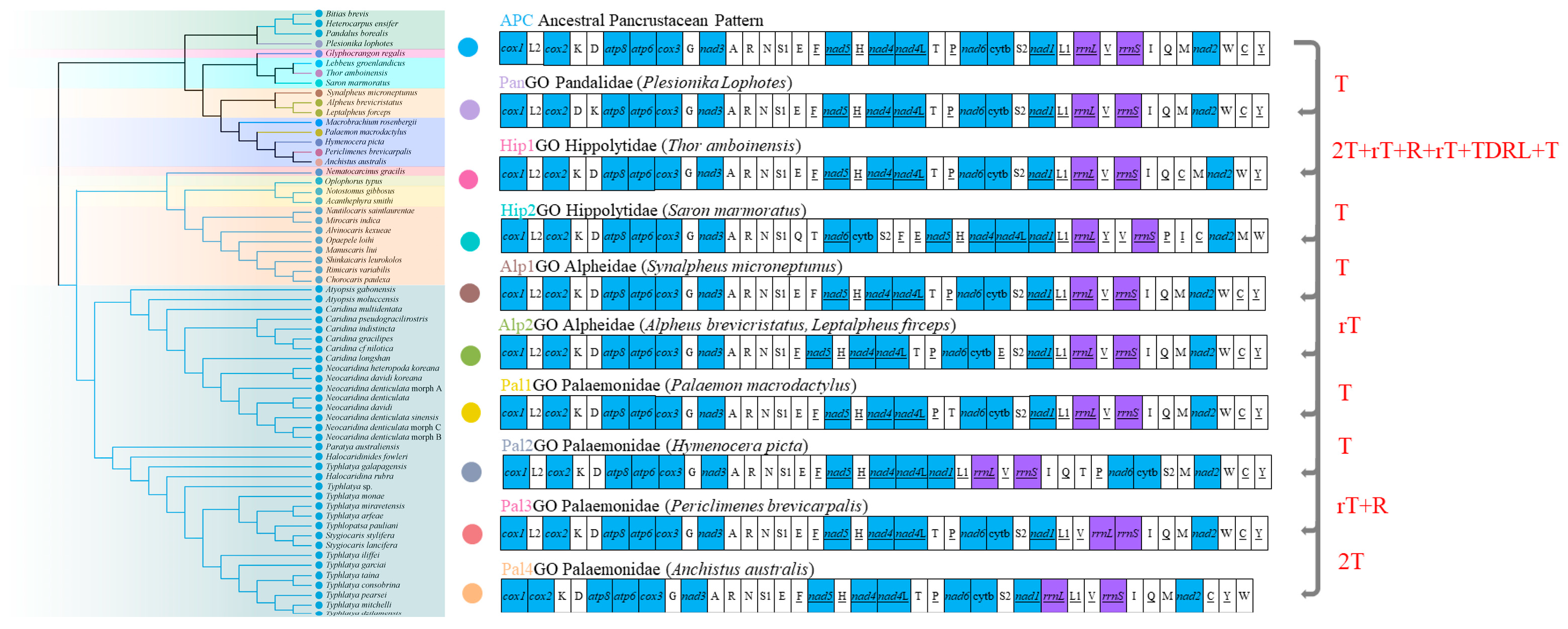
3.5. Mitochondrial Gene Order and Rearrangements
4. Discussion
4.1. Morphological Differences
4.2. Genetic Distances and Species Validity
4.3. Mitogenomic Phylogeny
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Grave, S.; Smith, K.G.; Adeler, N.A.; Allen, D.J.; Alvarez, F.; Anker, A.; Cai, Y.; Carrizo, S.F.; Klotz, W.; Mantelatto, F.L.; et al. Dead shrimp blues: A global assessment of extinction risk in freshwater shrimps (Crustacea: Decapoda: Caridea). PLoS ONE 2015, 10, e0120198. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.E.; Grave, S.D.; Daniels, S.R. Comparative evolutionary study reveals radically different scales of genetic structuring within two atyid shrimp species (Crustacea: Decapoda: Atyidae). Zool. J. Linn. Soc. 2019, 186, 200–212. [Google Scholar] [CrossRef]
- Liang, X.Q. Fauna Sinica. Invertebrata: Crustacea: Decapoda: Atyidae; Science Press: Beijing, China, 2004; pp. 1–375. [Google Scholar]
- Shih, H.T.; Cai, Y.; Niwa, N.; Nakahara, Y. A new species of land-locked freshwater shrimp of the genus Neocaridina (Decapoda: Caridea: Atyidae) from Iki Island, Kyushu. Japan. Zool. Stud. 2017, 56, 30. [Google Scholar] [CrossRef]
- Snyder, M.N.; Small, G.E.; Pringle, C.M. Diet-switching by omnivorous freshwater shrimp diminishes differences in nutrient recycling rates and body stoichiometry across a food quality gradient. Freshwater Biol. 2015, 60, 526–536. [Google Scholar] [CrossRef]
- Tang, C.H.; Chen, W.Y.; Wu, C.C.; Lu, E.; Shih, W.Y.; Chen, J.W.; Tsai, J.W. Ecosystem metabolism regulates seasonal bioaccumulation of metals in atyid shrimp (Neocaridina denticulata) in a tropical brackish wetland. Aquat. Toxicol. 2020, 225, 105522. [Google Scholar] [CrossRef]
- Pantaleão, J.A.F.; Gregati, R.A.; Costa, R.C.; López-Greco, L.S.; Negreiros-Fransozo, M.L. Post-hatching development of the ornamental ‘red cherry shrimp’ Neocaridina davidi (Bouvier, 1904) (Crustacea, Caridea, Atyidae) under laboratorial conditions. Aquac. Res. 2017, 48, 553–569. [Google Scholar] [CrossRef]
- Cai, Y. A revision of the genus Neocaridina (Crustacea, Decapoda, Atyidae). Acta Zootaxonomica Sin. 1996, 21, 129–160. [Google Scholar]
- Shih, H.T.; Cai, Y. Two new species of the land-locked freshwater shrimps genus, Neocaridina Kubo, 1938 (Decapoda: Caridea: Atyidae), from Taiwan, with notes on speciation on the island. Zool. Stud. 2007, 46, 680–694. [Google Scholar]
- Klotz, W.; Miesen, F.W.; Hüllen, S.; Herder, F. Two Asian fresh water shrimp species found in a thermally polluted stream system in north Rhine-Westphalia, Germany. Aquat. Invasions 2013, 8, 333–339. [Google Scholar] [CrossRef]
- Hung, M.S.; Chan, T.Y.; Yu, H.P. Atyid Shrimps (Decapoda: Caridea) of Taiwan, with descriptions of three new species. J. Crustac. Biol. 1993, 13, 481–503. [Google Scholar] [CrossRef]
- Englund, R.A.; Cai, Y. The occurrence and description of Neocaridina denticulata sinensis (Kemp, 1918) (Crustacea: Decapoda: Atyidae), a new introduction to the Hawaiian Islands. Bishop Mus. Occas. Pap. 1999, 58, 58–65. [Google Scholar]
- Liang, X.Q. On new species of atyid shrimps (Decapoda, Caridea) from China. Oceanol. Limnol. Sin. 2002, 33, 167–173. [Google Scholar]
- De Grave, S.; Fransen, C.H.J.M. Carideorum Catalogus: The Recent Species of the Dendrobranchiate, Stenopodidean, Procarididean and Caridean Shrimps (Crustacea: Decapoda). Zool. Meded. Leiden 2011, 85, 195–588. [Google Scholar]
- Han, C.C.; Hsu, K.C.; Fang, L.S.; Cheng, I.M.; Lin, H.D. Geographical and temporal origins of Neocaridina species (Decapoda: Caridea: Atyidae) in Taiwan. BMC Genet. 2019, 20, 86. [Google Scholar] [CrossRef]
- Levitt-Barmats, Y.; Yanai, Z.; Magory, C.T.; Shenkar, N. Life-history traits and ecological characteristics of the ornamental shrimp Neocaridina denticulata (De Haan, 1844), recently introduced into the freshwater systems of Israel. Aquat. Invasions 2019, 14, 684–702. [Google Scholar] [CrossRef]
- Bracken, H.D.; De Grave, S.; Felder, D.L. Decapod Crustacean Phylogenetics: Phylogeny of the Infraorder Caridea Based on Mitochondrial and Nuclear Genes (Crustacea: Decapoda); Taylor and Francis/CRC Press: Boca Raton, FL, USA, 2009; pp. 281–305. [Google Scholar]
- Tan, M.H.; Gan, H.M.; Lee, Y.P.; Poore, G.C.; Austin, C.M. Digging deeper: New gene order rearrangements and distinct patterns of codons usage in mitochondrial genomes among shrimps from the Axiidea, Gebiidea and Caridea (Crustacea: Decapoda). PeerJ 2017, 5, e2982. [Google Scholar] [CrossRef]
- Sun, Y.M.; Chen, J.; Ye, Y.Y.; Xu, K.D.; Li, J.J. Comparison of mitochondrial Genome Sequences between Two Palaemon Species of the Family Palaemonidae (Decapoda: Caridea): Gene Rearrangement and Phylogenetic Implications. Genes 2023, 14, 1499. [Google Scholar] [CrossRef]
- Li, C.P.; De Grave, S.; Chan, T.Y.; Lei, H.C.; Chu, K.H. Molecular systematics of caridean shrimps based on five nuclear genes: Implications for superfamily classification. Zool. Anz. 2011, 250, 270–279. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Z.Q.; Tang, D.; Xu, X.Y.; Tao, Y.T.; Ji, C.Y.; Wang, Z.F. Characterization and comparison of the mitochondrial genomes from two Alpheidae species and insights into the phylogeny of Caridea. Genomics 2020, 112, 65–70. [Google Scholar] [CrossRef]
- Ye, Y.Y.; Miao, J.; Guo, Y.H.; Gong, L.; Guo, B.Y. The first mitochondrial genome of the genus Exhippolysmata (Decapoda: Caridea: Lysmatidae), with gene rearrangements and phylogenetic associations in Caridea. Sci. Rep. 2021, 11, 14446. [Google Scholar] [CrossRef]
- Helfenbein, K.G.; Fourcade, H.M.; Vanjani, R.G.; Boore, J.L. The mitochondrial genome of Paraspadella gotoi is highlyreduced and reveals that chaetognaths are a sister group to protostomes. Proc. Natl. Acad. Sci. USA 2004, 101, 10639–10643. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Gong, L.; Sui, J.X.; Li, X.Z. The complete mitochondrial genome of Calyptogena marissinica (Heterodonta: Veneroida: Vesicomyidae): Insight into the deep-sea adaptive evolution of vesicomyids. PLoS ONE 2019, 14, e0217952. [Google Scholar] [CrossRef]
- Kou, Q.; Xu, P.; Poore, G.C.B.; Li, X.Z.; Wang, C.S. A new species of the deep-sea sponge-associated genus Eiconaxius (Crustacea: Decapoda: Axiidae), with new insights into the distribution, speciation, and mitogenomic phylogeny of axiidean shrimps. Front. Mar. Sci. 2020, 7, 469. [Google Scholar] [CrossRef]
- Li, M.; Chen, W.T.; Zhang, Q.L.; Liu, M.; Xing, C.W.; Cao, Y.; Luo, F.Z.; Yuan, M.L. Mitochondrial phylogenomics provides insights into the phylogeny and evolution of spiders (Arthropoda: Araneae). Zool. Res. 2011, 43, 566–584. [Google Scholar] [CrossRef]
- Yu, Y.; Kong, L.F.; Li, Q. Mitogenomic phylogeny of Muricidae (Gastropoda: Neogastropoda). Zool. Scr. 2023, 52, 413–425. [Google Scholar] [CrossRef]
- Boore, J.L.; Brown, W.M. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]
- Tan, M.H.; Gan, H.M.; Lee, Y.P.; Lintona, S.; Grandjeand, F.; BartholomeiSantos, M.L.; Miller, A.D.; Austin, C.M. ORDER Within the chaos: Insights into phylogenetic relationships within the Anomura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Mol. Phylogenet. Evol. 2018, 127, 320–331. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, J.; Xu, T.; Qiu, J.W.; Qian, P.Y. Phylogenetic relationships and adaptation in deep-sea mussels: Insights from mitochondrial genomes. Int. J. Mol. Sci. 2021, 22, 1900. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, L.; Lu, X.; Jiang, L.; Liu, B.; Liu, L.; Lü, Z.; Li, P.; Zhang, X. Gene rearrangements in the mitochondrial genome of Chiromantes eulimene (Brachyura: Sesarmidae) and phylogenetic implications for Brachyura. Int. J. Biol. Macromol. 2021, 162, 704–714. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; Depamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2007, 24, 172–175. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.Y.; Gao, F.L.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3. 2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Bernt, M.; Merkle, D.; Ramsch, K.; Fritzsch, G.; Perseke, M.; Bernhard, D.; Schlegel, M.; Stadler, P.F.; Middendorf, M. CREx: Inferring Genomic Rearrangements Based on Common Intervals. Bioinformatics 2007, 23, 2957–2958. [Google Scholar] [CrossRef] [PubMed]
- Castellana, S.; Vicario, S.; Saccone, C. Evolutionary patterns of the mitochondrial genome in Metazoa: Exploring the role of mutation and selection in mitochondrial protein-coding genes. Genome Biol. Evol. 2011, 3, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, Y.H.; Wang, X.; Zhang, H.X.; Lin, Q. The mitochondrial genome of a sea anemone Bolocera sp. exhibits novel genetic structures potentially involved in adaptation to the seep-sea environment. Ecol. Evol. 2017, 7, 4951–4962. [Google Scholar] [CrossRef]
- Kubo, I. On the Japanese atyid shrimps. J. Imp. Fish. Inst. 1938, 33, 67–100. [Google Scholar]
- Jugovic, J.; Prevorčnik, S.; Sket, B. Development of sexual characters in the cave shrimp genus Troglocaris (Crustacea: Decapoda: Atyidae) and their applicability in taxonomy. Zootaxa 2010, 2488, 1–21. [Google Scholar] [CrossRef]
- Ntakis, A.; Anastasiadou, C.; Liasko, R.; Leonardos, I.D. Larval development of the shrimp Hippolyte sapphica d’Udekem d’Acoz, 1993 forma A and B (Decapoda: Caridea: Hippolytidae) reared in the laboratory, confirmation of the conspecific status of the two forms. Zootaxa 2010, 2579, 45–58. [Google Scholar] [CrossRef]
- Yatsuya, M.; Ueno, M.; Yamashita, Y. Occurrence and distribution of freshwater shrimp in the Isazu and Yura Rivers, Kyoto, western Japan. Plankton Benthos Res. 2012, 7, 175–187. [Google Scholar] [CrossRef]
- Jabłońska, A.; Mamos, T.; Gruszka, P.; Szlauer-Łukaszewska, A.; Grabowski, M. First record and DNA barcodes of the aquarium shrimp, Neocaridina davidi, in Central Europe from thermally polluted River Oder canal, Poland. Knowl. Manag. Aquat. Ecosyst. 2018, 419, 14. [Google Scholar] [CrossRef]
- Weiperth, A.; Gábris, V.; Danyik, T.; Farkas, A.; Kuříková, P.; Kouba, A.; Patoka, J. Occurrence of non-native red cherry shrimp in European temperate waterbodies: A case study from Hungary. Knowl. Manag. Aquat. Ecosyst. 2018, 420, 9. [Google Scholar] [CrossRef]
- Andersson, M.; Iwasa, Y. Sexual selection. Trends Ecol. Evol. 1996, 11, 53–58. [Google Scholar] [CrossRef]
- Andersson, M.; Simmons, L.W. Sexual selection and mate choice. Trends Ecol. Evol. 2006, 21, 296–302. [Google Scholar] [CrossRef]
- Ráanan, Z.; Sagi, A. Alternative mating strategies in male morphotypes of the freshwater prawn Macrobrachium rosenbergii (de Man). Biol. Bull. 1985, 169, 592–601. [Google Scholar] [CrossRef]
- Rufino, M.M.; Jones, D.A. Observations on the function of the fifth pereiopod in late stage larvae of Lysmata debelius (Decapoda, Hippolytidae). Crustaceana 2001, 74, 977–990. [Google Scholar] [CrossRef]
- Wood, L.E.; Grave, S.D. Functional morphology of the first pereiopod in crangonoid shrimps (Crustacea, Decapoda, Caridea, Crangonoidea). Zoomorphology 2015, 134, 469–486. [Google Scholar] [CrossRef]
- Cabezas, P.; Macpherson, E.; Machordom, A. Morphological and molecular description of new species of squat lobster (Crustacea: Decapoda: Galatheidae) from the Solomon and Fiji Islands (South-West Pacific). Zool. J. Linn. Soc. 2009, 156, 465–493. [Google Scholar] [CrossRef]
- Lavery, S.D.; Farhadi, A.; Farahmand, H.; Chan, T.Y.; Azhdehakoshpour, A.; Thakur, V.; Jeffs, A.G. Evolutionary divergence of geographic subspecies within the scalloped spiny lobster Panulirus homarus (Linnaeus 1758). PLoS ONE 2014, 9, e97247. [Google Scholar] [CrossRef] [PubMed]
- Lefébure, T.; Douady, C.J.; Gouy, M.; Gibert, J. Relationship between morphological taxonomy and molecular divergence within Crustacea: Proposal of a molecular threshold to help species delimitation. Mol. Phylogenet. Evol. 2006, 40, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, 657–1663. [Google Scholar] [CrossRef]
- Robe, L.J.; Machadao, S.; Bartholomei-Santos, M.L. The DNA barcoding and the caveats with respect to its application to some species of Palaemonidae (Crustacea, Decapoda). Zool. Sci. 2012, 29, 714–724. [Google Scholar] [CrossRef]
- Wiley, E.O. The evolutionary species concept reconsidered. Syst. Zool. 1978, 27, 17–26. [Google Scholar] [CrossRef]
- von Rintelen, K.; Page, T.J.; Cai, Y.X.; Roe, K.; Stelbrink, B.; Kuhajda, B.R.; Iliffe, T.M.; Hughes, J.; von Rintelen, T. Drawn to the dark side: A molecular phylogeny of freshwater shrimps (Crustacea: Decapoda: Caridea: Atyidae) reveals frequent cave invasions and challenges current taxonomic hypotheses. Mol. Phylogenet. Evol. 2012, 63, 82–96. [Google Scholar] [CrossRef]
- Smith, M.J.; Williams, W.D. Infraspecific variation within the Atyidae: A study of morphological variation within a population of Paratya australiensis (Crustacea: Decapoda). Aust. J. Mar. Fresh. Res. 1980, 31, 397–407. [Google Scholar] [CrossRef]
- von Rintelen, K.; Cai, Y. Radiation of endemic species flocks in ancient lakes: Systematic revision of the freshwater shrimp Caridina H. Milne Edwards, 1837 (Crustacea: Decapoda: Atyidae) from the ancient lakes of Sulawesi, Indonesia, with the description of eight new species. Raffles Bull. Zool. 2009, 57, 343–452. [Google Scholar]
- de Mazancourt, V.; Klotz, W.; Marquet, G.; Mos, B.; Rogers, D.C.; Keith, P. The complex study of complexes: The first well-supported phylogeny of two species complexes within genus Caridina (Decapoda: Caridea: Atyidae) sheds light on evolution, biogeography, and habitat. Mol. Phylogenet. Evol. 2019, 131, 164–180. [Google Scholar] [CrossRef] [PubMed]
- Castelin, M.; Marquet, G.; Klotz, W. Elephantis, a new genus for Caridina natalensis Bouvier, 1925 from eastern rivers of Madagascar. Zootaxa 2013, 3702, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Zakšek, V.; Sket, B.; Trontelj, P. Phylogeny of the cave shrimp Troglocaris: Evidence of a young connection between Balkans and Caucasus. Mol. Phylogen. Evol. 2007, 42, 223–235. [Google Scholar] [CrossRef]
- Jurado-Rivera, J.A.; Pons, J.; Alvarez, F.; Botello, A.; Humphreys, W.F.; Page, T.J.; Iliffe, T.M.; Willassen, E.; Meland, K.; Juan, C.; et al. Phylogenetic evidence that both ancient vicariance and dispersal have contributed to the biogeographic patterns of anchialine cave shrimps. Sci. Rep. 2017, 7, 2852. [Google Scholar] [CrossRef]
- Park, J.; Kim, Y.; Kwon, W.; Xi, H.; Park, J. The complete mitochondrial genome of Neocaridina heteropoda koreana Kubo, 1938 (Decapoda: Atyidae). Mitochondrial DNA B Resour. 2019, 4, 2332–2334. [Google Scholar] [CrossRef]
- Chak, S.T.C.; Barden, P.; Baeza, J.A. The complete mitochondrial genome of the eusocial sponge-dwelling snapping shrimp Synalpheus microneptunus. Sci. Rep. 2020, 10, 7744. [Google Scholar] [CrossRef]
- Cronin, T.J.; Jones, S.J.M.; Antonio, B.J. The complete mitochondrial genome of the spot prawn, Pandalus platyceros Brandt in von Middendorf, 1851 (Decapoda: Caridea: Pandalidae), assembled from linked-reads sequencing. J. Crustac. Biol. 2022, 42, ruac003. [Google Scholar] [CrossRef]



| Gene | Strand | Position | Size | Codon | Intergenic Nucleotides | |||
|---|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | Anticodon | ||||
| cox1 | H | 1/1/1 | 1536/1536/1536 | 1536/1536/1536 | ATG/ATG/ATG | TAA/TAA/TAA | 0/0/0 | |
| trnL2 | H | 1539/1539/1539 | 1602/1602/1602 | 64/64/64 | TAA/TAA/TAA | 2/2/2 | ||
| cox2 | H | 1604/1604/1604 | 2291/2291/2291 | 688/688/688 | ATG/ATG/ATG | T--/T--/T-- | 1/1/1 | |
| trnK | H | 2292/2292/2292 | 2359/2359/2359 | 68/68/68 | TTT/TTT/TTT | 0/0/0 | ||
| trnD | H | 2365/2368/2368 | 2431/2435/2435 | 67/68/68 | GTC/GTC/GTC | 5/8/8 | ||
| atp8 | H | 2432/2436/2436 | 2590/2594/2594 | 159/159/159 | ATC/ATT/ATT | TAA/TAA/TAA | 0/0/0 | |
| atp6 | H | 2584/2588/2588 | 3258/3262/3262 | 675/675/675 | ATG/ATG/ATG | TAA/TAA/TAA | −7/−7/−7 | |
| cox3 | H | 3258/3262/3262 | 4043/4047/4047 | 786/786/786 | ATG/ATG/ATG | TAG/TAG/TAG | −1/−1/−1 | |
| trnG | H | 4047/4051/4051 | 4110/4115/4115 | 64/65/65 | TCC/TCC/TCC | 3/3/3 | ||
| nad3 | H | 4111/4116/4116 | 4464/4469/4469 | 354/354/354 | ATC/ATC/ATC | TAA/TAA/TAA | 0/0/0 | |
| trnA | H | 4463/4468/4468 | 4526/4531/4531 | 64/64/64 | TGC/TGC/TGC | −2/−2/−2 | ||
| trnR | H | 4527/4532/4532 | 4589/4594/4594 | 63/63/63 | TCG/TCG/TCG | 0/0/0 | ||
| trnN | H | 4594/4599/4599 | 4660/4665/4665 | 67/67/67 | GTT/GTT/GTT | 4/4/4 | ||
| trnS1 | H | 4661/4666/4666 | 4727/4732/4732 | 67/67/67 | TCT/TCT/TCT | 0/0/0 | ||
| trnE | H | 4728/4733/4733 | 4795/4800/4800 | 68/68/68 | TTC/TTC/TTC | 0/0/0 | ||
| trnF | L | 4794/4799/4799 | 4859/4864/4864 | 66/66/66 | GAA/GAA/GAA | −2/−2/−2 | ||
| nad5 | L | 4860/4865/4865 | 6587/6592/6592 | 1728/1728/1728 | ATG/ATG/ATG | TAA/TAA/TAA | 0/0/0 | |
| trnH | L | 6588/6593/6593 | 6653/6658/6658 | 66/66/66 | GTG/GTG/GTG | 0/0/0 | ||
| nad4 | L | 6654/6659/6659 | 7992/7997/7997 | 1339/1339/1339 | ATG/ATG/ATG | T--/T--/T-- | 0/0/0 | |
| nad4l | L | 7986/7991/7991 | 8288/8293/8293 | 303/303/303 | ATG/ATG/ATG | TAA/TAA/TAA | −7/−7/−7 | |
| trnT | H | 8291/8296/8296 | 8356/8361/8361 | 66/66/66 | TGT/TGT/TGT | 2/2/2 | ||
| trnP | L | 8357/8362/8362 | 8422/8427/8426 | 66/66/65 | TGG/TGG/TGG | 0/0/0 | ||
| nad6 | H | 8425/8430/8429 | 8940/8945/8944 | 516/516/516 | ATT/ATT/ATT | TAA/TAA/TAA | 2/2/2 | |
| cob | H | 8940/8945/8944 | 10076/10081/10080 | 1137/1137/1137 | ATG/ATG/ATG | TAG/TAG/TAG | −1/−1/−1 | |
| trnS2 | H | 10075/10080/10079 | 10144/10149/10148 | 70/70/70 | TGA/TGA/TGA | −2/−2/−2 | ||
| nad1 | L | 10163/10168/10167 | 11104/11109/11108 | 942/942/942 | ATT/ATT/ATT | TAA/TAA/TAA | 18/18/18 | |
| trnL1 | L | 11129/11134/11133 | 11196/11200/11199 | 68/67/67 | TAG/TAG/TAG | 24/24/24 | ||
| rrnL | L | 11197/11201/11200 | 12530/12534/12530 | 1334/1334/1331 | 0/0/0 | |||
| trnV | L | 12531/12535/12531 | 12597/12601/12597 | 67/67/67 | TAC/TAC/TAC | 0/0/0 | ||
| rrnS | L | 12598/12602/12598 | 13458/13463/13459 | 861/862/862 | 0/0/0 | |||
| CR | H | 13459/13464/13460 | 14137/14142/14137 | 679/679/678 | 0/0/0 | |||
| trnI | H | 14138/14143/14138 | 14202/14207/14202 | 65/65/65 | GAT/GAT/GAT | 0/0/0 | ||
| trnQ | L | 14211/14216/14211 | 14278/14283/14278 | 68/68/68 | TTG/TTG/TTG | 8/8/8 | ||
| trnM | H | 14284/14289/14284 | 14349/14354/14349 | 66/66/66 | CAT/CAT/CAT | 5/5/5 | ||
| nad2 | H | 14350/14355/14350 | 15354/15359/15354 | 1005/1005/1005 | ATT/ATT/ATT | TAA/TAA/TAA | 0/0/0 | |
| trnW | H | 15353/15358/15353 | 15422/15427/15423 | 70/70/71 | TCA/TCA/TCA | −2/−2/−2 | ||
| trnC | L | 15422/15427/15423 | 15486/15491/15487 | 65/65/65 | GCA/GCA/GCA | −1/−1/−1 | ||
| trnY | L | 15487/15492/15488 | 15553/15558/15554 | 67/67/67 | GTA/GTA/GTA | 0/0/0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Cui, X.; Li, X.; Dong, D.; Kang, X.; Gan, Z. Integrative Taxonomy Reveals New Insights into the Species Validity of the Neocaridina davidi-N. denticulata-N. heteropoda Complex and Mitogenomic Phylogeny of Caridean Shrimps. Curr. Issues Mol. Biol. 2024, 46, 12279-12298. https://doi.org/10.3390/cimb46110729
Yang M, Cui X, Li X, Dong D, Kang X, Gan Z. Integrative Taxonomy Reveals New Insights into the Species Validity of the Neocaridina davidi-N. denticulata-N. heteropoda Complex and Mitogenomic Phylogeny of Caridean Shrimps. Current Issues in Molecular Biology. 2024; 46(11):12279-12298. https://doi.org/10.3390/cimb46110729
Chicago/Turabian StyleYang, Mei, Xiaodong Cui, Xinzheng Li, Dong Dong, Xianjiang Kang, and Zhibin Gan. 2024. "Integrative Taxonomy Reveals New Insights into the Species Validity of the Neocaridina davidi-N. denticulata-N. heteropoda Complex and Mitogenomic Phylogeny of Caridean Shrimps" Current Issues in Molecular Biology 46, no. 11: 12279-12298. https://doi.org/10.3390/cimb46110729
APA StyleYang, M., Cui, X., Li, X., Dong, D., Kang, X., & Gan, Z. (2024). Integrative Taxonomy Reveals New Insights into the Species Validity of the Neocaridina davidi-N. denticulata-N. heteropoda Complex and Mitogenomic Phylogeny of Caridean Shrimps. Current Issues in Molecular Biology, 46(11), 12279-12298. https://doi.org/10.3390/cimb46110729






