A Medicago truncatula Autoregulation of Nodulation Mutant Transcriptome Analysis Reveals Disruption of the SUNN Pathway Causes Constitutive Expression Changes in Some Genes, but Overall Response to Rhizobia Resembles Wild-Type, Including Induction of TML1 and TML2
Abstract
1. Introduction
2. Materials and Methods
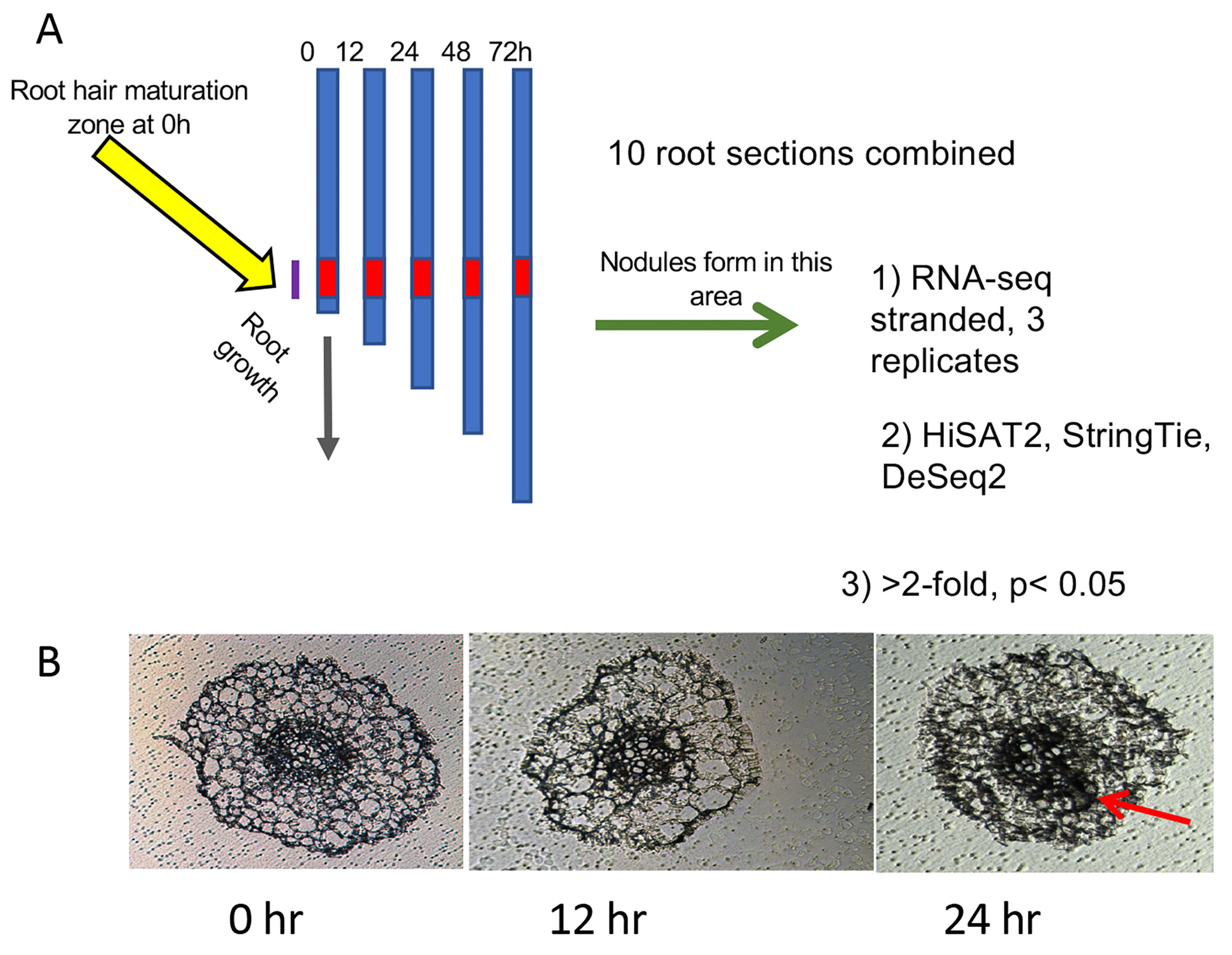
2.1. Plant Growth and Tissue Sampling
2.2. RNA Preparation, Libraries, and Sequencing
2.3. Analysis of Gene Expression
- Functional enrichment analysis was performed with the Medicago Classification Superviewer (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer_medicago.cgi accessed multiple times in July 2020) using the default settings and a significance threshold of p < 0.05 [18].
3. Results
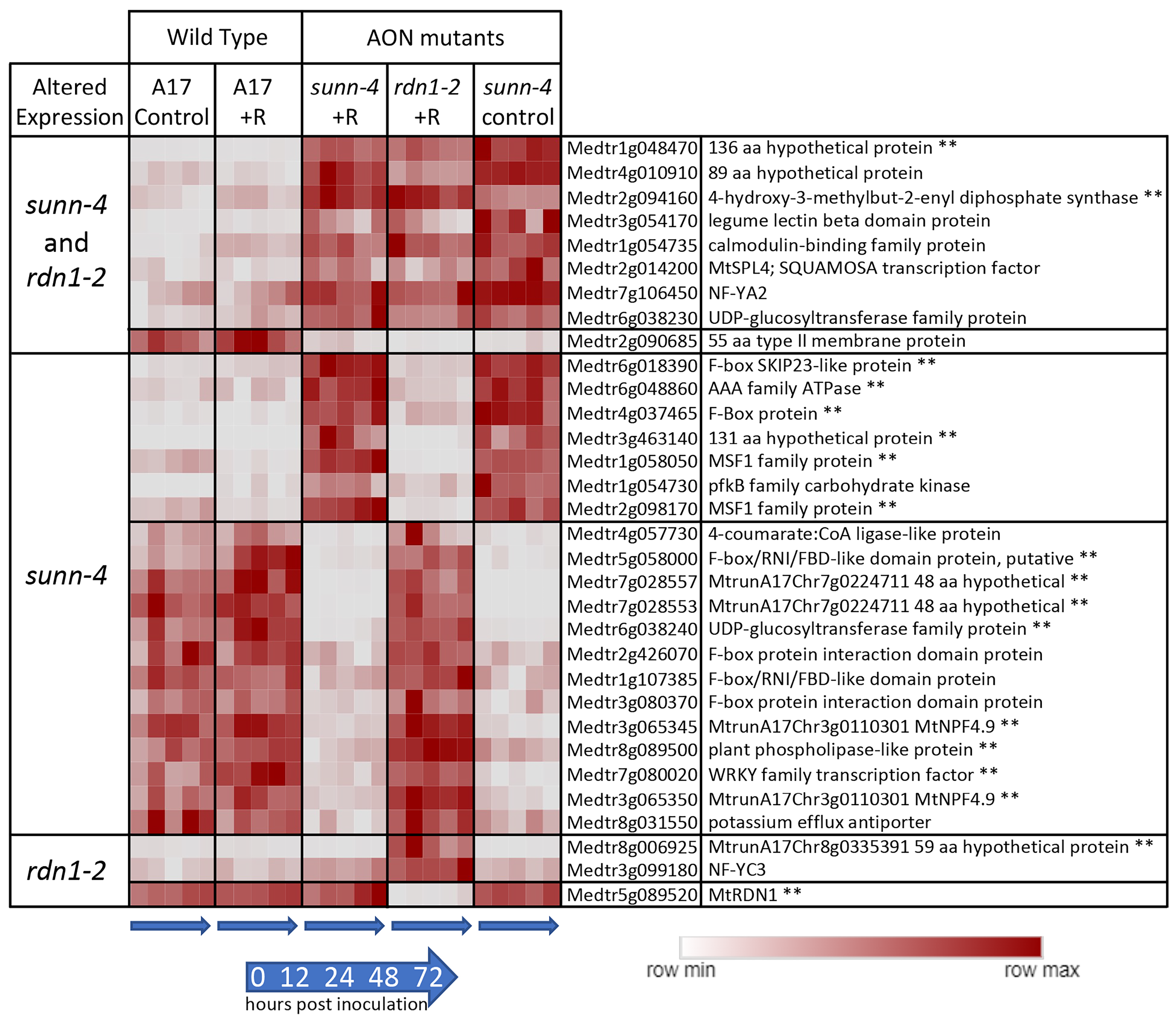
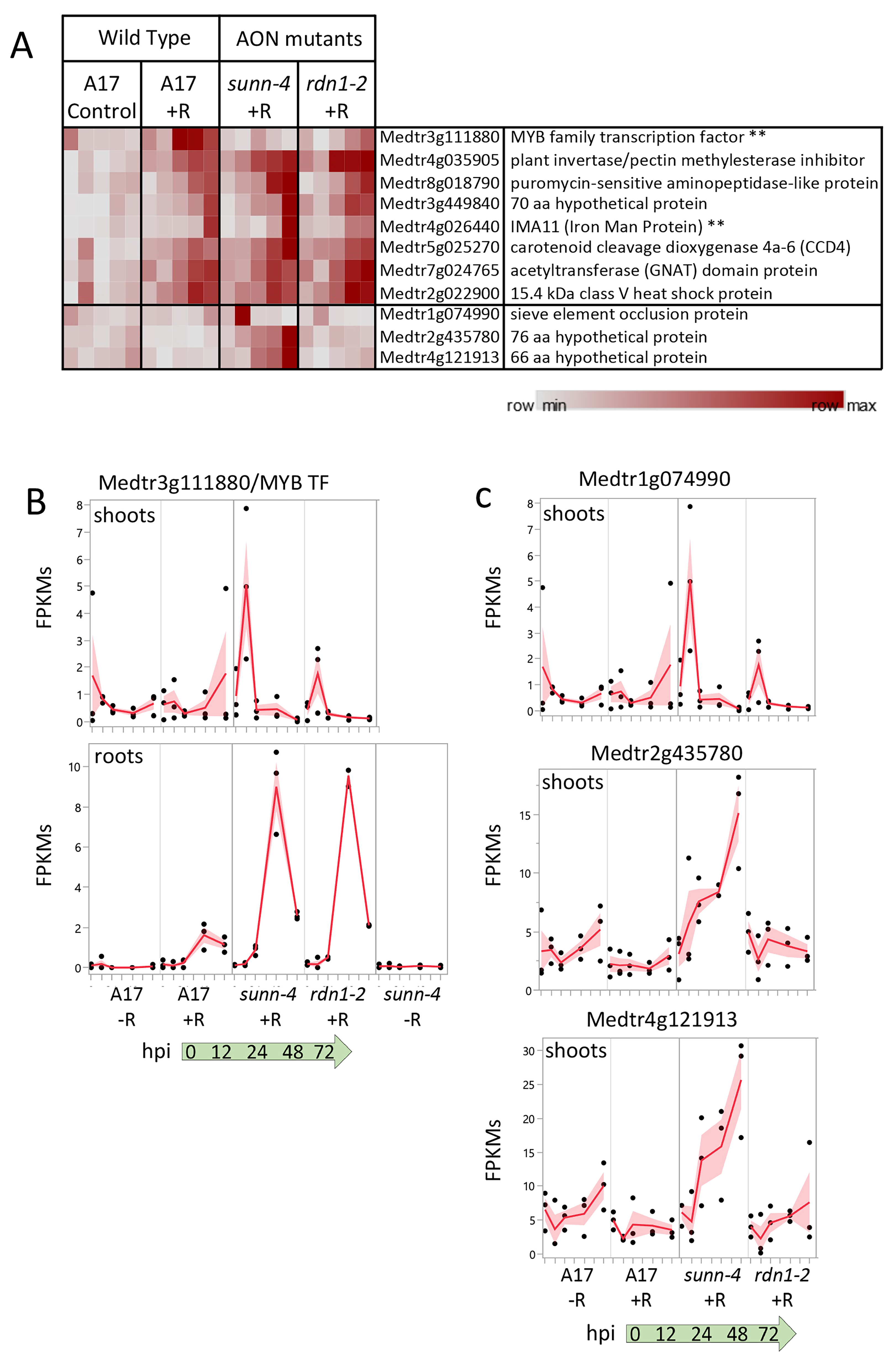
3.1. Constitutively Altered Gene Expression in sunn-4 Roots and Shoots
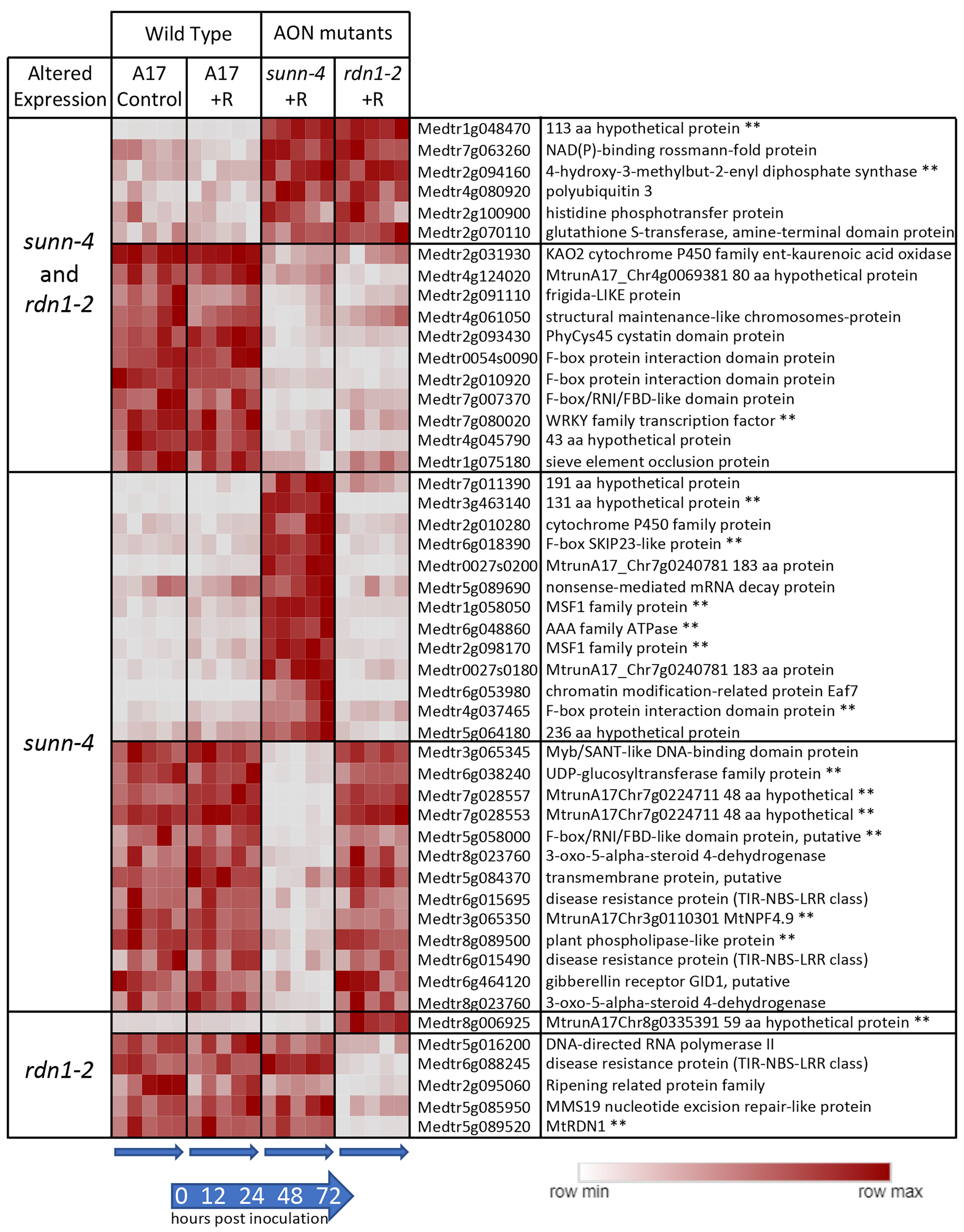
3.2. Response of Genes to Rhizobia in Roots in Wild-Type and sunn-4
3.2.1. Nodulation Pathway Genes
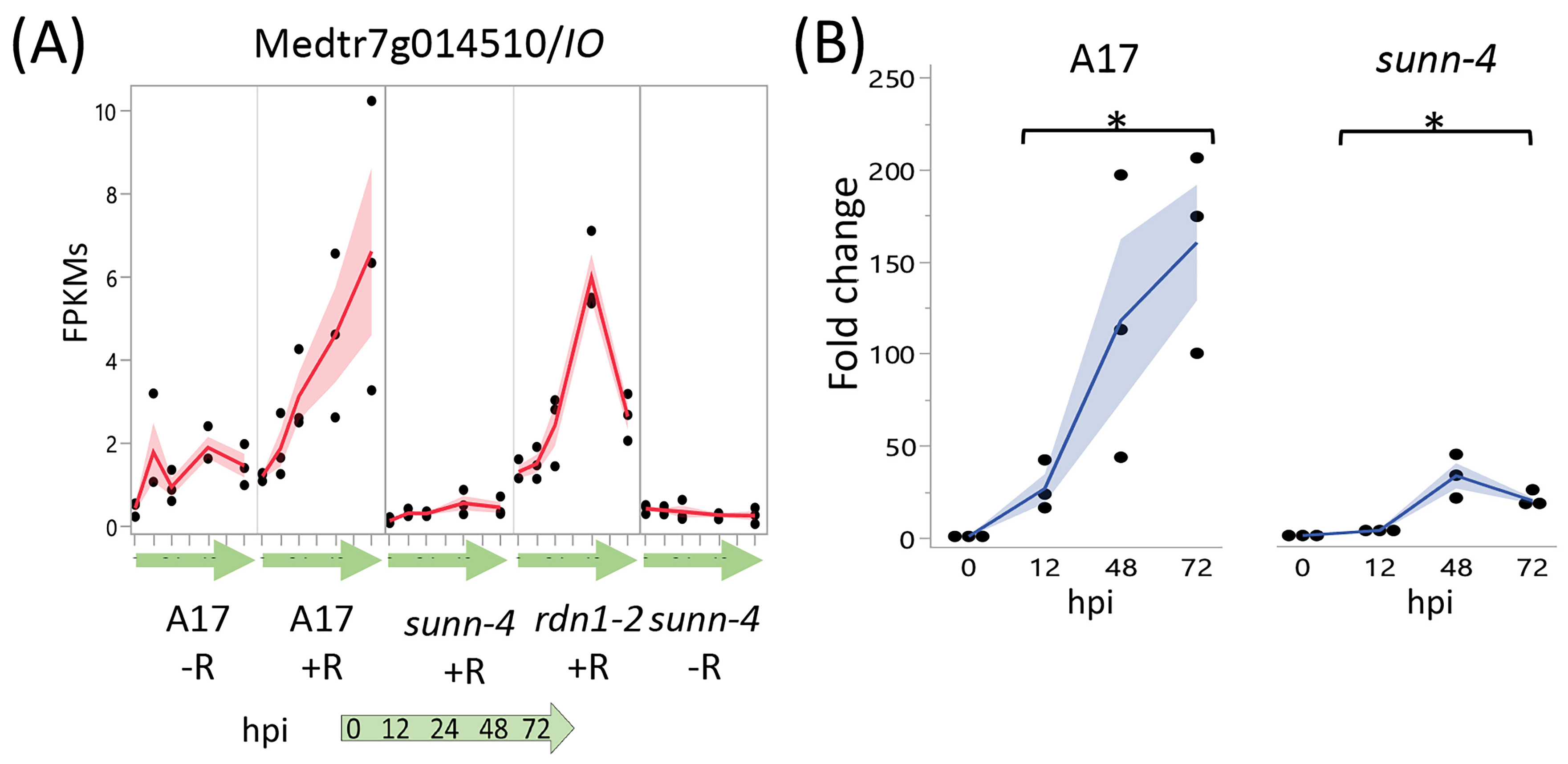
3.2.2. TMLs
3.2.3. Genes Unresponsive to Rhizobia in sunn-4 Mutants
3.2.4. Induction of Small Signaling Peptide Genes in Roots
3.3. Rhizobial Response in Shoots
4. Discussion
4.1. Expression Differences in AON Mutants
4.2. Peptide Responses to Rhizobia
4.3. Rhizobial Response in AON Mutants Includes Induction of TML Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Mens, C.; Hastwell, A.H.; Zhang, M.; Su, H.; Jones, C.H.; Chu, X.; Gresshoff, P.M. Legume nodulation: The host controls the party. Plant Cell Environ. 2019, 42, 41–51. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef]
- Gage, D.J. Infection and Invasion of Roots by Symbiotic, Nitrogen-Fixing Rhizobia during Nodulation of Temperate Legumes. Microbiol. Mol. Biol. Rev. 2004, 68, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Crawford, N.M.; Kahn, M.L.; Leustek, T.; Long, S.R. Nitrogen and sulfur. In Biochemistry and Molecular Biology of Plants; Buchanan, B.B., Gruissem, W., Jones, R.L., Eds.; American Association of Plant Physiologists: Rockville, MD, USA, 2000; pp. 787–849. [Google Scholar]
- Chaulagain, D.; Frugoli, J. The Regulation of Nodule Number in Legumes Is a Balance of Three Signal Transduction Pathways. Int. J. Mol. Sci. 2021, 22, 1171. [Google Scholar] [CrossRef] [PubMed]
- Kassaw, T.K.; Frugoli, J.A. Simple and efficient methods to generate split roots and grafted plants useful for long-distance signaling studies in Medicago truncatula and other small plants. Plant Methods 2012, 8, 38. [Google Scholar] [CrossRef]
- Müller, L.M.; Flokova, K.; Schnabel, E.; Sun, X.; Fei, Z.; Frugoli, J.; Bouwmeester, H.J.; Harrison, M.J. A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat. Plants 2019, 5, 933–939. [Google Scholar] [CrossRef]
- Penmetsa, R.V.; Frugoli, J.; Smith, L.; Long, S.R.; Cook, D. Genetic evidence for dual pathway control of nodule number in Medicago truncatula. Plant Physiol. 2003, 131, 998–1008. [Google Scholar] [CrossRef]
- Schnabel, E.; Journet, E.P.; Carvalho-Niebel, F.; Duc, G.; Frugoli, J. The Medicago truncatula SUNN gene encoding a CLV1-like leucine-rich repeat receptor kinase regulates both nodule number and root length. Plant Mol. Biol. 2005, 58, 809–822. [Google Scholar] [CrossRef]
- Schnabel, E.; Mukherjee, A.; Smith, L.; Kassaw, T.; Long, S.; Frugoli, J. The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiol. 2010, 154, 1390–1402. [Google Scholar] [CrossRef]
- Kassaw, T.; Nowak, S.; Schnabel, E.; Frugoli, J. ROOT DETERMINED NODULATION1 Is Required for M. truncatula CLE12, But Not CLE13, Peptide Signaling through the SUNN Receptor Kinase. Plant Physiol. 2017, 174, 2445–2456. [Google Scholar] [CrossRef]
- Schnabel, E.; Kassaw, T.; Smith, L.; Marsh, J.; Oldroyd, G.; Long, S.; Frugoli, J. The ROOT DETERMINED NODULATION 1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol. 2011, 157, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Poehlman, W.L.; Schnabel, E.L.; Chavan, S.A.; Frugoli, J.A.; Feltus, F.A. Identifying Temporally Regulated Root Nodulation Biomarkers Using Time Series Gene Co-Expression Network Analysis. Front. Plant Sci. 2019, 10, 1409. [Google Scholar] [CrossRef]
- Cai, J.; Veerappan, V.; Arildsen, K.; Sullivan, C.; Piechowicz, M.; Frugoli, J.; Dickstein, R. A Modified Aeroponic System for Growing Plants to Study Root Systems. Plant Methods 2023, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Gautrat, P.; Mortier, V.; Laffont, C.; De Keyser, A.; Fromentin, J.; Frugier, F.; Goormachtig, S. Unraveling new molecular players involved in the autoregulation of nodulation in Medicago truncatula. J. Exp. Bot. 2019, 70, 1407–1417. [Google Scholar] [CrossRef]
- Gao, Y.; Selee, B.; Schnabel, E.L.; Poehlman, W.L.; Chavan, S.A.; Frugoli, J.A.; Feltus, F.A. Time Series Transcriptome Analysis in Medicago truncatula Shoot and Root Tissue During Early Nodulation. Front. Plant Sci. 2022, 13, 861639. [Google Scholar] [CrossRef] [PubMed]
- Herrbach, V.; Chirinos, X.; Rengel, D.; Agbevenou, K.; Vincent, R.; Pateyron, S.; Huguet, S.; Balzergue, S.; Pasha, A.; Provart, N. Nod factors potentiate auxin signaling for transcriptional regulation and lateral root formation in Medicago truncatula. J. Exp. Bot. 2017, 68, 569–583. [Google Scholar]
- Laloum, T.; Baudin, M.; Frances, L.; Lepage, A.; Billault-Penneteau, B.; Cerri, M.R.; Ariel, F.; Jardinaud, M.F.; Gamas, P.; de Carvalho-Niebel, F. Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 2014, 79, 757–768. [Google Scholar] [CrossRef]
- Baudin, M.; Laloum, T.; Lepage, A.; Ripodas, C.; Ariel, F.; Frances, L.; Crespi, M.; Gamas, P.; Blanco, F.A.; Zanetti, M.E.; et al. A phylogenetically conserved group of NF-Y transcription factors interact to control nodulation in legumes. Plant Physiol. 2015, 169, 2761–2773. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [PubMed]
- Wang, Y.; Wang, Z.S.; Amyot, L.; Tian, L.N.; Xu, Z.Q.; Gruber, M.Y.; Hannoufa, A. Ectopic expression of miR156 represses nodulation and causes morphological and developmental changes in Lotus japonicus. Mol. Genet. Genom. 2015, 290, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Gavrin, A.; Kulikova, O.; Bisseling, T.; Fedorova, E.E. Interface Symbiotic Membrane Formation in Root Nodules of Medicago truncatula: The Role of Synaptotagmins MtSyt1, MtSyt2 and MtSyt3. Front. Plant Sci. 2017, 8, 201. [Google Scholar] [CrossRef] [PubMed]
- Pislariu, C.I.; Sinharoy, S.; Torres-Jerez, I.; Nakashima, J.; Blancaflor, E.B.; Udvardi, M.K. The nodule-specific PLAT domain protein NPD1 is required for nitrogen-fixing symbiosis. Plant Physiol. 2019, 180, 1480–1497. [Google Scholar] [CrossRef]
- Franssen, H.J.; Xiao, T.T.; Kulikova, O.; Wan, X.; Bisseling, T.; Scheres, B.; Heidstra, R. Root developmental programs shape the Medicago truncatula nodule meristem. Development 2015, 142, 2941–2950. [Google Scholar] [PubMed]
- Okamoto, S.; Ohnishi, E.; Sato, S.; Takahashi, H.; Nakazono, M.; Tabata, S.; Kawaguchi, M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009, 50, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Mortier, V.; Den Herder, G.; Whitford, R.; Van de Velde, W.; Rombauts, S.; D’Haeseleer, K.; Holsters, M.; Goormachtig, S. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010, 153, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Boschiero, C.; Dai, X.; Lundquist, P.K.; Roy, S.; Christian De Bang, T.; Zhang, S.; Zhuang, Z.; Torres-Jerez, I.; Udvardi, M.K.; Scheible, W.-R.; et al. MtSSPdb: The Medicago truncatula Small Secreted Peptide Database. Plant Physiol. 2020, 183, 399–413. [Google Scholar] [CrossRef]
- Grillet, L.; Lan, P.; Li, W.; Mokkapati, G.; Schmidt, W. IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 2018, 4, 953–963. [Google Scholar] [CrossRef]
- Van de Velde, W.; Zehirov, G.; Szatmari, A.; Debreczeny, M.; Ishihara, H.; Kevei, Z.; Farkas, A.; Mikulass, K.; Nagy, A.; Tiricz, H. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 2010, 327, 1122–1126. [Google Scholar] [CrossRef]
- Larrainzar, E.; Riely, B.K.; Kim, S.C.; Carrasquilla-Garcia, N.; Yu, H.-J.; Hwang, H.-J.; Oh, M.; Kim, G.B.; Surendrarao, A.K.; Chasman, D.; et al. Deep Sequencing of the Medicago truncatula Root Transcriptome Reveals a Massive and Early Interaction between Nodulation Factor and Ethylene Signals. Plant Physiol. 2015, 169, 233–265. [Google Scholar] [CrossRef]
- Schiessl, K.; Lilley, J.L.S.; Lee, T.; Tamvakis, I.; Kohlen, W.; Bailey, P.C.; Thomas, A.; Luptak, J.; Ramakrishnan, K.; Carpenter, M.D.; et al. NODULE INCEPTION Recruits the Lateral Root Developmental Program for Symbiotic Nodule Organogenesis in Medicago truncatula. Curr. Biol. 2019, 29, 3657–3668.e5. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, E.; Smith, L.; Long, S.; Frugoli, J. Transcript profiling in M. truncatula lss and sunn-1 mutants reveals different expression profiles despite disrupted SUNN gene function in both mutants. Plant Signal. Behav. 2010, 5, 1657–1659. [Google Scholar] [CrossRef]
- Tarkowská, D.; Strnad, M. Isoprenoid-derived plant signaling molecules: Biosynthesis and biological importance. Planta 2018, 247, 1051–1066. [Google Scholar] [CrossRef]
- Deavours, B.E.; Liu, C.-J.; Naoumkina, M.A.; Tang, Y.; Farag, M.A.; Sumner, L.W.; Noel, J.P.; Dixon, R.A. Functional analysis of members of the isoflavone and isoflavanone O-methyltransferase enzyme families from the model legume Medicago truncatula. Plant Mol. Biol. 2006, 62, 715–733. [Google Scholar] [CrossRef] [PubMed]
- Paiva, N.L.; Oommen, A.; Harrison, M.J.; Dixon, R.A. Regulation of isoflavonoid metabolism in alfalfa. In Primary and Secondary Metabolism of Plants and Cell Cultures III: Proceedings of the Workshop Held in Leiden, The Netherlands, 4–7 April 1993; Springer: Berlin/Heidelberg, Germany, 1995. [Google Scholar]
- He, X.Z.; Dixon, R.A. Genetic manipulation of isoflavone 7-O-methyltransferase enhances biosynthesis of 4’-O-methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell 2000, 12, 1689–1702. [Google Scholar] [PubMed]
- Subramanian, S.; Stacey, G.; Yu, O. Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhisobium japonicum. Plant J. 2006, 48, 261–273. [Google Scholar] [CrossRef] [PubMed]
- van Noorden, G.E.; Ross, J.J.; Reid, J.B.; Rolfe, B.J.; Mathesius, U. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 2006, 140, 1494–1506. [Google Scholar] [CrossRef] [PubMed]
- de Bang, T.C.; Lundquist, P.K.; Dai, X.; Boschiero, C.; Zhuang, Z.; Pant, P.; Torres-Jerez, I.; Roy, S.; Nogales, J.; Veerappan, V. Genome-wide identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 2017, 175, 1669–1689. [Google Scholar] [CrossRef]
- Jeon, B.W.; Kim, M.-J.; Pandey, S.K.; Oh, E.; Seo, P.J.; Kim, J. Recent advances in peptide signaling during Arabidopsis root development. J. Exp. Bot. 2021, 72, 2889–2902. [Google Scholar] [CrossRef]
- Kim, J.S.; Jeon, B.W.; Kim, J. Signaling peptides regulating abiotic stress responses in plants. Front. Plant Sci. 2021, 12, 704490. [Google Scholar] [CrossRef] [PubMed]
- Guefrachi, I.; Nagymihaly, M.; Pislariu, C.I.; Van de Velde, W.; Ratet, P.; Mars, M.; Udvardi, M.K.; Kondorosi, E.; Mergaert, P.; Alunni, B. Extreme specificity of NCR gene expression in Medicago truncatula. BMC Genom. 2014, 15, 712. [Google Scholar] [CrossRef]
- Maróti, G.; Downie, J.A.; Kondorosi, É. Plant cysteine-rich peptides that inhibit pathogen growth and control rhizobial differentiation in legume nodules. Curr. Opin. Plant Biol. 2015, 26, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Jardinaud, M.F.; Boivin, S.; Rodde, N.; Catrice, O.; Kisiala, A.; Lepage, A.; Moreau, S.; Roux, B.; Cottret, L.; Sallet, E.; et al. A Laser Dissection-RNAseq Analysis Highlights the Activation of Cytokinin Pathways by Nod Factors in the Medicago truncatula Root Epidermis. Plant Physiol. 2016, 171, 2256–2276. [Google Scholar] [CrossRef] [PubMed]
- Mergaert, P.; Kereszt, A.; Kondorosi, E. Gene Expression in Nitrogen-Fixing Symbiotic Nodule Cells in Medicago truncatula and Other Nodulating Plants. Plant Cell 2020, 32, 46–68. [Google Scholar] [CrossRef]
- Gautrat, P.; Laffont, C.; Frugier, F. Compact Root Architecture 2 Promotes Root Competence for Nodulation through the miR2111 Systemic Effector. Curr. Biol. 2020, 30, 1339–1345.e3. [Google Scholar] [CrossRef] [PubMed]
- Moreau, C.; Gautrat, P.; Frugier, F. Nitrate-induced CLE35 signaling peptides inhibit nodulation through the SUNN receptor and miR2111 repression. Plant Physiol. 2021, 185, 1216–1228. [Google Scholar] [CrossRef]
- Chaulagain, D. Genetic and miRNA Transcriptomic Analysis of Autoregulation of Nodulation Signaling in Medicago truncatula. Ph.D. Thesis, Clemson University, Clemson, SC, USA, December 2020. TigerPrints. Available online: https://tigerprints.clemson.edu/all_dissertations/3252/ (accessed on 4 April 2023).
- Okuma, N.; Kawaguchi, M. Systemic optimization of legume nodulation: A shoot-derived regulator, miR2111. Front. Plant Sci. 2021, 12, 682486. [Google Scholar] [CrossRef]


| Peptide Family | Induced Genes /Total Genes (v4 Genome) | Increase 12 hpi | Increase 24 hpi | Increase 48–72 hpi | Peptide Family Description |
|---|---|---|---|---|---|
| BBPI | 1/16 | BBPI16 | Bowman–Birk Peptidase Inhibitor | ||
| CAPE | 4/21 | CAPE1 | CAPE2, CAPE16, CAPE18 | CAP-derived Peptide | |
| CEP | 1/10 | CEP14 | C-terminally Encoded Peptide | ||
| CLE | 10/46 | CLE13, CLE53 | CLE29, CLE35 | CLE12, CLE34, CLE37, CLE41, CLE44, CLE45 | Clavata/Embryo Surrounding Region |
| EPFL | 5/21 | EPFL1, EPFL14, EPFL19, EPFL9, | Epidermal Patterning Factor-Like | ||
| GASA | 4/28 | GASA25 | GASA17, GASA22, GASA29 | Gibberellic Acid Stimulated in Arabidopsis | |
| GLV | 3/15 | GLV9, GLV10 | GLV8 | Golven/Root Growth Factor | |
| IDA | 1/38 | IDA15 | Inflorescence Deficient in Abscission | ||
| IMA | 14/14 | IMA1, IMA2, IMA3, IMA5, IMA6, IMA7, IMA8, IMA9, IMA10, IMA11, IMA12, IMA13, IMA14, IMA15 | Iron Man | ||
| Kunitz | 2/48 | Kunitz13, Kunitz18 | Kunitz-P trypsin inhibitor | ||
| LAT52-POE | 3/40 | LAT52/POE1, LAT52/POE12 | LAT52/POE21 | LAT52/Pollen Ole e 1 Allergen | |
| LCR | 1/89 | LCR64 | Low-molecular weight Cys-rich | ||
| Legin | 8/48 | Legin20 | Legin32, Legin37, Legin38, Legin42, Legin43, Legin44, Legin47 | Leginsulin | |
| LP | 4/20 | LP8, LP9, LP14, LP15 | LEED..PEED | ||
| N26 | 2/4 | N26-3, N26-4 | Nodulin26 | ||
| NCR-A | 13/327 | NCR025, NCR037, NCR267, NCR279, NCR323, NCR376, NCR396, NCR547, NCR639, NCR685/NCR686 | Nodule-Specific Cysteine RichGroup A | ||
| NCR-B | 36/365 | NCR150 | NCR031, NCR051, NCR057, NCR117, NCR157, NCR158, NCR209, NCR223, NCR229, NCR235, NCR252, NCR308, NCR386, NCR415, NCR454, NCR455, NCR465, NCR507, NCR527, NCR529, NCR567, NCR568, NCR573, NCR648, NCR657, NCR673, NCR678, NCR708, NCR713, NCR730, NCR736, NCR737, NCR738, NCR757, NCR793 | Nodule-Specific Cysteine RichGroup B | |
| NodGRP | 14/54 | NodGRP15, NodGRP45 | NodGRP1B, NodGRP3C, NodGRP4, NodGRP12, NodGRP23, NodGRP30, NodGRP32, NodGRP33, NodGRP34, NodGRP35, NodGRP36 | Nodule-Specific Glycine-rich Protein | |
| nsLTP | 18/132 | nsLTP53, nsLTP61, nsLTP62 | nsLTP100 | nsLTP25, nsLTP49, nsLTP50, nsLTP51, nsLTP52, nsLTP54, nsLTP72, nsLTP75, nsLTP76, nsLTP81, nsLTP83, nsLTP84, nsLTP102, nsLTP110 | Non-Specific Lipid Transfer Protein |
| PCY | 11/86 | PCY16, PCY27, PCY33 | PCY47, PCY59 | PCY19, PCY35, PCY64, PCY68, PCY72, PCY78 | Plantcyanin/Chemocyanin |
| 16/16 | PDF5, PDF6, PDF7, PDF10, PDF13, PDF14, PDF39 | PDF2, PDF9, PDF11, PDF27, PDF36, PDF38, PDF44, PDF45, PDF57 | Plant Defensin | ||
| PSK | 1/10 | PSK8 | Phytosulfokine | ||
| RTFL/DVL | 2/15 | RTFL/DVL1 | RTFL/DVL13 | Rotundifolia/Devil | |
| STIG-GRI | 1/18 | STIG/GRI4 | Stigma1/GRI |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schnabel, E.L.; Chavan, S.A.; Gao, Y.; Poehlman, W.L.; Feltus, F.A.; Frugoli, J.A. A Medicago truncatula Autoregulation of Nodulation Mutant Transcriptome Analysis Reveals Disruption of the SUNN Pathway Causes Constitutive Expression Changes in Some Genes, but Overall Response to Rhizobia Resembles Wild-Type, Including Induction of TML1 and TML2. Curr. Issues Mol. Biol. 2023, 45, 4612-4631. https://doi.org/10.3390/cimb45060293
Schnabel EL, Chavan SA, Gao Y, Poehlman WL, Feltus FA, Frugoli JA. A Medicago truncatula Autoregulation of Nodulation Mutant Transcriptome Analysis Reveals Disruption of the SUNN Pathway Causes Constitutive Expression Changes in Some Genes, but Overall Response to Rhizobia Resembles Wild-Type, Including Induction of TML1 and TML2. Current Issues in Molecular Biology. 2023; 45(6):4612-4631. https://doi.org/10.3390/cimb45060293
Chicago/Turabian StyleSchnabel, Elise L., Suchitra A. Chavan, Yueyao Gao, William L. Poehlman, Frank Alex Feltus, and Julia A. Frugoli. 2023. "A Medicago truncatula Autoregulation of Nodulation Mutant Transcriptome Analysis Reveals Disruption of the SUNN Pathway Causes Constitutive Expression Changes in Some Genes, but Overall Response to Rhizobia Resembles Wild-Type, Including Induction of TML1 and TML2" Current Issues in Molecular Biology 45, no. 6: 4612-4631. https://doi.org/10.3390/cimb45060293
APA StyleSchnabel, E. L., Chavan, S. A., Gao, Y., Poehlman, W. L., Feltus, F. A., & Frugoli, J. A. (2023). A Medicago truncatula Autoregulation of Nodulation Mutant Transcriptome Analysis Reveals Disruption of the SUNN Pathway Causes Constitutive Expression Changes in Some Genes, but Overall Response to Rhizobia Resembles Wild-Type, Including Induction of TML1 and TML2. Current Issues in Molecular Biology, 45(6), 4612-4631. https://doi.org/10.3390/cimb45060293






