Methionine Sources Differently Affect Production of Reactive Oxygen Species, Mitochondrial Bioenergetics, and Growth of Murine and Quail Myoblasts In Vitro
Abstract
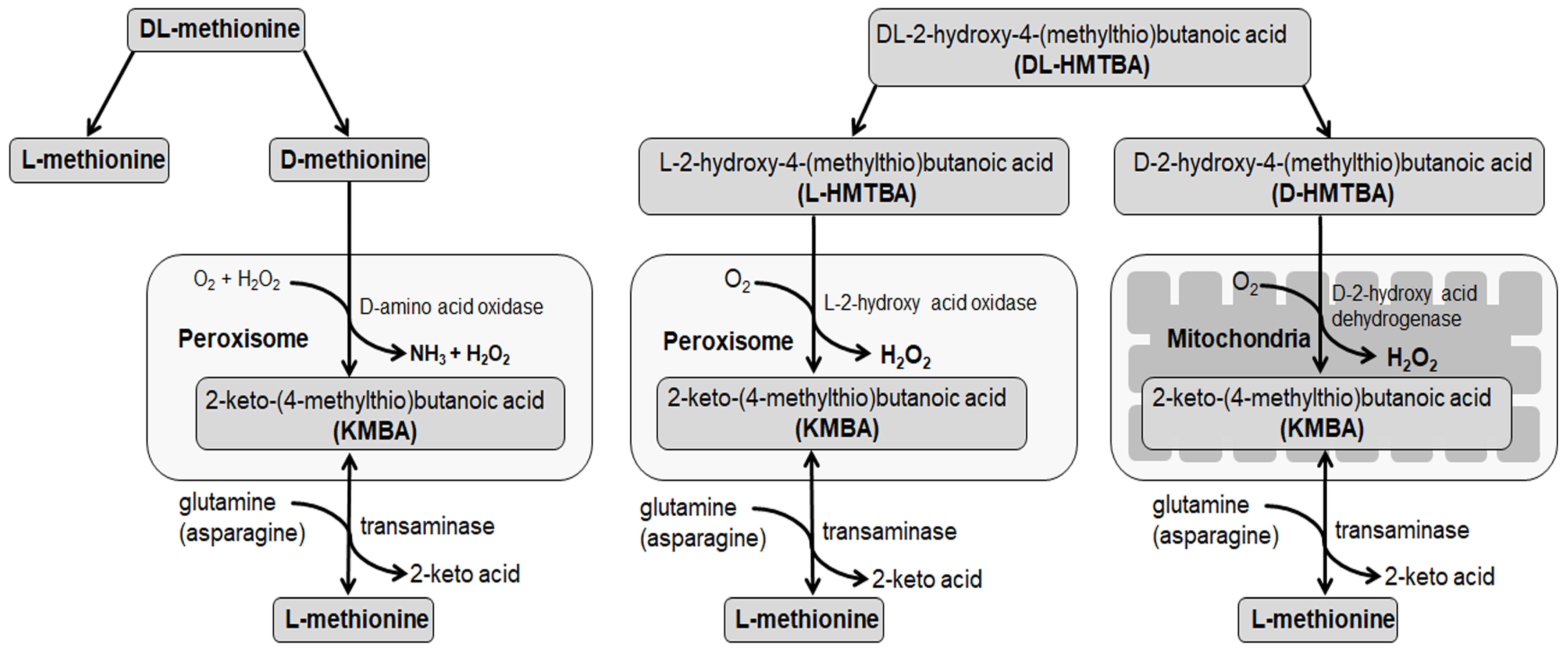
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
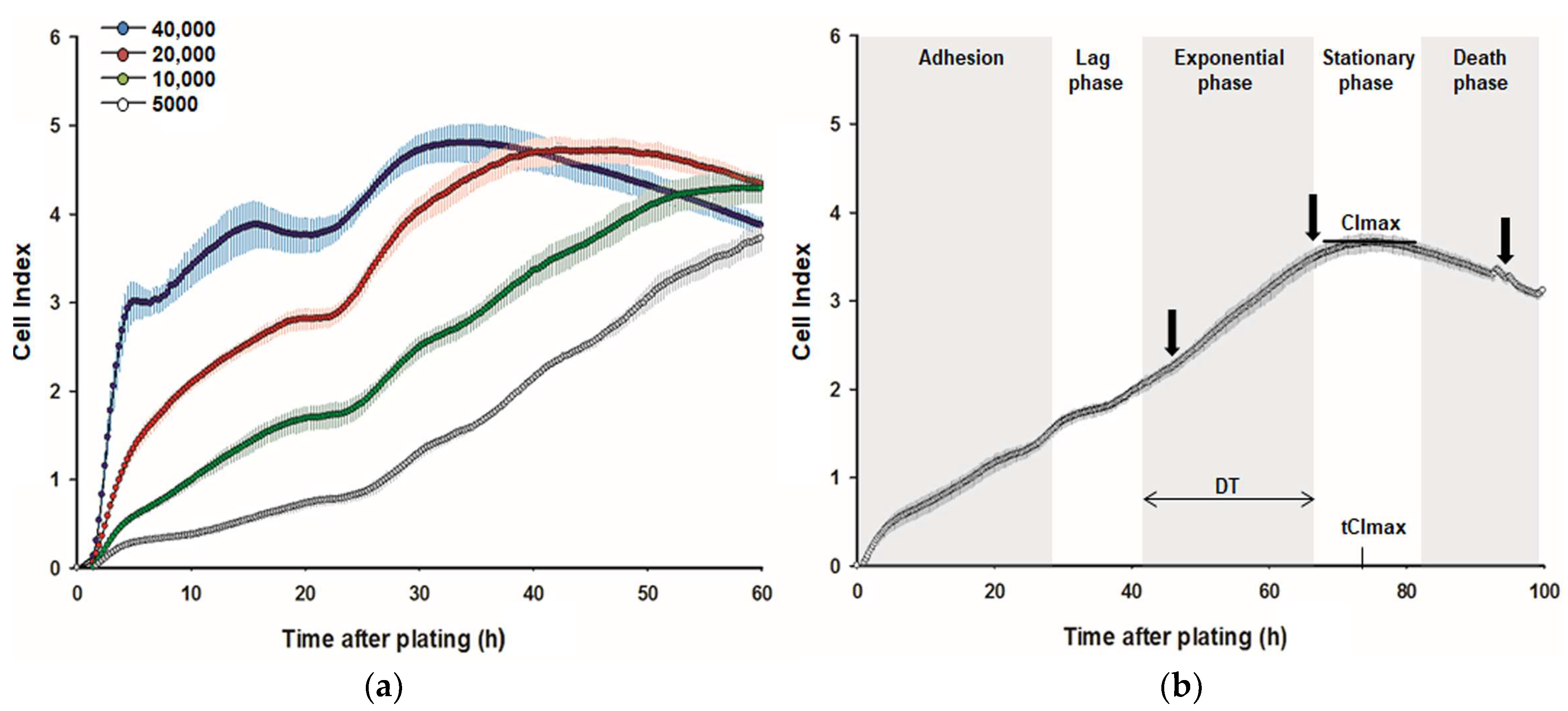
2.2. Impedance-Based Recording and Analysis of Real-Time Kinetic Growth Curves
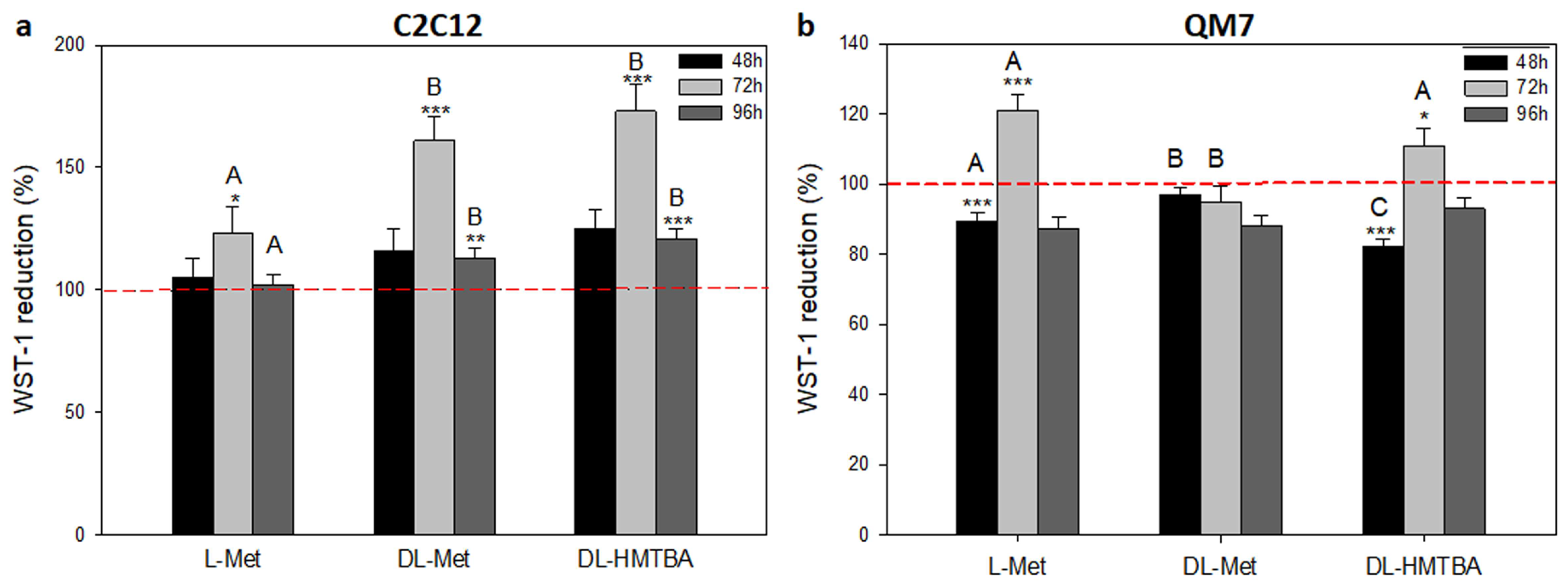
2.3. Assessment of Cell Viability and Metabolic Activity
2.4. Determination of DNA and Protein Concentration
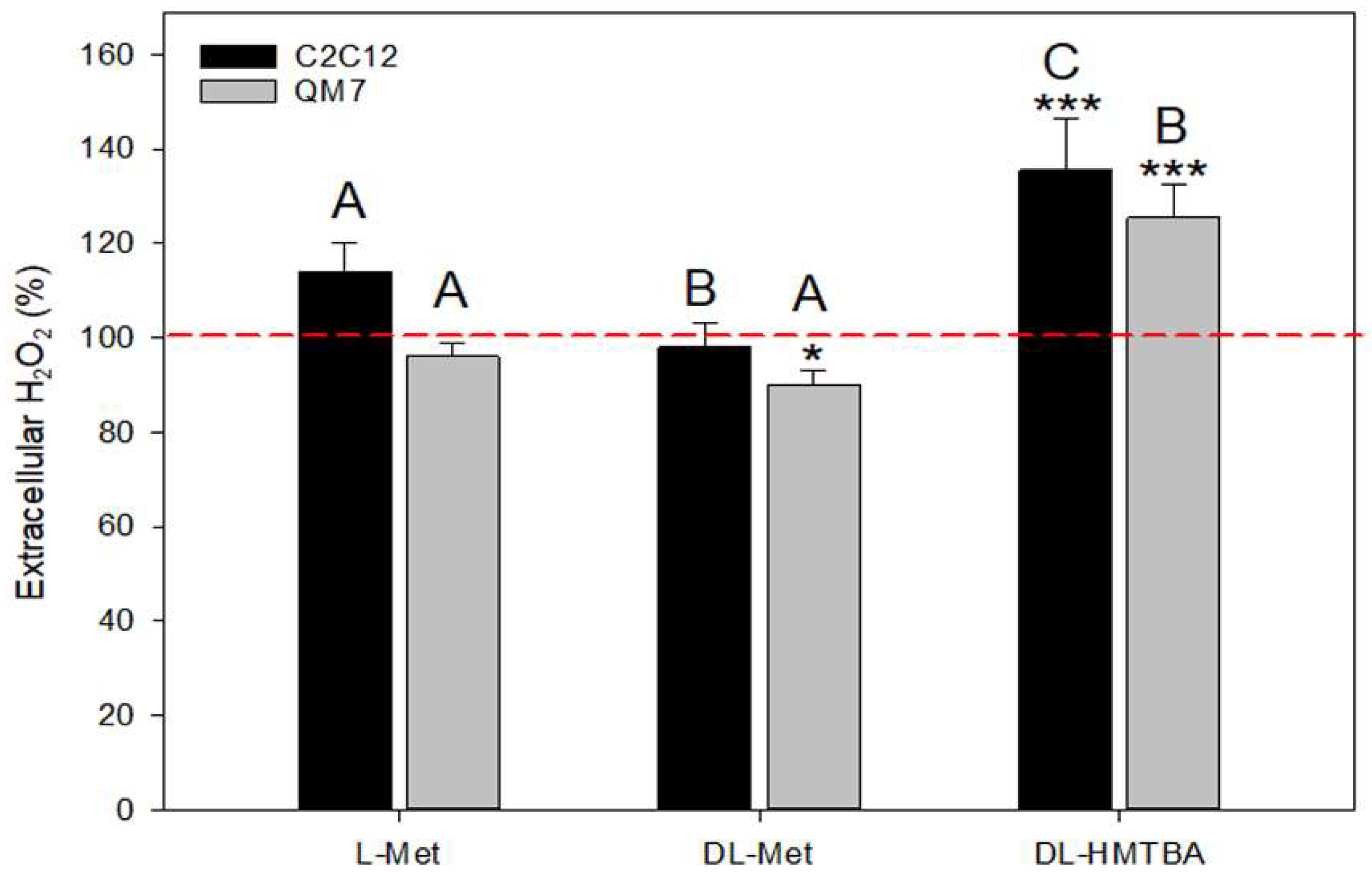
2.5. Hydrogen Peroxide Assay
2.6. Determination of Oxygen Consumption Rate (OCR) and Mitochondrial Bioenergetics
2.7. Statistical Analysis
3. Results
3.1. Effect of L-Met, DL-Met, and DL-HMTBA on Cellular Growth Kinetics and Proliferation
| Growth Parameters | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Line | Supplement | Concentration | CImax | tCImax | DT | |||||
| [µM] | (AU) | (h) | (h) | |||||||
| C2C12 | Control | 0 | 3.69 ± 0.11 | 74.3 ± 1.3 | 30.9 ± 0.4 | |||||
| L-Met | 100.0 | 4.11 ± 0.15 * | 75.2 ± 0.9 | a | 26.8 ± 0.9 | |||||
| DL-Met | 100.0 | 4.10 ± 0.11 | 73.0 ± 0.5 | 28.2 ± 1.0 | ||||||
| DL-HMTBA | 100.0 | 4.22 ± 0.13 ** | 75.2 ± 1.4 | a | 27.2 ± 1.3 | |||||
| L-Met | 1000.0 | 4.00 ± 0.12 | 88.0 ± 1.2 *** | A | b | 28.6 ± 1.4 | ||||
| DL-Met | 1000.0 | 4.15 ± 0.06 * | 73.8 ± 0.7 | B | 27.3 ± 0.6 | |||||
| DL-HMTBA | 1000.0 | 4.09 ± 0.08 | 93.3 ± 2.7 *** | C | b | 27.1 ± 0.6 | ||||
| QM7 | Control | 0 | 6.04 ± 0.12 | 84.5 ± 0.8 | 15.7 ± 0.6 | |||||
| L-Met | 100.0 | 6.64 ± 0.16 ** | 88.7 ± 0.6 * | 14.0 ± 0.2 * | ||||||
| DL-Met | 100.0 | 6.96 ± 0.08 ** | 89.0 ± 0.6 * | 14.1 ± 0.1 * | ||||||
| DL-HMTBA | 100.0 | 7.21 ± 0.19 *** | 88.8 ± 0.7 * | a | 15.8 ± 0.1 | a | ||||
| L-Met | 1000.0 | 6.70 ± 0.08 | 87.3 ± 1.2 | A | 14.3 ± 0.2 | A | ||||
| DL-Met | 1000.0 | 7.23 ± 0.12 *** | 87.8 ± 1.4 | A | 13.9 ± 0.1 * | A | ||||
| DL-HMTBA | 1000.0 | 6.91 ± 0.26 | 94.5 ± 0.9 ** | B | b | 19.4 ± 0.3 *** | B | b | ||
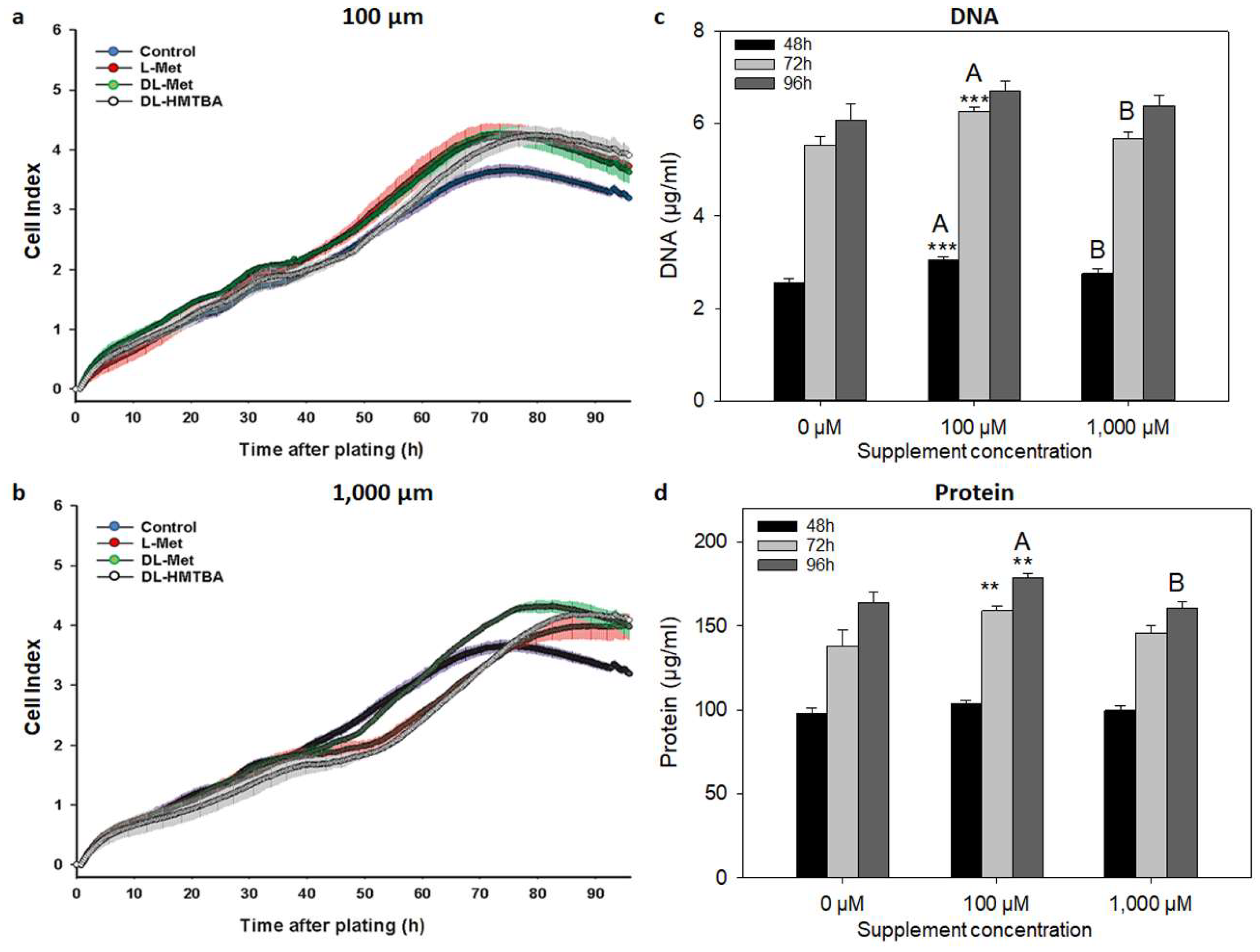
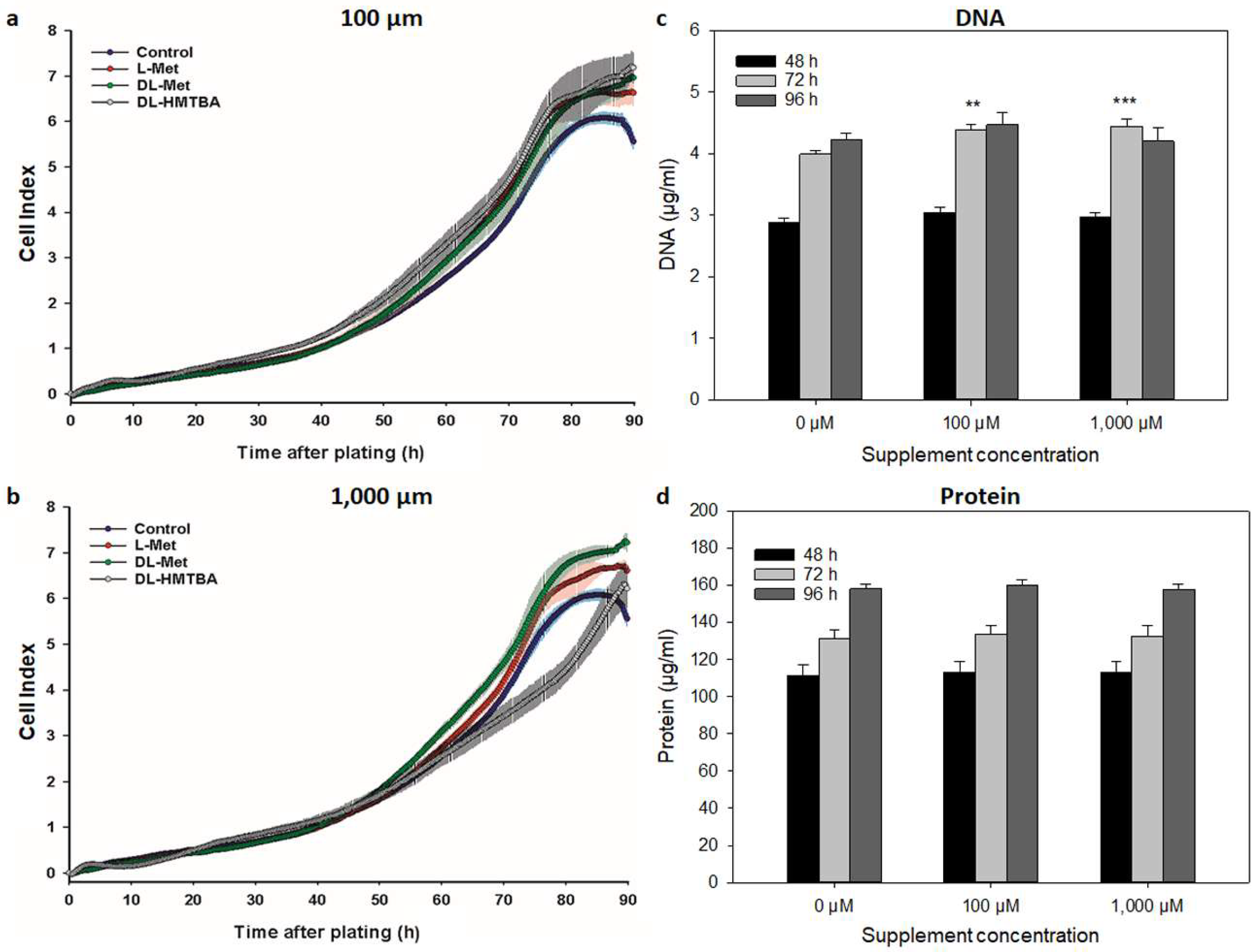
3.2. Effect of L-Met, DL-Met, and DL-HMTBA on Viability and Metabolic Activity
3.3. Effect of L-Met, DL-Met, and DL-HMTBA on Extracellular Hydrogen Peroxide Production
3.4. Metabolic Rate and Mitochondrial Function after Treatment with L-Met, DL-Met, or DL HMTBA
| Cell Line | Group | Nonmitochondrial Respiration | Mitochondrial Respiration | ||||
|---|---|---|---|---|---|---|---|
| Concen-tration | OCRbas | ATP-LR | PL | ||||
| [µM] | (fmol/min) | (fmol/min) | (fmol/min) | (fmol/min) | |||
| C2C12 | Control | 0 | 1682 ± 92 | 4832 ± 358 | 4253 ± 355 | 579 ± 48 | |
| L-Met | 1000 | 1736 ± 39 | 4039 ± 129 | 3630 ± 153 | 409 ± 80 | ||
| DL-Met | 1000 | 1693 ± 51 | 4048 ± 105 | 3583 ± 32 | 465 ± 77 # | ||
| DL-HMTBA | 1000 | 1808 ± 255 | 3482 ± 398 *** | 3012 ± 387 *** | 448 ± 105 | ||
| QM7 | Control | 0 | 2925 ± 145 | 7962 ± 218 | 6647 ± 278 | 1315 ± 150 | |
| L-Met | 1000 | 2419 ± 98 | 8325 ± 209 | A | 6964 ± 426 | 1360 ± 296 | |
| DL-Met | 1000 | 2394 ± 111 | 8394 ± 286 | A | 6744 ± 435 | 1651 ± 238 | |
| DL-HMTBA | 1000 | 4058 ± 281 ** | 6544 ± 248 ** | B | 5773 ± 344 | 771 ± 142 | |
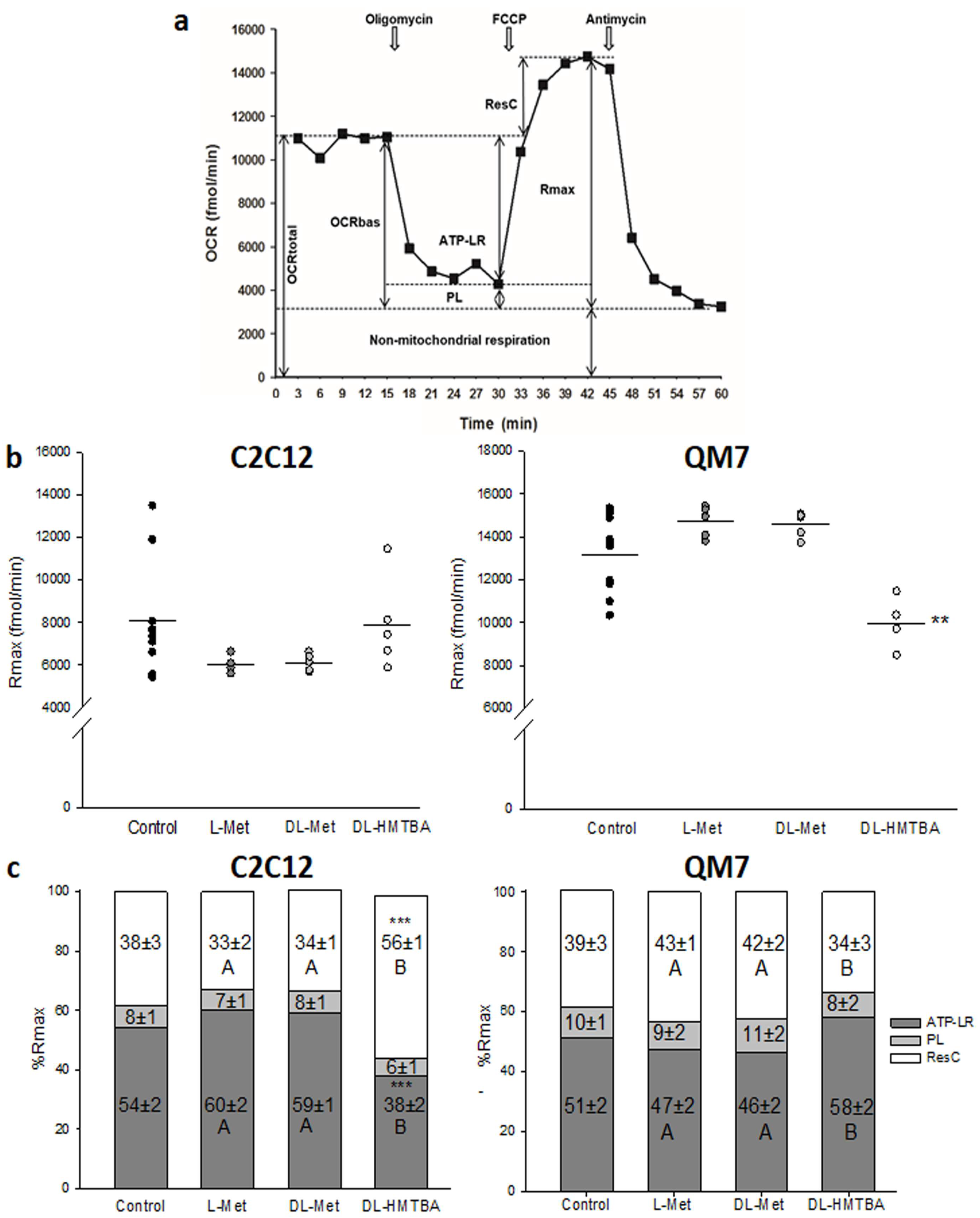
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moss, F.P.; Leblond, C.P. Satellite cells as the source of nuclei in muscles of growing rats. Anat. Rec. 1971, 170, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.C.; Lawson, J.A.; Flygare, S.D.; Fox, Z.D.; Colasanto, M.P.; Mathew, S.J.; Yandell, M.; Kardon, G. Muscle stem cells contribute to myofibres in sedentary adult mice. Nat. Commun. 2015, 6, 7087. [Google Scholar] [CrossRef]
- Schultz, E.; Gibson, M.C.; Champion, T. Satellite cells are mitotically quiescent in mature mouse muscle: An EM and radioautographic study. J. Exp. Zool. 1978, 206, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Lepper, C.; Conway, S.J.; Fan, C.M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 2009, 460, 627–631. [Google Scholar] [CrossRef]
- Barbieri, E.; Sestili, P. Reactive oxygen species in skeletal muscle signaling. J. Signal. Transduct. 2012, 2012, 982794. [Google Scholar] [CrossRef]
- Kosmidou, I.; Xagorari, A.; Roussos, C.; Papapetropoulos, A. Reactive oxygen species stimulate VEGF production from C(2)C(12) skeletal myotubes through a PI3K/Akt pathway. Am. J. Physiol. Lung Cell Mol. Physiol. 2001, 280, L585–L592. [Google Scholar] [CrossRef]
- Long, X.; Goldenthal, M.J.; Wu, G.M.; Marin-Garcia, J. Mitochondrial Ca2+ flux and respiratory enzyme activity decline are early events in cardiomyocyte response to H2O2. J. Mol. Cell Cardiol. 2004, 37, 63–70. [Google Scholar] [CrossRef]
- Ito, K.; Suda, T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell Biol. 2014, 15, 243–256. [Google Scholar] [CrossRef]
- Rocheteau, P.; Gayraud-Morel, B.; Siegl-Cachedenier, I.; Blasco, M.A.; Tajbakhsh, S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 2012, 148, 112–125. [Google Scholar] [CrossRef]
- Liang, R.; Ghaffari, S. Stem cells, redox signaling, and stem cell aging. Antioxid. Redox Signal 2014, 20, 1902–1916. [Google Scholar] [CrossRef]
- Deldicque, L.; Bertrand, L.; Patton, A.; Francaux, M.; Baar, K. ER stress induces anabolic resistance in muscle cells through PKB-induced blockade of mTORC1. PLoS ONE 2011, 6, e20993. [Google Scholar] [CrossRef]
- Rodgers, J.T.; King, K.Y.; Brett, J.O.; Cromie, M.J.; Charville, G.W.; Maguire, K.K.; Brunson, C.; Mastey, N.; Liu, L.; Tsai, C.R.; et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 2014, 510, 393–396. [Google Scholar] [CrossRef]
- Schieke, S.M.; Phillips, D.; McCoy, J.P., Jr.; Aponte, A.M.; Shen, R.F.; Balaban, R.S.; Finkel, T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 2006, 281, 27643–27652. [Google Scholar] [CrossRef]
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef]
- Moon, M.K.; Kim, M.; Chung, S.S.; Lee, H.J.; Koh, S.H.; Svovoda, P.; Jung, M.H.; Cho, Y.M.; Park, Y.J.; Choi, S.H.; et al. S-Adenosyl-L-methionine ameliorates TNFalpha-induced insulin resistance in 3T3-L1 adipocytes. Exp. Mol. Med. 2010, 42, 345–352. [Google Scholar] [CrossRef]
- Nierobisz, L.S.; Felts, V.; Mozdziak, P.E. The effect of early dietary amino acid levels on muscle satellite cell dynamics in turkeys. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007, 148, 286–294. [Google Scholar] [CrossRef]
- Metayer-Coustard, S.; Mameri, H.; Seiliez, I.; Crochet, S.; Crepieux, P.; Mercier, Y.; Geraert, P.A.; Tesseraud, S. Methionine deprivation regulates the S6K1 pathway and protein synthesis in avian QM7 myoblasts without activating the GCN2/eIF2 alpha cascade. J. Nutr. 2010, 140, 1539–1545. [Google Scholar] [CrossRef]
- Caso, G.; Garlick, P.J. Control of muscle protein kinetics by acid-base balance. Curr. Opin. Clin. Nutr. Metab. Care 2005, 8, 73–76. [Google Scholar] [CrossRef]
- Jin, C.J.; Park, H.K.; Cho, Y.M.; Pak, Y.K.; Lee, K.U.; Kim, M.S.; Friso, S.; Choi, S.W.; Park, K.S.; Lee, H.K. S-adenosyl-L-methionine increases skeletal muscle mitochondrial DNA density and whole body insulin sensitivity in OLETF rats. J. Nutr 2007, 137, 339–344. [Google Scholar] [CrossRef]
- Harden, L.M.; Neveling, N.; Rossouw, F.; Semple, S.J.; Marx, F.E.; Rossouw, J.; Rogers, G. The effects of an L-methionine combination supplement on symptoms of upper respiratory tract infections and performance in ultramarathon runners before, during and after ultra-endurance exercise. South. Afr. J. Sport. Med. 2004, 16, 10–16. [Google Scholar] [CrossRef]
- Nunes, A.J.P.; Sá, M.V.C.; Browdy, C.L.; Vazquez-Anon, M. Practical supplementation of shrimp and fish feeds with crystalline amino acids. Aquaculture 2014, 431, 20–27. [Google Scholar] [CrossRef]
- Pennings, B.; Groen, B.; de Lange, A.; Gijsen, A.P.; Zorenc, A.H.; Senden, J.M.; van Loon, L.J. Amino acid absorption and subsequent muscle protein accretion following graded intakes of whey protein in elderly men. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E992–E999. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Scientific Opinion on DL-methionine, DL-methionine sodium salt, the hydroxy analogue of methionine and the calcium salt of methionine hydroxy analogue in all animal species; on the isopropyl ester of methionine hydroxy analogue and DL-methionine technically pure protected with copolymer vinylpyridine/styrene in dairy cows; and on DL-methionine technically pure protected with ethylcellulose in ruminants. EFSA J. 2012, 10, 2623. [Google Scholar] [CrossRef]
- Hunter, E.A.; Grimble, R.F. Dietary sulphur amino acid adequacy influences glutathione synthesis and glutathione-dependent enzymes during the inflammatory response to endotoxin and tumour necrosis factor-alpha in rats. Clin. Sci. 1997, 92, 297–305. [Google Scholar] [CrossRef]
- Kerasioti, E.; Stagos, D.; Priftis, A.; Aivazidis, S.; Tsatsakis, A.M.; Hayes, A.W.; Kouretas, D. Antioxidant effects of whey protein on muscle C2C12 cells. Food Chem. 2014, 155, 271–278. [Google Scholar] [CrossRef]
- Delrieu, F.; Ferrand, B.; Amor, B. Preliminary study of L-methionine in the treatment of rheumatoid polyarthritis. Rev. Rhum. Mal. Osteoartic 1988, 55, 995–997. [Google Scholar]
- Grimble, R.F. The effects of sulfur amino acid intake on immune function in humans. J. Nutr. 2006, 136, 1660S–1665S. [Google Scholar] [CrossRef]
- Brooks, N.; Cloutier, G.J.; Cadena, S.M.; Layne, J.E.; Nelsen, C.A.; Freed, A.M.; Roubenoff, R.; Castaneda-Sceppa, C. Resistance training and timed essential amino acids protect against the loss of muscle mass and strength during 28 days of bed rest and energy deficit. J. Appl. Physiol. 2008, 105, 241–248. [Google Scholar] [CrossRef]
- Kreider, R.B.; Miriel, V.; Bertun, E. Amino acid supplementation and exercise performance. Analysis of the proposed ergogenic value. Sports Med. 1993, 16, 190–209. [Google Scholar] [CrossRef]
- Canh, T.T.; Aarnink, A.J.A.; Schutte, J.B.; Sutton, A.; Langhout, D.J.; Verstegen, M.W.A. Dietary protein affects nitrogen excretion and ammonia emission from slurry of growing-finishing pigs. Livest. Prod. Sci. 1998, 56, 181–191. [Google Scholar] [CrossRef]
- Zhang, S.; Wong, E.A.; Gilbert, E.R. Bioavailability of different dietary supplemental methionine sources in animals. Front. Biosci. 2015, 7, 478–490. [Google Scholar] [CrossRef]
- Forster, I.P.; Dominy, W.G. Efficacy of Three Methionine Sources in Diets for Pacific White Shrimp, Litopenaeus vannamei. J. World Aquac. Soc. 2006, 37, 474–480. [Google Scholar] [CrossRef]
- Hickling, D.; Guenter, W.; Jackson, M.E. The effects of dietary methionine and lysine on broiler chicken performance and breast meat yield. Can. J. Anim. Sci. 1990, 70, 673–678. [Google Scholar] [CrossRef]
- Garlick, P.J. Toxicity of methionine in humans. J. Nutr. 2006, 136, 1722S–1725S. [Google Scholar] [CrossRef]
- Benavides, M.A.; Oelschlager, D.K.; Zhang, H.G.; Stockard, C.R.; Vital-Reyes, V.S.; Katkoori, V.R.; Manne, U.; Wang, W.; Bland, K.I.; Grizzle, W.E. Methionine inhibits cellular growth dependent on the p53 status of cells. Am. J. Surg. 2007, 193, 274–283. [Google Scholar] [CrossRef]
- Gomez, J.; Caro, P.; Sanchez, I.; Naudi, A.; Jove, M.; Portero-Otin, M.; Lopez-Torres, M.; Pamplona, R.; Barja, G. Effect of methionine dietary supplementation on mitochondrial oxygen radical generation and oxidative DNA damage in rat liver and heart. J. Bioenerg. Biomembr. 2009, 41, 309–321. [Google Scholar] [CrossRef]
- Wei, Y.H.; Lu, C.Y.; Lee, H.C.; Pang, C.Y.; Ma, Y.S. Oxidative damage and mutation to mitochondrial DNA and age-dependent decline of mitochondrial respiratory function. Ann. NY Acad. Sci. 1998, 854, 155–170. [Google Scholar] [CrossRef]
- Xie, L.; Zhu, X.; Hu, Y.; Li, T.; Gao, Y.; Shi, Y.; Tang, S. Mitochondrial DNA oxidative damage triggering mitochondrial dysfunction and apoptosis in high glucose-induced HRECs. Invest. Ophthalmol. Vis. Sci. 2008, 49, 4203–4209. [Google Scholar] [CrossRef]
- Barazzoni, R. Skeletal muscle mitochondrial protein metabolism and function in ageing and type 2 diabetes. Curr. Opin. Clin. Nutr. Metab. Care 2004, 7, 97–102. [Google Scholar] [CrossRef]
- Sanchez-Roman, I.; Barja, G. Regulation of longevity and oxidative stress by nutritional interventions: Role of methionine restriction. Exp. Gerontol. 2013, 48, 1030–1042. [Google Scholar] [CrossRef]
- Sanz, A.; Caro, P.; Ayala, V.; Portero-Otin, M.; Pamplona, R.; Barja, G. Methionine restriction decreases mitochondrial oxygen radical generation and leak as well as oxidative damage to mitochondrial DNA and proteins. FASEB J. 2006, 20, 1064–1073. [Google Scholar] [CrossRef]
- Richie, J.P., Jr.; Leutzinger, Y.; Parthasarathy, S.; Malloy, V.; Orentreich, N.; Zimmerman, J.A. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994, 8, 1302–1307. [Google Scholar] [CrossRef]
- Laner, B.W., Jr. The Biochemical Conversion of 2-Hydroxy-4-Methylthiobutyric Acid into Methionine by the Rat in Vitro. Biochem. J. 1965, 95, 683–687. [Google Scholar] [CrossRef]
- Rangel-Lugo, M.; Austic, R.E. Transamination of 2-oxo-4-[methylthio]butanoic acid in chicken tissues. Poult. Sci. 1998, 77, 98–104. [Google Scholar] [CrossRef]
- Stipanuk, M.H. The keto acid of methionine is a safe and efficacious substitute for dietary L-methionine: The answer from chick bioassays. J. Nutr. 2007, 137, 1844–1845. [Google Scholar] [CrossRef]
- Phaneuf, S.; Leeuwenburgh, C. Apoptosis and exercise. Med. Sci. Sports Exerc. 2001, 33, 393–396. [Google Scholar] [CrossRef]
- Dargelos, E.; Brule, C.; Stuelsatz, P.; Mouly, V.; Veschambre, P.; Cottin, P.; Poussard, S. Up-regulation of calcium-dependent proteolysis in human myoblasts under acute oxidative stress. Exp. Cell Res. 2010, 316, 115–125. [Google Scholar] [CrossRef]
- Burattini, S.; Ferri, P.; Battistelli, M.; Curci, R.; Luchetti, F.; Falcieri, E. C2C12 murine myoblasts as a model of skeletal muscle development: Morpho-functional characterization. Eur. J. Histochem. 2004, 48, 223–233. [Google Scholar]
- Xi, B.; Yu, N.; Wang, X.; Xu, X.; Abassi, Y.A. The application of cell-based label-free technology in drug discovery. Biotechnol. J. 2008, 3, 484–495. [Google Scholar] [CrossRef]
- Rago, R.; Mitchen, J.; Wilding, G. DNA fluorometric assay in 96-well tissue culture plates using Hoechst 33258 after cell lysis by freezing in distilled water. Anal. Biochem. 1990, 191, 31–34. [Google Scholar] [CrossRef]
- Peterson, G.L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 1977, 83, 346–356. [Google Scholar] [CrossRef]
- Nitta, Y.; Muraoka-Hirayama, S.; Sakurai, K. Catalase is required for peroxisome maintenance during adipogenesis. Biochim Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158726. [Google Scholar] [CrossRef]
- Miersch, C.; Stange, K.; Hering, S.; Kolisek, M.; Viergutz, T.; Rontgen, M. Molecular and functional heterogeneity of early postnatal porcine satellite cell populations is associated with bioenergetic profile. Sci. Rep. 2017, 7, 45052. [Google Scholar] [CrossRef]
- Cook, C.C.; Kim, A.; Terao, S.; Gotoh, A.; Higuchi, M. Consumption of oxygen: A mitochondrial-generated progression signal of advanced cancer. Cell Death Dis. 2012, 3, e258. [Google Scholar] [CrossRef]
- Kornasio, R.; Riederer, I.; Butler-Browne, G.; Mouly, V.; Uni, Z.; Halevy, O. Beta-hydroxy-beta-methylbutyrate (HMB) stimulates myogenic cell proliferation, differentiation and survival via the MAPK/ERK and PI3K/Akt pathways. Biochim. Biophys. Acta 2009, 1793, 755–763. [Google Scholar] [CrossRef]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metabol. 2014, 19, 780–794. [Google Scholar] [CrossRef]
- Harper, J.M.; Wang, M.; Galecki, A.T.; Ro, J.; Williams, J.B.; Miller, R.A. Fibroblasts from long-lived bird species are resistant to multiple forms of stress. J. Exp. Biol. 2011, 214, 1902–1910. [Google Scholar] [CrossRef]
- Holmes, D.J.; Ottinger, M.A. Birds as long-lived animal models for the study of aging. Exp. Gerontol. 2003, 38, 1365–1375. [Google Scholar] [CrossRef]
- Brosnan, J.T.; Wijekoon, E.P.; Warford-Woolgar, L.; Trottier, N.L.; Brosnan, M.E.; Brunton, J.A.; Bertolo, R.F. Creatine synthesis is a major metabolic process in neonatal piglets and has important implications for amino acid metabolism and methyl balance. J. Nutr. 2009, 139, 1292–1297. [Google Scholar] [CrossRef]
- Finkelstein, J.D.; Martin, J.J. Methionine metabolism in mammals. Adaptation to methionine excess. J. Biol. Chem. 1986, 261, 1582–1587. [Google Scholar] [CrossRef]
- McBreairty, L.E.; Bertolo, R.F. The dynamics of methionine supply and demand during early development. Appl. Physiol. Nutr. Metabol. 2016, 41, 581–587. [Google Scholar] [CrossRef]
- Smith, H.J.; Greenberg, N.A.; Tisdale, M.J. Effect of eicosapentaenoic acid, protein and amino acids on protein synthesis and degradation in skeletal muscle of cachectic mice. Br. J. Cancer 2004, 91, 408–412. [Google Scholar] [CrossRef]
- Atherton, P.J.; Smith, K.; Etheridge, T.; Rankin, D.; Rennie, M.J. Distinct anabolic signalling responses to amino acids in C2C12 skeletal muscle cells. Amino Acids 2010, 38, 1533–1539. [Google Scholar] [CrossRef]
- Bohe, J.; Low, A.; Wolfe, R.R.; Rennie, M.J. Human muscle protein synthesis is modulated by extracellular, not intramuscular amino acid availability: A dose-response study. J. Physiol. 2003, 552, 315–324. [Google Scholar] [CrossRef]
- Wolfe, R.R. Regulation of muscle protein by amino acids. J. Nutr. 2002, 132, 3219S–3224S. [Google Scholar] [CrossRef]
- Quent, V.M.; Loessner, D.; Friis, T.; Reichert, J.C.; Hutmacher, D.W. Discrepancies between metabolic activity and DNA content as tool to assess cell proliferation in cancer research. J. Cell Mol. Med. 2010, 14, 1003–1013. [Google Scholar] [CrossRef]
- Basoah, A.; Matthews, P.M.; Morten, K.J. Rapid rates of newly synthesized mitochondrial protein degradation are significantly affected by the generation of mitochondrial free radicals. FEBS Lett. 2005, 579, 6511–6517. [Google Scholar] [CrossRef]
- Lennicke, C.; Rahn, J.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Hydrogen peroxide—Production, fate and role in redox signaling of tumor cells. Cell Commun. Signal. 2015, 13, 39. [Google Scholar] [CrossRef]
- Mailloux, R.J. Mitochondrial Antioxidants and the Maintenance of Cellular Hydrogen Peroxide Levels. Oxid Med. Cell Longev. 2018, 2018, 7857251. [Google Scholar] [CrossRef]
- Tomalin, L.E.; Day, A.M.; Underwood, Z.E.; Smith, G.R.; Dalle Pezze, P.; Rallis, C.; Patel, W.; Dickinson, B.C.; Bahler, J.; Brewer, T.F.; et al. Increasing extracellular H2O2 produces a bi-phasic response in intracellular H2O2, with peroxiredoxin hyperoxidation only triggered once the cellular H2O2-buffering capacity is overwhelmed. Free Radic. Biol. Med. 2016, 95, 333–348. [Google Scholar] [CrossRef]
- Buettner, G.R. Superoxide dismutase in redox biology: The roles of superoxide and hydrogen peroxide. Anticancer Agents Med. Chem. 2011, 11, 341–346. [Google Scholar] [CrossRef]
- Koningsberger, J.C.; van Asbeck, B.S.; van Faassen, E.; Wiegman, L.J.; van Hattum, J.; van Berge Henegouwen, G.P.; Marx, J.J. Copper, zinc-superoxide dismutase and hydrogen peroxide: A hydroxyl radical generating system. Clin. Chim. Acta 1994, 230, 51–61. [Google Scholar] [CrossRef]
- Sin, J.; Andres, A.M.; Taylor, D.J.; Weston, T.; Hiraumi, Y.; Stotland, A.; Kim, B.J.; Huang, C.; Doran, K.S.; Gottlieb, R.A. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 2016, 12, 369–380. [Google Scholar] [CrossRef]
- Orzechowski, A.; Lokociejewska, M.; Muras, P.; Hocquette, J.F. Preconditioning with millimolar concentrations of vitamin C or N-acetylcysteine protects L6 muscle cells insulin-stimulated viability and DNA synthesis under oxidative stress. Life Sci. 2002, 71, 1793–1808. [Google Scholar] [CrossRef]
- Wagatsuma, A.; Sakuma, K. Mitochondria as a potential regulator of myogenesis. Sci. World J. 2013, 2013, 593267. [Google Scholar] [CrossRef]
- Lopez-Bernardo, E.; Anedda, A.; Sanchez-Perez, P.; Acosta-Iborra, B.; Cadenas, S. 4-Hydroxynonenal induces Nrf2-mediated UCP3 upregulation in mouse cardiomyocytes. Free Radic. Biol. Med. 2015, 88, 427–438. [Google Scholar] [CrossRef]
- Santamaria, E.; Avila, M.A.; Latasa, M.U.; Rubio, A.; Martin-Duce, A.; Lu, S.C.; Mato, J.M.; Corrales, F.J. Functional proteomics of nonalcoholic steatohepatitis: Mitochondrial proteins as targets of S-adenosylmethionine. Proc. Natl. Acad. Sci. USA 2003, 100, 3065–3070. [Google Scholar] [CrossRef]
- Nakamura, J.; Fujikawa, M.; Yoshida, M. IF1, a natural inhibitor of mitochondrial ATP synthase, is not essential for the normal growth and breeding of mice. Biosci. Rep. 2013, 33, e00067. [Google Scholar] [CrossRef]
- Sanchez-Cenizo, L.; Formentini, L.; Aldea, M.; Ortega, A.D.; Garcia-Huerta, P.; Sanchez-Arago, M.; Cuezva, J.M. Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J. Biol. Chem. 2010, 285, 25308–25313. [Google Scholar] [CrossRef]
- Shin, Y.K.; Yoo, B.C.; Chang, H.J.; Jeon, E.; Hong, S.H.; Jung, M.S.; Lim, S.J.; Park, J.G. Down-regulation of mitochondrial F1F0-ATP synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res. 2005, 65, 3162–3170. [Google Scholar] [CrossRef]
- Shchepina, L.A.; Pletjushkina, O.Y.; Avetisyan, A.V.; Bakeeva, L.E.; Fetisova, E.K.; Izyumov, D.S.; Saprunova, V.B.; Vyssokikh, M.Y.; Chernyak, B.V.; Skulachev, V.P. Oligomycin, inhibitor of the F0 part of H+-ATP-synthase, suppresses the TNF-induced apoptosis. Oncogene 2002, 21, 8149–8157. [Google Scholar] [CrossRef] [PubMed]
- Sansbury, B.E.; Jones, S.P.; Riggs, D.W.; Darley-Usmar, V.M.; Hill, B.G. Bioenergetic function in cardiovascular cells: The importance of the reserve capacity and its biological regulation. Chem. Biol. Interact. 2011, 191, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Morino, K.; Mengistu, L.; Ishibashi, T.; Kiriyama, K.; Ikami, T.; Maegawa, H. Amla Enhances Mitochondrial Spare Respiratory Capacity by Increasing Mitochondrial Biogenesis and Antioxidant Systems in a Murine Skeletal Muscle Cell Line. Oxid Med. Cell Longev. 2016, 2016, 1735841. [Google Scholar] [CrossRef] [PubMed]
- Acin-Perez, R.; Benador, I.Y.; Petcherski, A.; Veliova, M.; Benavides, G.A.; Lagarrigue, S.; Caudal, A.; Vergnes, L.; Murphy, A.N.; Karamanlidis, G.; et al. A novel approach to measure mitochondrial respiration in frozen biological samples. EMBO J. 2020, 39, e104073. [Google Scholar] [CrossRef]
- Herst, P.M.; Berridge, M.V. Cell surface oxygen consumption: A major contributor to cellular oxygen consumption in glycolytic cancer cell lines. Biochim. Biophys. Acta 2007, 1767, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Barja, G.; Cadenas, S.; Rojas, C.; Perez-Campo, R.; Lopez-Torres, M. Low mitochondrial free radical production per unit O2 consumption can explain the simultaneous presence of high longevity and high aerobic metabolic rate in birds. Free Radic. Res. 1994, 21, 317–327. [Google Scholar] [CrossRef]
- Ogburn, C.E.; Austad, S.N.; Holmes, D.J.; Kiklevich, J.V.; Gollahon, K.; Rabinovitch, P.S.; Martin, G.M. Cultured renal epithelial cells from birds and mice: Enhanced resistance of avian cells to oxidative stress and DNA damage. J. Gerontol. Biol. Sci. Med. Sci. 1998, 53, B287–B292. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stange, K.; Schumacher, T.; Miersch, C.; Whelan, R.; Klünemann, M.; Röntgen, M. Methionine Sources Differently Affect Production of Reactive Oxygen Species, Mitochondrial Bioenergetics, and Growth of Murine and Quail Myoblasts In Vitro. Curr. Issues Mol. Biol. 2023, 45, 2661-2680. https://doi.org/10.3390/cimb45040174
Stange K, Schumacher T, Miersch C, Whelan R, Klünemann M, Röntgen M. Methionine Sources Differently Affect Production of Reactive Oxygen Species, Mitochondrial Bioenergetics, and Growth of Murine and Quail Myoblasts In Vitro. Current Issues in Molecular Biology. 2023; 45(4):2661-2680. https://doi.org/10.3390/cimb45040174
Chicago/Turabian StyleStange, Katja, Toni Schumacher, Claudia Miersch, Rose Whelan, Martina Klünemann, and Monika Röntgen. 2023. "Methionine Sources Differently Affect Production of Reactive Oxygen Species, Mitochondrial Bioenergetics, and Growth of Murine and Quail Myoblasts In Vitro" Current Issues in Molecular Biology 45, no. 4: 2661-2680. https://doi.org/10.3390/cimb45040174
APA StyleStange, K., Schumacher, T., Miersch, C., Whelan, R., Klünemann, M., & Röntgen, M. (2023). Methionine Sources Differently Affect Production of Reactive Oxygen Species, Mitochondrial Bioenergetics, and Growth of Murine and Quail Myoblasts In Vitro. Current Issues in Molecular Biology, 45(4), 2661-2680. https://doi.org/10.3390/cimb45040174





