Constructing Lipoparticles Capable of Endothelial Cell-Derived Exosome-Mediated Delivery of Anti-miR-33a-5p to Cultured Macrophages
Abstract
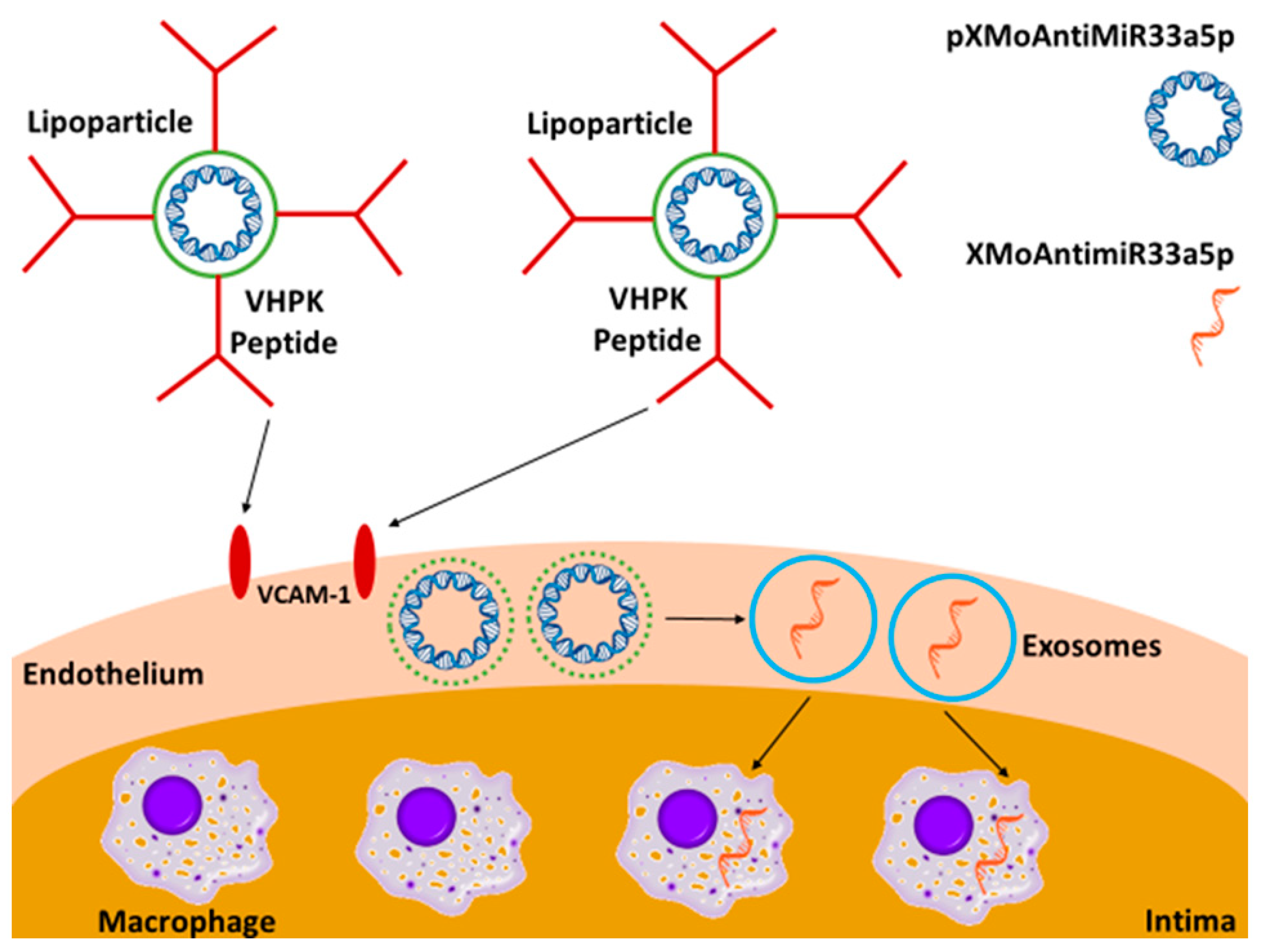
1. Introduction
2. Materials and Methods
2.1. Tissue Culture
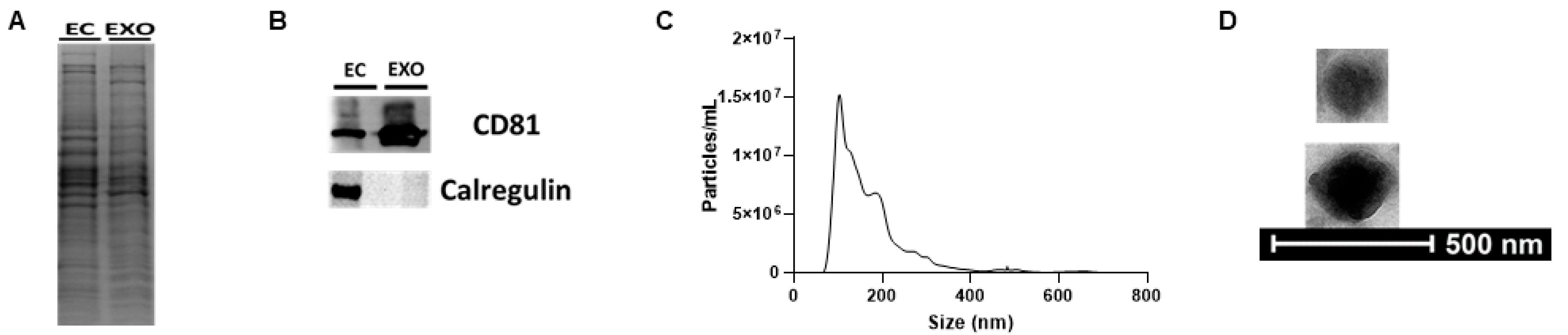
2.2. iMAEC-Derived Exosome Isolation and Characterization
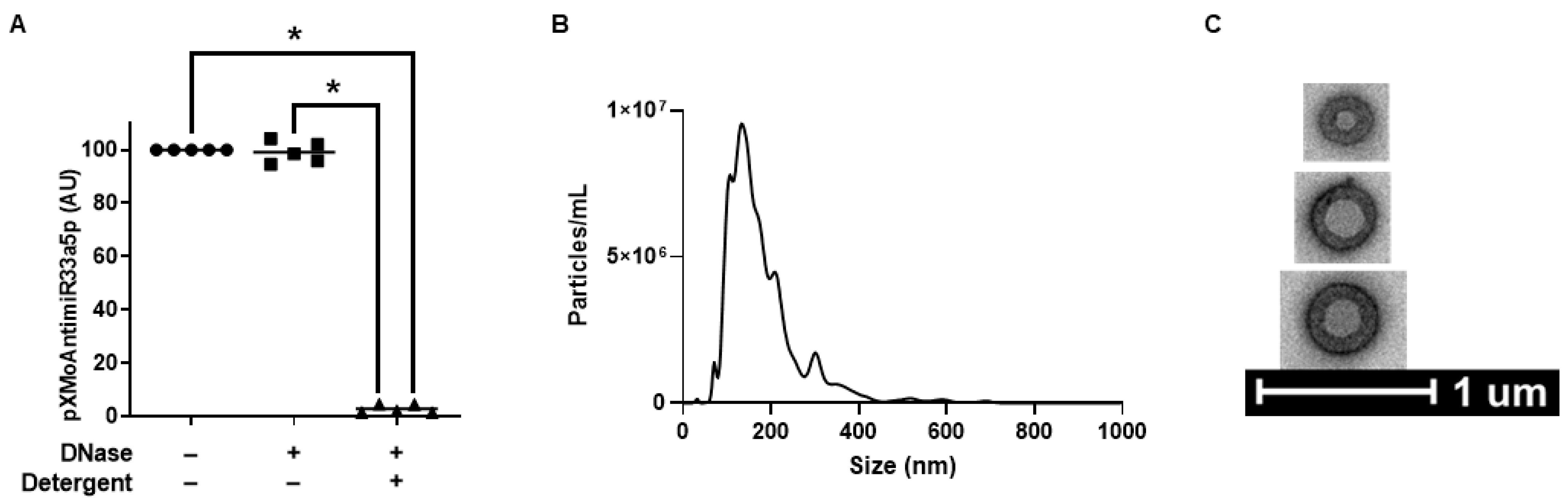
2.3. Generation and Characterization of LP-pXMoAntimiR33a5p
2.4. Transwell Assays
2.5. qPCR and RT-qPCR
2.6. SDS-PAGE and Immunoblotting
2.7. ApoAI-Mediated Cholesterol Efflux
2.8. Statistical Analysis
3. Results
3.1. iMAECs Secrete Exosomes
3.2. Characterization of LP-pXMoAntimiR33a5p
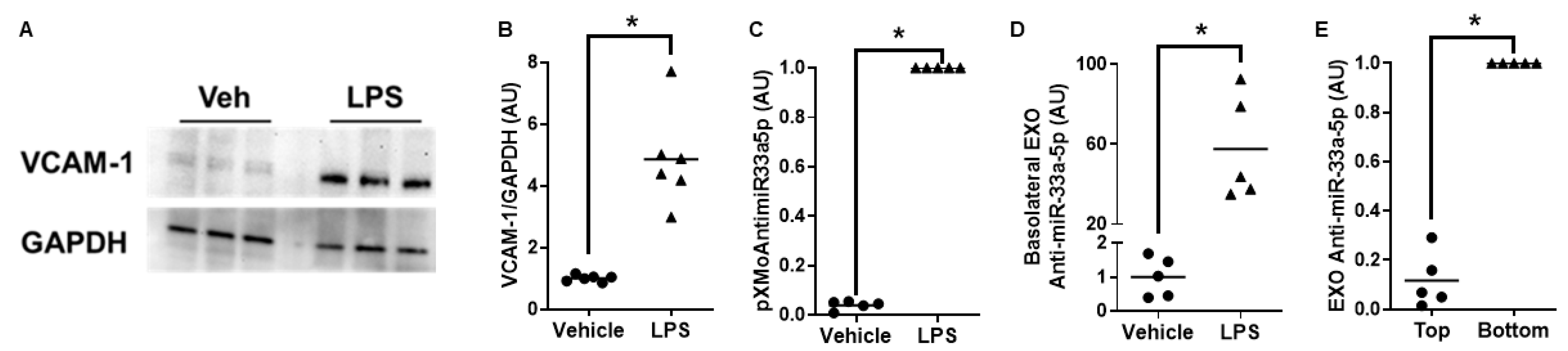
3.3. Proinflammatory iMAECs Exposed to LP-pXMoAntimiR33a5p Primarily Release Anti-miR-33a-5p-Loaded Exosomes Basolaterally
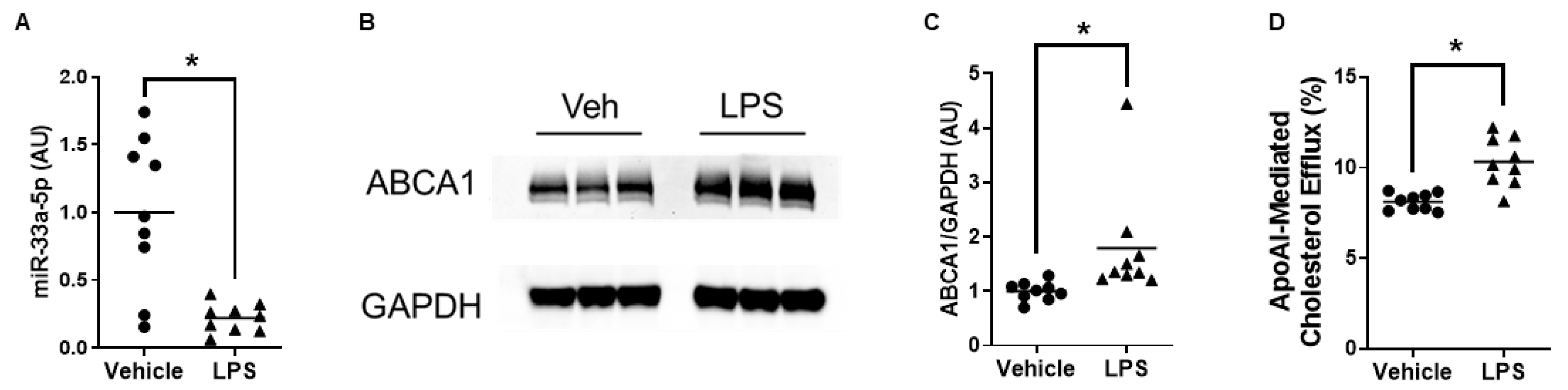
3.4. ABCA1-Dependent Cholesterol Efflux Is Enhanced in Macrophages Exposed to Exosomes Secreted Basolaterally by Proinflammatory Endothelial Cells Incubated with LP-pXMoAntimiR33a5p
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pahwa, R.; Jialal, I. StatPearls (Atherosclerosis). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Lusis, A.J. Atherosclerosis. Nature 2000, 407, 233–241. [Google Scholar] [CrossRef]
- Naghavi, M.; Libby, P.; Falk, E.; Casscells, S.W.; Litovsky, S.; Rumberger, J.; Badimon, J.J.; Stefanadis, C.; Moreno, P.; Pasterkamp, G. From vulnerable plaque to vulnerable patient: A call for new definitions and risk assessment strategies: Part I. Circulation 2003, 108, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.K.; Libby, P.; Tabas, I. Inflammation and plaque vulnerability. J. Intern. Med. 2015, 278, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-Y.; Li, C.-J.; Hou, M.-F.; Chu, P.-Y. New Insights into the Role of Inflammation in the Pathogenesis of Atherosclerosis. Int. J. Mol. Sci. 2017, 18, 2034. [Google Scholar] [CrossRef]
- Ohashi, R.; Mu, H.; Wang, X.; Yao, Q.; Chen, C. Reverse cholesterol transport and cholesterol efflux in atherosclerosis. Qjm Int. J. Med. 2005, 98, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, A.; Albarrán-Juárez, J.; Liang, G.; Roquid, K.A.; Iring, A.; Tonack, S.; Chen, M.; Müller, O.J.; Weinstein, L.S.; Offermanns, S. Disturbed flow–induced Gs-mediated signaling protects against endothelial inflammation and atherosclerosis. J. Clin. Investig. 2020, 5, e140485. [Google Scholar] [CrossRef]
- Chiu, J.-J.; Chien, S.; Venturini, G.; Malagrino, P.A.; Padilha, K.; Tanaka, L.Y.; Laurindo, F.R.; Dariolli, R.; Carvalho, V.M.; Cardozo, K.H.M.; et al. Effects of Disturbed Flow on Vascular Endothelium: Pathophysiological Basis and Clinical Perspectives. Physiol. Rev. 2011, 91, 327–387. [Google Scholar] [CrossRef]
- Liao, J.K. Linking endothelial dysfunction with endothelial cell activation. J. Clin. Investig. 2013, 123, 540–541. [Google Scholar] [CrossRef]
- Boulanger, C.M. Highlight on Endothelial Activation and Beyond. Arter. Thromb. Vasc. Biol. 2018, 38, e198–e201. [Google Scholar] [CrossRef]
- Davignon, J.; Ganz, P. Role of endothelial dysfunction in atherosclerosis. Circulation 2004, 109, III27–III32. [Google Scholar] [CrossRef]
- Gimbrone, M.A., Jr.; Garcia-Cardena, G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016, 118, 620–636. [Google Scholar] [CrossRef]
- Cook-Mills, J.M.; Marchese, M.E.; Abdala-Valencia, H. Vascular Cell Adhesion Molecule-1 Expression and Signaling During Disease: Regulation by Reactive Oxygen Species and Antioxidants. Antioxid. Redox Signal. 2011, 15, 1607–1638. [Google Scholar] [CrossRef]
- Meerschaert, J.; Furie, M.B. The adhesion molecules used by monocytes for migration across endothelium include CD11a/CD18, CD11b/CD18, and VLA-4 on monocytes and ICAM-1, VCAM-1, and other ligands on endothelium. J. Immunol. 1995, 154, 4099–4112. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, T.; Ley, K. Monocyte trafficking across the vessel wall. Cardiovasc. Res. 2015, 107, 321–330. [Google Scholar] [CrossRef]
- Libby, P.; Ridker, P.M.; Maseri, A. Inflammation and atherosclerosis. Circulation 2002, 105, 1135–1143. [Google Scholar] [CrossRef]
- Libby, P. Inflammation in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2045–2051. [Google Scholar] [CrossRef]
- Libby, P. Inflammation in atherosclerosis. Nature 2002, 420, 868–874. [Google Scholar] [CrossRef]
- Thayse, K.; Kindt, N.; Laurent, S.; Carlier, S. VCAM-1 Target in Non-Invasive Imaging for the Detection of Atherosclerotic Plaques. Biology 2020, 9, 368. [Google Scholar] [CrossRef] [PubMed]
- Khodabandehlou, K.; Masehi-Lano, J.J.; Poon, C.; Wang, J.; Chung, E.J. Targeting cell adhesion molecules with nanoparticles using in vivo and flow-based in vitro models of atherosclerosis. Exp. Biol. Med. 2017, 242, 799–812. [Google Scholar] [CrossRef]
- Kheirolomoom, A.; Kim, C.W.; Seo, J.W.; Kumar, S.; Son, D.J.; Gagnon, M.K.; Ingham, E.S.; Ferrara, K.W.; Jo, H. Multifunctional Nanoparticles Facilitate Molecular Targeting and miRNA Delivery to Inhibit Atherosclerosis in ApoE−/− Mice. ACS Nano 2015, 9, 8885–8897. [Google Scholar] [CrossRef] [PubMed]
- Dosta, P.; Tamargo, I.; Ramos, V.; Kumar, S.; Kang, D.W.; Borros, S.; Jo, H. Delivery of Anti-microRNA-712 to Inflamed Endothelial Cells Using Poly(beta-amino ester) Nanoparticles Conjugated with VCAM-1 Targeting Peptide. Adv. Healthc. Mater. 2021, 10, 2001894. [Google Scholar]
- Moore, K.J.; Tabas, I. The Cellular Biology of Macrophages in Atherosclerosis. Cell 2011, 145, 341–355. [Google Scholar] [CrossRef]
- Bi, L.; Wacker, B.K.; Stamatikos, A.; Sethuraman, M.; Komandur, K.; Dichek, D.A. Jugular Vein Injection of High-Titer Lentiviral Vectors Does Not Transduce the Aorta—Brief Report. Arter. Thromb. Vasc. Biol. 2021, 41, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Qian, K.; Du, L.; Luttrell, I.; Chitaley, K.; Dichek, D.A. Helper-Dependent Adenovirus is Superior to First-Generation Adenovirus for Expressing Transgenes in Atherosclerosis-Prone Arteries. Arter. Thromb. Vasc. Biol. 2011, 31, 1317–1325. [Google Scholar] [CrossRef]
- Peterson, M.F.; Otoc, N.; Sethi, J.K.; Gupta, A.; Antes, T.J. Integrated systems for exosome investigation. Methods 2015, 87, 31–45. [Google Scholar] [CrossRef]
- Fujii, N.; Hirata, H.; Ueno, K.; Mori, J.; Oka, S.; Shimizu, K.; Kawai, Y.; Inoue, R.; Yamamoto, Y.; Matsumoto, H.; et al. Extracellular miR-224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget 2017, 8, 109877–109888. [Google Scholar] [CrossRef] [PubMed]
- Stamatikos, A.; Knight, E.; Vojtech, L.; Bi, L.; Wacker, B.K.; Tang, C.; Dichek, D.A. Exosome-Mediated Transfer of Anti-miR-33a-5p from Transduced Endothelial Cells Enhances Macrophage and Vascular Smooth Muscle Cell Cholesterol Efflux. Hum. Gene Ther. 2020, 31, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Näär, A.M. MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef]
- Rayner, K.J.; Sheedy, F.J.; Esau, C.C.; Hussain, F.N.; Temel, R.E.; Parathath, S.; van Gils, J.M.; Rayner, A.J.; Chang, A.N.; Suarez, Y.; et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Investig. 2011, 121, 2921–2931. [Google Scholar] [CrossRef]
- Ni, C.-W.; Kumar, S.; Ankeny, C.J.; Jo, H. Development of immortalized mouse aortic endothelial cell lines. Vasc. Cell 2014, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Jo, H.; Echesabal-Chen, J.; Stamatikos, A. Combined LXR and RXR Agonist Therapy Increases ABCA1 Protein Expression and Enhances ApoAI-Mediated Cholesterol Efflux in Cultured Endothelial Cells. Metabolites 2021, 11, 640. [Google Scholar] [CrossRef] [PubMed]
- Bazban-Shotorbani, S.; Khare, H.A.; Kajtez, J.; Basak, S.; Lee, J.H.; Kamaly, N. Effect of Nanoparticle Biophysicochemical Properties on Binding and Transport across Cardiovascular Endothelial Dysfunction Models. ACS Appl. Nano Mater. 2021, 4, 4077–4091. [Google Scholar] [CrossRef]
- Huang, K.; Garimella, S.; Clay-Gilmour, A.; Vojtech, L.; Armstrong, B.; Bessonny, M.; Stamatikos, A. Comparison of Human Urinary Exosomes Isolated via Ultracentrifugation Alone versus Ultracentrifugation Followed by SEC Column-Purification. J. Pers. Med. 2022, 12, 340. [Google Scholar] [CrossRef]
- Wang, X.; Wilkinson, R.; Kildey, K.; Ungerer, J.P.J.; Hill, M.M.; Shah, A.K.; Mohamed, A.; Dutt, M.; Molendijk, J.; Healy, H.; et al. Molecular and functional profiling of apical versus basolateral small extracellular vesicles derived from primary human proximal tubular epithelial cells under inflammatory conditions. J. Extracell. Vesicles 2021, 10, e12064. [Google Scholar] [CrossRef]
- Thayanithy, V.; O’hare, P.; Wong, P.; Zhao, X.; Steer, C.J.; Subramanian, S.; Lou, E. A transwell assay that excludes exosomes for assessment of tunneling nanotube-mediated intercellular communication. Cell Commun. Signal. 2017, 15, 46. [Google Scholar] [CrossRef] [PubMed]
- Esobi, I.C.; Barksdale, C.; Heard-Tate, C.; Powell, R.R.; Bruce, T.F.; Stamatikos, A. MOVAS Cells: A Versatile Cell Line for Studying Vascular Smooth Muscle Cell Cholesterol Metabolism. Lipids 2021, 56, 413–422. [Google Scholar] [CrossRef]
- Stamatikos, A.; Dronadula, N.; Ng, P.; Palmer, D.; Knight, E.; Wacker, B.K.; Tang, C.; Kim, F.; Dichek, D.A. ABCA1 Overexpression in Endothelial Cells In Vitro Enhances ApoAI-Mediated Cholesterol Efflux and Decreases Inflammation. Hum. Gene Ther. 2019, 30, 236–248. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Fujimoto, S.; Fujita, Y.; Kadota, T.; Araya, J.; Kuwano, K. Intercellular Communication by Vascular Endothelial Cell-Derived Extracellular Vesicles and Their MicroRNAs in Respiratory Diseases. Front. Mol. Biosci. 2021, 7, 619697. [Google Scholar] [CrossRef]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and Comprehensive Proteome Profiling of Exosomes Secreted by Hepatocytes. J. Proteome Res. 2008, 7, 5157–5166. [Google Scholar] [CrossRef] [PubMed]
- Kotrbová, A.; Štěpka, K.; Maška, M.; Pálenik, J.J.; Ilkovics, L.; Klemová, D.; Kravec, M.; Hubatka, F.; Dave, Z.; Hampl, A.; et al. TEM ExosomeAnalyzer: A computer-assisted software tool for quantitative evaluation of extracellular vesicles in transmission electron microscopy images. J. Extracell. Vesicles 2019, 8, 1560808. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.; Woo, S.; Hughes, S.; Levy, C.; Ballweber, L.; Sauteraud, R.P.; Strobl, J.; Westerberg, K.; Gottardo, R.; Tewari, M.; et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014, 42, 7290–7304. [Google Scholar] [CrossRef]
- Wong, D.; Dorovini-Zis, K. Expression of Vascular Cell Adhesion Molecule-1 (VCAM-1) by Human Brain Microvessel Endothelial Cells in Primary Culture. Microvasc. Res. 1995, 49, 325–339. [Google Scholar] [CrossRef]
- Kaur, G.; Dufour, J.M. Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef]
- Barquera, S.; Pedroza-Tobías, A.; Medina, C.; Hernández-Barrera, L.; Bibbins-Domingo, K.; Lozano, R.; Moran, A.E. Global Overview of the Epidemiology of Atherosclerotic Cardiovascular Disease. Arch. Med Res. 2015, 46, 328–338. [Google Scholar] [CrossRef]
- Waters, D.D.; Hsue, P.Y. PCSK9 Inhibition to Reduce Cardiovascular Risk: Tempering Expectations. Circ. Res. 2017, 120, 1537–1539. [Google Scholar] [CrossRef]
- Thompson, P.L.; Nidorf, S.M. Anti-inflammatory therapy with canakinumab for atherosclerotic disease: Lessons from the CANTOS trial. J. Thorac. Dis. 2018, 10, 695–698. [Google Scholar] [CrossRef]
- Sabatine, M.S.; Giugliano, R.P.; Keech, A.C.; Honarpour, N.; Wiviott, S.D.; Murphy, S.A.; Kuder, J.F.; Wang, H.; Liu, T.; Wasserman, S.M.; et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N. Engl. J. Med. 2017, 376, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Ridker, P.M.; Everett, B.M.; Thuren, T.; MacFadyen, J.G.; Chang, W.H.; Ballantyne, C.; Fonseca, F.; Nicolau, J.; Koenig, W.; Anker, S.D.; et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N. Engl. J. Med. 2017, 377, 1119–1131. [Google Scholar] [CrossRef]
- Tabas, I.; Bornfeldt, K.E. Macrophage Phenotype and Function in Different Stages of Atherosclerosis. Circ. Res. 2016, 118, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Mundi, S.; Massaro, M.; Scoditti, E.; Carluccio, M.A.; van Hinsbergh, V.W.M.; Iruela-Arispe, M.L.; De Caterina, R. Endothelial permeability, LDL deposition, and cardiovascular risk factors-a review. Cardiovasc. Res. 2018, 114, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Hossaini Nasr, S.; Huang, X. Nanotechnology for Targeted Therapy of Atherosclerosis. Front. Pharmacol. 2021, 12, 755569. [Google Scholar] [CrossRef]
- Darwitan, A.; Wong, Y.S.; Nguyen, L.T.H.; Czarny, B.; Vincent, A.; Nedumaran, A.M.; Tan, Y.F.; Muktabar, A.; Tang, J.K.; Ng, K.W.; et al. Liposomal Nanotherapy for Treatment of Atherosclerosis. Adv. Health Mater. 2020, 9, 2000465. [Google Scholar] [CrossRef]
- Nie, S.; Zhang, J.; Martinez-Zaguilán, R.; Sennoune, S.; Hossen, M.N.; Lichtenstein, A.H.; Cao, J.; Meyerrose, G.E.; Paone, R.; Soontrapa, S.; et al. Detection of atherosclerotic lesions and intimal macrophages using CD36-targeted nanovesicles. J. Control. Release 2015, 220, 61–70. [Google Scholar] [CrossRef]
- Zhang, J.; Nie, S.; Zu, Y.; Abbasi, M.; Cao, J.; Li, C.; Wu, D.; Labib, S.; Brackee, G.; Shen, C.-L.; et al. Anti-atherogenic effects of CD36-targeted epigallocatechin gallate-loaded nanoparticles. J. Control. Release 2019, 303, 263–273. [Google Scholar] [CrossRef]
- Rodrigues, S.F.; Granger, D.N. Blood cells and endothelial barrier function. Tissue Barriers 2015, 3, e978720. [Google Scholar] [CrossRef]
- Peng, Z.; Shu, B.; Zhang, Y.; Wang, M. Endothelial Response to Pathophysiological Stress. Arter. Thromb. Vasc. Biol. 2019, 39, e233–e243. [Google Scholar] [CrossRef]
- Claesson-Welsh, L.; Dejana, E.; McDonald, D.M. Permeability of the Endothelial Barrier: Identifying and Reconciling Controversies. Trends Mol. Med. 2021, 27, 314–331. [Google Scholar] [CrossRef] [PubMed]
- Baumer, Y.; McCurdy, S.; Weatherby, T.M.; Mehta, N.N.; Halbherr, S.; Halbherr, P.; Yamazaki, N.; Boisvert, W.A. Hyperlipidemia-induced cholesterol crystal production by endothelial cells promotes atherogenesis. Nat. Commun. 2017, 8, 1129. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.; Azzam, K.M.; Lin, W.-C.; Rai, P.; Lowe, J.M.; Gabor, K.A.; Madenspacher, J.H.; Aloor, J.J.; Parks, J.S.; Näär, A.M.; et al. MicroRNA-33 Regulates the Innate Immune Response via ATP Binding Cassette Transporter-mediated Remodeling of Membrane Microdomains. J. Biol. Chem. 2016, 291, 19651–19660. [Google Scholar] [CrossRef] [PubMed]
- Esobi, I.; Olanrewaju, O.; Echesabal-Chen, J.; Stamatikos, A. Utilizing the LoxP-Stop-LoxP System to Control Transgenic ABC-Transporter Expression In Vitro. Biomolecules 2022, 12, 679. [Google Scholar] [CrossRef]
- Heinecke, J.W. Small HDL promotes cholesterol efflux by the ABCA1 pathway in macrophages: Implications for therapies targeted to HDL. Circ. Res. 2015, 116, 1101–1103. [Google Scholar] [CrossRef] [PubMed]
- Du, X.-M.; Kim, M.-J.; Hou, L.; Le Goff, W.; Chapman, M.J.; Van Eck, M.; Curtiss, L.K.; Burnett, J.R.; Cartland, S.P.; Quinn, C.M.; et al. HDL Particle Size Is a Critical Determinant of ABCA1-Mediated Macrophage Cellular Cholesterol Export. Circ. Res. 2015, 116, 1133–1142. [Google Scholar] [CrossRef]
- Feig, J.E.; Fisher, E.A. Laser Capture Microdissection for Analysis of Macrophage Gene Expression from Atherosclerotic Lesions. Laser Capt. Microdissect. Methods Protoc. 2013, 1027, 123–135. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Echesabal-Chen, J.; Huang, K.; Vojtech, L.; Oladosu, O.; Esobi, I.; Sachdeva, R.; Vyavahare, N.; Jo, H.; Stamatikos, A. Constructing Lipoparticles Capable of Endothelial Cell-Derived Exosome-Mediated Delivery of Anti-miR-33a-5p to Cultured Macrophages. Curr. Issues Mol. Biol. 2023, 45, 5631-5644. https://doi.org/10.3390/cimb45070355
Echesabal-Chen J, Huang K, Vojtech L, Oladosu O, Esobi I, Sachdeva R, Vyavahare N, Jo H, Stamatikos A. Constructing Lipoparticles Capable of Endothelial Cell-Derived Exosome-Mediated Delivery of Anti-miR-33a-5p to Cultured Macrophages. Current Issues in Molecular Biology. 2023; 45(7):5631-5644. https://doi.org/10.3390/cimb45070355
Chicago/Turabian StyleEchesabal-Chen, Jing, Kun Huang, Lucia Vojtech, Olanrewaju Oladosu, Ikechukwu Esobi, Rakesh Sachdeva, Naren Vyavahare, Hanjoong Jo, and Alexis Stamatikos. 2023. "Constructing Lipoparticles Capable of Endothelial Cell-Derived Exosome-Mediated Delivery of Anti-miR-33a-5p to Cultured Macrophages" Current Issues in Molecular Biology 45, no. 7: 5631-5644. https://doi.org/10.3390/cimb45070355
APA StyleEchesabal-Chen, J., Huang, K., Vojtech, L., Oladosu, O., Esobi, I., Sachdeva, R., Vyavahare, N., Jo, H., & Stamatikos, A. (2023). Constructing Lipoparticles Capable of Endothelial Cell-Derived Exosome-Mediated Delivery of Anti-miR-33a-5p to Cultured Macrophages. Current Issues in Molecular Biology, 45(7), 5631-5644. https://doi.org/10.3390/cimb45070355




