ΔNp63 Regulates Radioresistance in Human Head and Neck Squamous Carcinoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture and Treatment
2.3. siRNA Transfection
2.4. In Vitro X-ray Irradiation
2.5. Clonogenic Survival Assay
2.6. Apoptosis Analysis
2.7. SDS-PAGE and Western Blot Analysis
2.8. Subcellular Fraction
2.9. Bioinformatics and Data Analysis
2.10. Statistical Analysis
3. Results
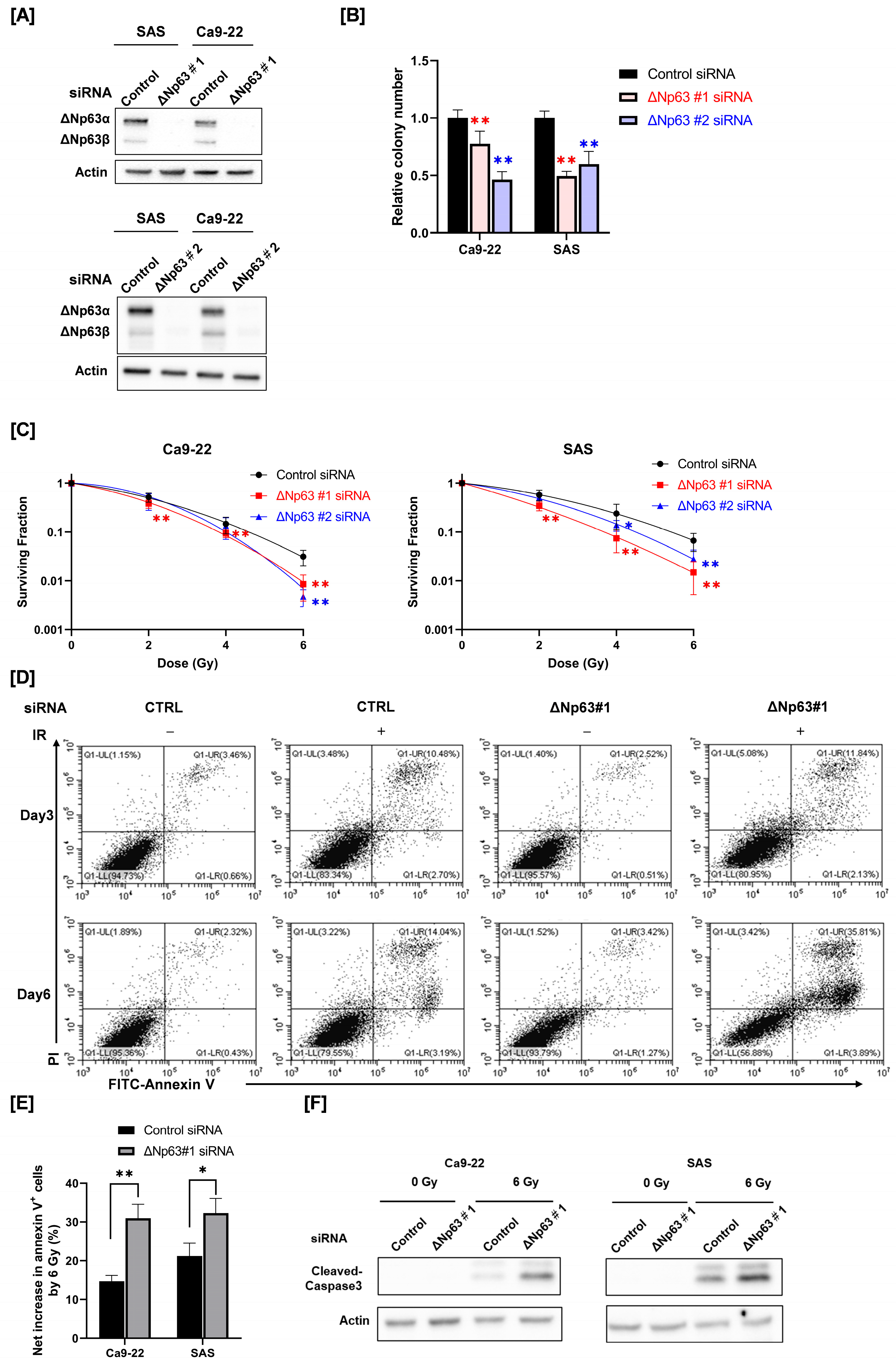
3.1. Effect of ΔNp63 Knockdown on Radiosensitivity and Radiation-Induced Apoptosis of HNSCC Cells
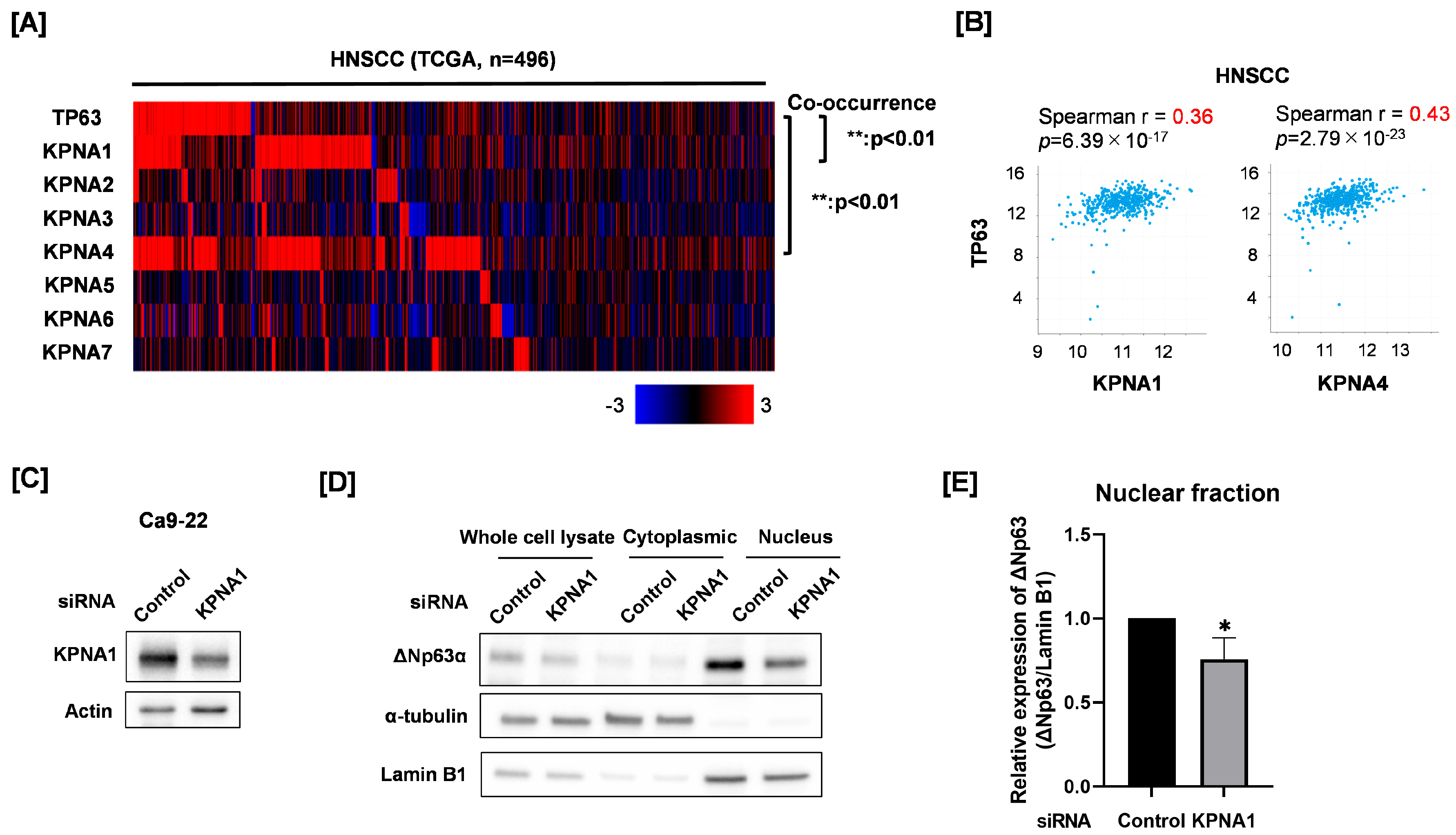
3.2. Relationship of Nuclear ΔNp63 Protein Expression and KPNA1 in Ca9-22 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Perri, F.; Pacelli, R.; Della Vittoria Scarpati, G.; Cella, L.; Giuliano, M.; Caponigro, F.; Pepe, S. Radioresistance in head and neck squamous cell carcinoma: Biological bases and therapeutic implications. Head Neck 2015, 37, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.M.; Hong, Y.; Lee, S.; Liu, P.; Lim, J.H.; Lee, Y.H.; Lee, T.H.; Chang, K.T.; Hong, Y. Therapeutic Implications for Overcoming Radiation Resistance in Cancer Therapy. Int. J. Mol. Sci. 2015, 16, 26880–26913. [Google Scholar] [CrossRef] [PubMed]
- Olivares-Urbano, M.A.; Griñán-Lisón, C.; Marchal, J.A.; Núñez, M.I. CSC Radioresistance: A Therapeutic Challenge to Improve Radiotherapy Effectiveness in Cancer. Cells 2020, 9, 1651. [Google Scholar] [CrossRef]
- Huang, R.-X.; Zhou, P.-K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [PubMed]
- Hazawa, M.; Sakai, K.; Kobayashi, A.; Yoshino, H.; Iga, Y.; Iwashima, Y.; Lim, K.S.; Voon, D.C.-C.; Jiang, Y.-Y.; Horike, S.-I.; et al. Disease-specific alteration of karyopherin-α subtype establishes feed-forward oncogenic signaling in head and neck squamous cell carcinoma. Oncogene 2019, 39, 2212–2223. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, Y.; Lu, C.; Zhang, R.; Wang, Y.; Luo, L.; Zhang, Y.; Chu, C.H.; Wang, K.J.; Obbad, S.; et al. Inhibition of Karyopherin beta 1 suppresses prostate cancer growth. Oncogene 2019, 38, 4700–4714. [Google Scholar] [CrossRef]
- Chi, R.-P.A.; van der Watt, P.; Wei, W.; Birrer, M.J.; Leaner, V.D. Inhibition of Kpnβ1 mediated nuclear import enhances cisplatin chemosensitivity in cervical cancer. BMC Cancer 2021, 21, 106. [Google Scholar] [CrossRef] [PubMed]
- Hazawa, M.; Yoshino, H.; Nakagawa, Y.; Shimizume, R.; Nitta, K.; Sato, Y.; Sato, M.; Wong, R.W.; Kashiwakura, I. Karyopherin-β1 Regulates Radioresistance and Radiation-Increased Programmed Death-Ligand 1 Expression in Human Head and Neck Squamous Cell Carcinoma Cell Lines. Cancers 2020, 12, 908. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.L.; Balinth, S.; Mills, A.A. p63-related signaling at a glance. J. Cell Sci. 2020, 133, jcs228015. [Google Scholar] [CrossRef] [PubMed]
- King, K.E.; Ha, L.; Camilli, T.; Weinberg, W.C. Delineating Molecular Mechanisms of Squamous Tissue Homeostasis and Neoplasia: Focus on p63. J. Ski. Cancer 2013, 2013, 632028. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, X.; Xiong, Q.; Han, J.; Zhu, Q. The dual role of p63 in cancer. Front. Oncol. 2023, 13, 1116061. [Google Scholar] [CrossRef]
- Gatti, V.; Fierro, C.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. ΔNp63 in squamous cell carcinoma: Defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol. Oncol. 2019, 13, 981–1001. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Tan, Y.; Wang, L.; Cai, J.; Li, X.; Zeng, Z.; Xiong, W.; Li, G.; Li, X.; Tan, P.; et al. TP63 links chromatin remodeling and enhancer reprogramming to epidermal differentiation and squamous cell carcinoma development. Cell. Mol. Life Sci. 2020, 77, 4325–4346. [Google Scholar] [CrossRef]
- Leonard, M.K.; Kommagani, R.; Payal, V.; Mayo, L.D.; Shamma, H.N.; Kadakia, M.P. ΔNp63α regulates keratinocyte proliferation by controlling PTEN expression and localization. Cell Death Differ. 2011, 18, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liang, S.; Bergholz, J.; He, H.; Walsh, E.M.; Zhang, Y.; Xiao, Z.-X. ΔNp63α activates CD82 metastasis suppressor to inhibit cancer cell invasion. Cell Death Dis. 2014, 5, e1280. [Google Scholar] [CrossRef] [PubMed]
- Danilov, A.V.; Neupane, D.; Nagaraja, A.S.; Feofanova, E.V.; Humphries, L.A.; DiRenzo, J.; Korc, M. DeltaNp63alpha-Mediated Induction of Epidermal Growth Factor Receptor Promotes Pancreatic Cancer Cell Growth and Chemoresistance. PLoS ONE 2011, 6, e26815. [Google Scholar] [CrossRef]
- Westfall, M.D.; Mays, D.J.; Sniezek, J.C.; Pietenpol, J.A. The ΔNp63α Phosphoprotein Binds the p21 and 14-3-3σ Promoters In Vivo and Has Transcriptional Repressor Activity That Is Reduced by Hay-Wells Syndrome-Derived Mutations. Mol. Cell. Biol. 2003, 23, 2264–2276. [Google Scholar] [CrossRef] [PubMed]
- Rocco, J.W.; Leong, C.-O.; Kuperwasser, N.; DeYoung, M.P.; Ellisen, L.W. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 2006, 9, 45–56. [Google Scholar] [CrossRef]
- Bretz, A.C.; Gittler, M.P.; Charles, J.P.; Gremke, N.; Eckhardt, I.; Mernberger, M.; Mandic, R.; Thomale, J.; Nist, A.; Wanzel, M.; et al. ΔNp63 activates the Fanconi anemia DNA repair pathway and limits the efficacy of cisplatin treatment in squamous cell carcinoma. Nucleic Acids Res. 2016, 44, 3204–3218. [Google Scholar] [CrossRef] [PubMed]
- Kudo, K.-I.; Tsuyama, N.; Nagata, K.; Imaoka, T.; Iizuka, D.; Sugai-Takahashi, M.; Muramatsu, M.; Sakai, A. ΔNp63α transcriptionally represses p53 target genes involved in the radiation-induced DNA damage response: ΔNp63α may cause genomic instability in epithelial stem cells. Radiat. Oncol. 2022, 17, 183. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshino, H.; Kashiwakura, I.; Tsuruga, E. DAP3 Is Involved in Modulation of Cellular Radiation Response by RIG-I-Like Receptor Agonist in Human Lung Adenocarcinoma Cells. Int. J. Mol. Sci. 2021, 22, 420. [Google Scholar] [CrossRef] [PubMed]
- Nyui, T.; Yoshino, H.; Nunota, T.; Sato, Y.; Tsuruga, E. cGAS Regulates the Radioresistance of Human Head and Neck Squamous Cell Carcinoma Cells. Cells 2022, 11, 1434. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H.; Iwabuchi, M.; Kazama, Y.; Furukawa, M.; Kashiwakura, I. Effects of retinoic acid-inducible gene-I-like receptors activations and ionizing radiation cotreatment on cytotoxicity against human non-small cell lung cancer in vitro. Oncol. Lett. 2018, 15, 4697–4705. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H.; Kumai, Y.; Kashiwakura, I. Effects of endoplasmic reticulum stress on apoptosis induction in radioresistant macrophages. Mol. Med. Rep. 2017, 15, 2867–2872. [Google Scholar] [CrossRef]
- Kadota, S.; Ou, J.; Shi, Y.; Lee, J.T.; Sun, J.; Yildirim, E. Nucleoporin 153 links nuclear pore complex to chromatin architecture by mediating CTCF and cohesin binding. Nat. Commun. 2020, 11, 2606. [Google Scholar] [CrossRef]
- Cheng, G.; Kong, D.; Hou, X.; Liang, B.; He, M.; Liang, N.; Ma, S.; Liu, X.; Zhang, W.-W.; Li, L.; et al. The Tumor Suppressor, p53, Contributes to Radiosensitivity of Lung Cancer Cells by Regulating Autophagy and Apoptosis. Cancer Biother. Radiopharm. 2013, 28, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Xiaoyun, H.; Haifeng, Q.; Jing, L.; Weixu, H.; Ruofan, D.; Jinjin, Y.; Zongji, S. MicroRNA-218 Enhances the Radiosensitivity of Human Cervical Cancer via Promoting Radiation Induced Apoptosis. Int. J. Med. Sci. 2014, 11, 691–696. [Google Scholar] [CrossRef]
- Silva, F.F.V.e.; Padín-Iruegas, M.E.; Caponio, V.C.A.; Lorenzo-Pouso, A.I.; Saavedra-Nieves, P.; Chamorro-Petronacci, C.M.; Suaréz-Peñaranda, J.; Pérez-Sayáns, M. Caspase 3 and Cleaved Caspase 3 Expression in Tumorogenesis and Its Correlations with Prognosis in Head and Neck Cancer: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2022, 23, 11937. [Google Scholar] [CrossRef]
- Hazawa, M.; Lin, D.; Kobayashi, A.; Jiang, Y.; Xu, L.; Dewi, F.R.P.; Mohamed, M.S.; Hartono; Nakada, M.; Meguro-Horike, M.; et al. ROCK-dependent phosphorylation of NUP 62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 2017, 19, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Moergel, M.; Abt, E.; Stockinger, M.; Kunkel, M. Overexpression of p63 is associated with radiation resistance and prognosis in oral squamous cell carcinoma. Oral Oncol. 2010, 46, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, Y.; Li, L.; Baba, T.; Nakagawa, H.; Shimura, T.; Yamamoto, Y.; Ohkubo, Y.; Fukumoto, M. Clinically relevant radioresistant cells efficiently repair DNA double-strand breaks induced by X-rays. Cancer Sci. 2009, 100, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, S.; Sakata, K.-I.; Liang, S.; Yoshikawa, K.; Iizasa, H.; Tada, M.; Hamada, J.-I.; Kashiwazaki, H.; Kitagawa, Y.; Yamazaki, Y. Dominant-negative p53 mutant R248Q increases the motile and invasive activities of oral squamous cell carcinoma cells. Biomed. Res. 2019, 40, 37–49. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, W.; Chen, X. P63 regulates tubular formation via epithelial-to-mesenchymal transition. Oncogene 2013, 33, 1548–1557. [Google Scholar] [CrossRef][Green Version]
- Yu, J.; Zhang, L. PUMA, a potent killer with or without p53. Oncogene 2008, 27 (Suppl. S1), S71–S83. [Google Scholar] [CrossRef]
- Sun, Q.; Sakaida, T.; Yue, W.; Gollin, S.M.; Yu, J. Chemosensitization of head and neck cancer cells by PUMA. Mol. Cancer Ther. 2007, 6, 3180–3188. [Google Scholar] [CrossRef]
- Yang, X.; Lu, H.; Yan, B.; Romano, R.-A.; Bian, Y.; Friedman, J.; Duggal, P.; Allen, C.; Chuang, R.; Ehsanian, R.; et al. ΔNp63 Versatilely Regulates a Broad NF-κB Gene Program and Promotes Squamous Epithelial Proliferation, Migration, and Inflammation. Cancer Res. 2011, 71, 3688–3700. [Google Scholar] [CrossRef]
- Hikisz, P.; Kiliańska, Z. Puma, a critical mediator of cell death—One decade on from its discovery. Cell. Mol. Biol. Lett. 2012, 17, 646–669. [Google Scholar] [CrossRef]
- Mattes, K.; Vellenga, E.; Schepers, H. Differential redox-regulation and mitochondrial dynamics in normal and leukemic hematopoietic stem cells: A potential window for leukemia therapy. Crit. Rev. Oncol. 2019, 144, 102814. [Google Scholar] [CrossRef] [PubMed]
- Heiden, M.G.; Chandel, N.S.; Schumacker, P.T.; Thompson, C.B. Bcl-xL Prevents Cell Death following Growth Factor Withdrawal by Facilitating Mitochondrial ATP/ADP Exchange. Mol. Cell 1999, 3, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Mehrbod, P.; Ande, S.R.; Alizadeh, J.; Rahimizadeh, S.; Shariati, A.; Malek, H.; Hashemi, M.; Glover, K.K.M.; Sher, A.A.; Coombs, K.M.; et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence 2019, 10, 376–413. [Google Scholar] [CrossRef] [PubMed]
- Sekimoto, T.; Imamoto, N.; Nakajima, K.; Hirano, T.; Yoneda, Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997, 16, 7067–7077. [Google Scholar] [CrossRef] [PubMed]
- Takeda, E.; Murakami, T.; Matsuda, G.; Murakami, H.; Zako, T.; Maeda, M.; Aida, Y. Nuclear Exportin Receptor CAS Regulates the NPI-1–Mediated Nuclear Import of HIV-1 Vpr. PLoS ONE 2011, 6, e27815. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, K.; Yoshino, H.; Sato, Y.; Nakano, M.; Tsuruga, E. ΔNp63 Regulates Radioresistance in Human Head and Neck Squamous Carcinoma Cells. Curr. Issues Mol. Biol. 2023, 45, 6262-6271. https://doi.org/10.3390/cimb45080394
Sato K, Yoshino H, Sato Y, Nakano M, Tsuruga E. ΔNp63 Regulates Radioresistance in Human Head and Neck Squamous Carcinoma Cells. Current Issues in Molecular Biology. 2023; 45(8):6262-6271. https://doi.org/10.3390/cimb45080394
Chicago/Turabian StyleSato, Kota, Hironori Yoshino, Yoshiaki Sato, Manabu Nakano, and Eichi Tsuruga. 2023. "ΔNp63 Regulates Radioresistance in Human Head and Neck Squamous Carcinoma Cells" Current Issues in Molecular Biology 45, no. 8: 6262-6271. https://doi.org/10.3390/cimb45080394
APA StyleSato, K., Yoshino, H., Sato, Y., Nakano, M., & Tsuruga, E. (2023). ΔNp63 Regulates Radioresistance in Human Head and Neck Squamous Carcinoma Cells. Current Issues in Molecular Biology, 45(8), 6262-6271. https://doi.org/10.3390/cimb45080394




