Immunotherapeutic Approaches in Ovarian Cancer
Abstract
1. Introduction
2. NGS-Based Identification of New Pathways and Targets in OC
2.1. Bulk RNA-Sequencing Analysis (RNA-Seq)
2.2. Single-Cell RNA-Sequencing Analysis (scRNA-Seq)
2.3. Transposase-Accessible Chromatin Analysis by Sequencing (ATAC-Seq)
2.4. Combined Sequencing Analysis
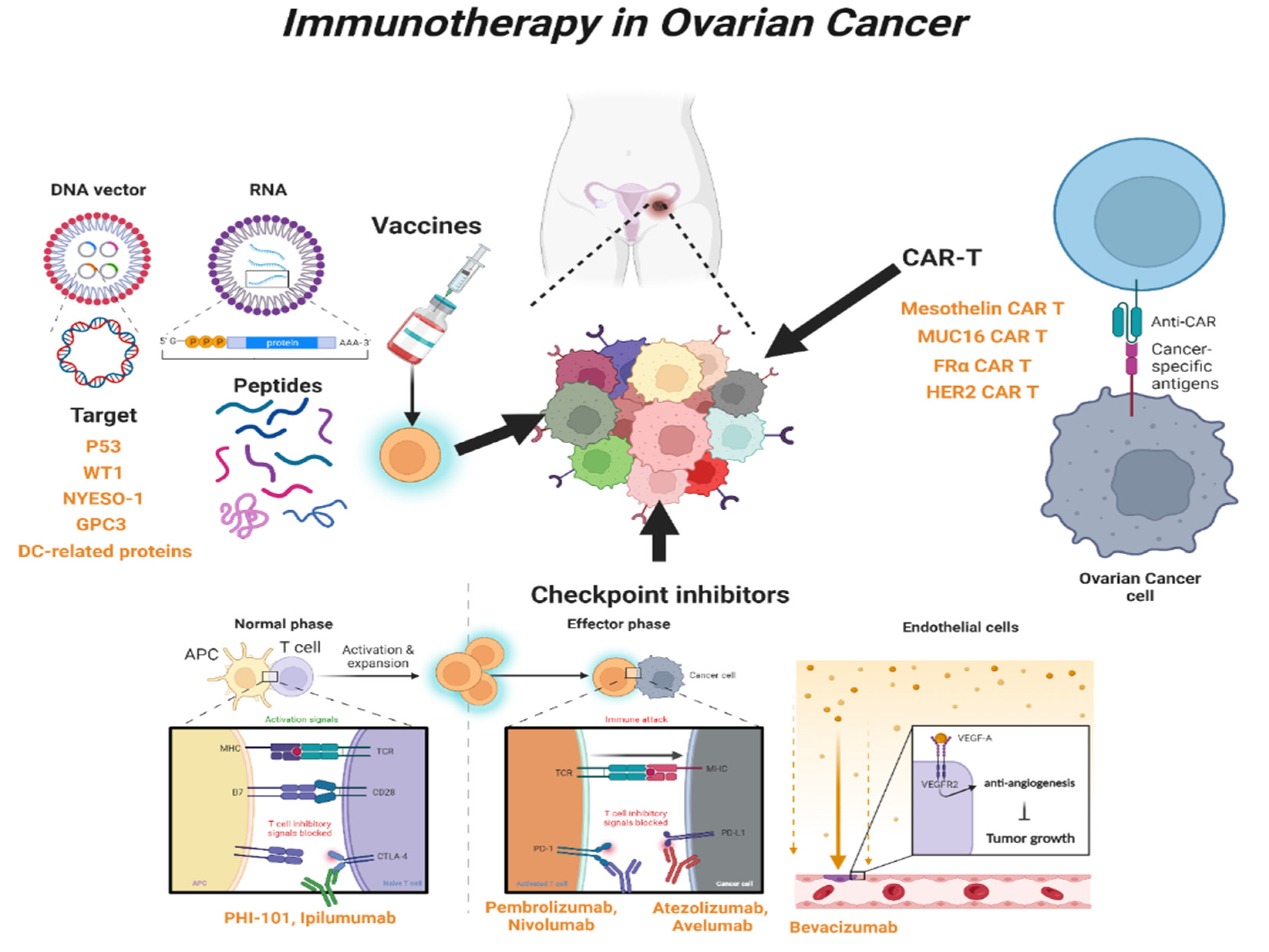
3. Immunotherapy in OC
3.1. Immune-Stimulating Peptide Vaccines
3.1.1. P53 Peptide Vaccine
3.1.2. Wilms’ Tumor 1 (WT1) Peptide Vaccine
3.1.3. New York Esophageal Squamous Cell Carcinoma-1 (NYESO-1) Peptide Vaccine
3.1.4. Glypican-3 (GPC3)-Derived Peptide Vaccine
3.1.5. Dendritic Cell (DC)-Based Peptide Vaccine
3.2. Blockade of Checkpoint
3.2.1. Cytotoxic T-lymphocyte Antigen 4 (CTLA-4) Inhibitor
3.2.2. Programed Cell Death Protein 1 (PD-1) and Programed Cell Death-Ligand 1 (PD-L1) Inhibitors
3.3. Chimeric Antigen Receptor T (CAR-T) Cells
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Q.; Peng, H.; Qi, X.; Wu, M.; Zhao, X. Targeted therapies in gynecological cancers: A comprehensive review of clinical evidence. Signal Transduct. Target. Ther. 2020, 5, 137. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Webb, P.M.; Jordan, S.J. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.; Sood, A.; Fallowfield, L.; Howitt, B.; Sehouli, J.; Karlan, B. Ovarian cancer. Nat. Rev. Dis. Prim. 2016, 2, 16061. [Google Scholar] [CrossRef] [PubMed]
- Prat, J. Ovarian carcinomas: Five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012, 460, 237–249. [Google Scholar] [CrossRef]
- Singer, G.; Oldt, R., 3rd; Cohen, Y.; Wang, B.G.; Sidransky, D.; Kurman, R.J.; Shih Ie, M. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J. Natl. Cancer Inst. 2003, 95, 484–486. [Google Scholar] [CrossRef]
- Singer, G.; Stohr, R.; Cope, L.; Dehari, R.; Hartmann, A.; Cao, D.F.; Wang, T.L.; Kurman, R.J.; Shih Ie, M. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: A mutational analysis with immunohistochemical correlation. Am. J. Surg. Pathol. 2005, 29, 218–224. [Google Scholar] [CrossRef]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef]
- Katsumata, N.; Yasuda, M.; Isonishi, S.; Takahashi, F.; Michimae, H.; Kimura, E.; Aoki, D.; Jobo, T.; Kodama, S.; Terauchi, F.; et al. Long-term results of dose-dense paclitaxel and carboplatin versus conventional paclitaxel and carboplatin for treatment of advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer (JGOG 3016): A randomised, controlled, open-label trial. Lancet Oncol. 2013, 14, 1020–1026. [Google Scholar] [CrossRef]
- Colombo, N.; Peiretti, M.; Parma, G.; Lapresa, M.; Mancari, R.; Carinelli, S.; Sessa, C.; Castiglione, M.; Group, E.G.W. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2010, 21, v23–v30. [Google Scholar] [CrossRef]
- Ledermann, J.A.; Raja, F.A.; Fotopoulou, C.; Gonzalez-Martin, A.; Colombo, N.; Sessa, C.; Group, E.G.W. Corrections to “Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up”. Ann. Oncol. 2018, 29, iv259. [Google Scholar] [CrossRef]
- Christie, E.L.; Bowtell, D.D.L. Acquired chemotherapy resistance in ovarian cancer. Ann. Oncol. 2017, 28, viii13–viii15. [Google Scholar] [CrossRef]
- Pignata, S.; Pisano, C.; Di Napoli, M.; Cecere, S.C.; Tambaro, R.; Attademo, L. Treatment of recurrent epithelial ovarian cancer. Cancer 2019, 125, 4609–4615. [Google Scholar] [CrossRef]
- Conteduca, V.; Scarpi, E.; Farolfi, A.; Brighi, N.; Rossi, L.; Gurioli, G.; Lolli, C.; Schepisi, G.; Bleve, S.; Gianni, C.; et al. Melphalan as a Promising Treatment for BRCA-Related Ovarian Carcinoma. Front. Oncol. 2021, 11, 716467. [Google Scholar] [CrossRef]
- Kaern, J.; Baekelandt, M.; Trope, C.G. A phase II study of weekly paclitaxel in platinum and paclitaxel-resistant ovarian cancer patients. Eur. J. Gynaecol. Oncol. 2002, 23, 383–389. [Google Scholar]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.L.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014, 15, 852–861. [Google Scholar] [CrossRef]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J.Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef]
- Gonzalez-Martin, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Lau, C.H.; Seow, K.M.; Chen, K.H. The Molecular Mechanisms of Actions, Effects, and Clinical Implications of PARP Inhibitors in Epithelial Ovarian Cancers: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 8125. [Google Scholar] [CrossRef]
- McMullen, M.; Karakasis, K.; Madariaga, A.; Oza, A.M. Overcoming Platinum and PARP-Inhibitor Resistance in Ovarian Cancer. Cancers 2020, 12, 1607. [Google Scholar] [CrossRef]
- Klotz, D.M.; Wimberger, P. Overcoming PARP inhibitor resistance in ovarian cancer: What are the most promising strategies? Arch. Gynecol. Obstet. 2020, 302, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome. Med. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, A.; Sholl, L.; Reardon, B.; Taylor-Weiner, A.; Amin-Mansour, A.; Miao, D.; Liu, D.; Oliver, N.; MacConaill, L.; Ducar, M.; et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome. Med. 2016, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Shyr, D.; Liu, Q. Next generation sequencing in cancer research and clinical application. Biol. Proced. Online 2013, 15, 4. [Google Scholar] [CrossRef]
- Morash, M.; Mitchell, H.; Beltran, H.; Elemento, O.; Pathak, J. The Role of Next-Generation Sequencing in Precision Medicine: A Review of Outcomes in Oncology. J. Pers. Med. 2018, 8, 30. [Google Scholar] [CrossRef]
- Tattersall, A.; Ryan, N.; Wiggans, A.J.; Rogozinska, E.; Morrison, J. Poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer. Cochrane Database Syst. Rev. 2022, 2, CD007929. [Google Scholar] [CrossRef]
- Kyo, S.; Kanno, K.; Takakura, M.; Yamashita, H.; Ishikawa, M.; Ishibashi, T.; Sato, S.; Nakayama, K. Clinical Landscape of PARP Inhibitors in Ovarian Cancer: Molecular Mechanisms and Clues to Overcome Resistance. Cancers 2022, 14, 2504. [Google Scholar] [CrossRef]
- Palleschi, M.; Tedaldi, G.; Sirico, M.; Virga, A.; Ulivi, P.; De Giorgi, U. Moving beyond PARP Inhibition: Current State and Future Perspectives in Breast Cancer. Int. J. Mol. Sci. 2021, 22, 7884. [Google Scholar] [CrossRef]
- Gurioli, G.; Tedaldi, G.; Farolfi, A.; Petracci, E.; Casanova, C.; Comerci, G.; Danesi, R.; Arcangeli, V.; Ravegnani, M.; Calistri, D.; et al. Clinical Impact of Next-Generation Sequencing Multi-Gene Panel Highlighting the Landscape of Germline Alterations in Ovarian Cancer Patients. Int. J. Mol. Sci. 2022, 23, 5789. [Google Scholar] [CrossRef]
- Kuksin, M.; Morel, D.; Aglave, M.; Danlos, F.X.; Marabelle, A.; Zinovyev, A.; Gautheret, D.; Verlingue, L. Applications of single-cell and bulk RNA sequencing in onco-immunology. Eur. J. Cancer 2021, 149, 193–210. [Google Scholar] [CrossRef]
- Medrano, M.; Communal, L.; Brown, K.R.; Iwanicki, M.; Normand, J.; Paterson, J.; Sircoulomb, F.; Krzyzanowski, P.; Novak, M.; Doodnauth, S.A.; et al. Interrogation of Functional Cell-Surface Markers Identifies CD151 Dependency in High-Grade Serous Ovarian Cancer. Cell Rep. 2017, 18, 2343–2358. [Google Scholar] [CrossRef]
- Mosig, R.A.; Lin, L.; Senturk, E.; Shah, H.; Huang, F.; Schlosshauer, P.; Cohen, S.; Fruscio, R.; Marchini, S.; D’Incalci, M.; et al. Application of RNA-Seq transcriptome analysis: CD151 is an Invasion/Migration target in all stages of epithelial ovarian cancer. J. Ovarian. Res. 2012, 5, 4. [Google Scholar] [CrossRef]
- Nagasawa, S.; Ikeda, K.; Horie-Inoue, K.; Sato, S.; Takeda, S.; Hasegawa, K.; Inoue, S. Identification of novel mutations of ovarian cancer-related genes from RNA-sequencing data for Japanese epithelial ovarian cancer patients. Endocr. J. 2020, 67, 219–229. [Google Scholar] [CrossRef]
- Saleh, A.; Perets, R. Mutated p53 in HGSC-From a Common Mutation to a Target for Therapy. Cancers 2021, 13, 3465. [Google Scholar] [CrossRef]
- Pal, S.; Garg, M.; Pandey, A.K. Deciphering the Mounting Complexity of the p53 Regulatory Network in Correlation to Long Non-Coding RNAs (lncRNAs) in Ovarian Cancer. Cells 2020, 9, 527. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Takeiwa, T.; Mitobe, Y.; Ikeda, K.; Hasegawa, K.; Horie, K.; Inoue, S. Long Intergenic Noncoding RNA OIN1 Promotes Ovarian Cancer Growth by Modulating Apoptosis-Related Gene Expression. Int. J. Mol. Sci. 2021, 22, 1242. [Google Scholar] [CrossRef]
- Earp, M.A.; Raghavan, R.; Li, Q.; Dai, J.; Winham, S.J.; Cunningham, J.M.; Natanzon, Y.; Kalli, K.R.; Hou, X.; Weroha, S.J.; et al. Characterization of fusion genes in common and rare epithelial ovarian cancer histologic subtypes. Oncotarget 2017, 8, 46891–46899. [Google Scholar] [CrossRef]
- Wang, J.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. RNA sequencing (RNA-Seq) and its application in ovarian cancer. Gynecol. Oncol. 2019, 152, 194–201. [Google Scholar] [CrossRef]
- Tang, Z.; Li, J.; Shen, Q.; Feng, J.; Liu, H.; Wang, W.; Xu, L.; Shi, G.; Ye, X.; Ge, M.; et al. Contribution of upregulated dipeptidyl peptidase 9 (DPP9) in promoting tumoregenicity, metastasis and the prediction of poor prognosis in non-small cell lung cancer (NSCLC). Int. J. Cancer 2017, 140, 1620–1632. [Google Scholar] [CrossRef]
- Deng, J.; Wang, L.; Chen, H.; Li, L.; Ma, Y.; Ni, J.; Li, Y. The role of tumour-associated MUC1 in epithelial ovarian cancer metastasis and progression. Cancer Metastasis Rev. 2013, 32, 535–551. [Google Scholar] [CrossRef]
- Fan, Q.; Zhang, W.; Emerson, R.E.; Xu, Y. ZIP4 Is a Novel Cancer Stem Cell Marker in High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 3692. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Li, L.; Wang, T.L.; Emerson, R.E.; Xu, Y. A Novel ZIP4-HDAC4-VEGFA Axis in High-Grade Serous Ovarian Cancer. Cancers 2021, 13, 3821. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Cai, Q.; Li, P.; Wang, W.; Wang, J.; Gerry, E.; Wang, T.L.; Shih, I.M.; Nephew, K.P.; Xu, Y. The novel ZIP4 regulation and its role in ovarian cancer. Oncotarget 2017, 8, 90090–90107. [Google Scholar] [CrossRef] [PubMed]
- Ediriweera, M.K.; Tennekoon, K.H.; Samarakoon, S.R. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. Semin. Cancer Biol. 2019, 59, 147–160. [Google Scholar] [CrossRef]
- Hanrahan, A.J.; Schultz, N.; Westfal, M.L.; Sakr, R.A.; Giri, D.D.; Scarperi, S.; Janakiraman, M.; Olvera, N.; Stevens, E.V.; She, Q.B.; et al. Genomic complexity and AKT dependence in serous ovarian cancer. Cancer Discov. 2012, 2, 56–67. [Google Scholar] [CrossRef]
- Bai, H.; Li, H.; Li, W.; Gui, T.; Yang, J.; Cao, D.; Shen, K. The PI3K/AKT/mTOR pathway is a potential predictor of distinct invasive and migratory capacities in human ovarian cancer cell lines. Oncotarget 2015, 6, 25520–25532. [Google Scholar] [CrossRef]
- Svensson, V.; Natarajan, K.N.; Ly, L.H.; Miragaia, R.J.; Labalette, C.; Macaulay, I.C.; Cvejic, A.; Teichmann, S.A. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 2017, 14, 381–387. [Google Scholar] [CrossRef]
- Kossai, M.; Leary, A.; Scoazec, J.Y.; Genestie, C. Ovarian Cancer: A Heterogeneous Disease. Pathobiology 2018, 85, 41–49. [Google Scholar] [CrossRef]
- Shih, A.J.; Menzin, A.; Whyte, J.; Lovecchio, J.; Liew, A.; Khalili, H.; Bhuiya, T.; Gregersen, P.K.; Lee, A.T. Identification of grade and origin specific cell populations in serous epithelial ovarian cancer by single cell RNA-seq. PLoS ONE 2018, 13, e0206785. [Google Scholar] [CrossRef]
- Mao, T.L.; Fan, K.F.; Liu, C.L. Targeting the CXCR4/CXCL12 axis in treating epithelial ovarian cancer. Gene Ther. 2017, 24, 621–629. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell Biochem. 2019, 120, 2782–2790. [Google Scholar] [CrossRef]
- Bareke, H.; Akbuga, J. Complement system’s role in cancer and its therapeutic potential in ovarian cancer. Scand. J. Immunol. 2018, 88, e12672. [Google Scholar] [CrossRef]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Wang, F.; Gao, C.; Cao, Y.; Wang, J. Identification of Specific Cell Subpopulations and Marker Genes in Ovarian Cancer Using Single-Cell RNA Sequencing. Biomed. Res. Int. 2021, 2021, 1005793. [Google Scholar] [CrossRef]
- Li, Z.; Schulz, M.H.; Look, T.; Begemann, M.; Zenke, M.; Costa, I.G. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019, 20, 45. [Google Scholar] [CrossRef]
- Bysani, M.; Agren, R.; Davegardh, C.; Volkov, P.; Ronn, T.; Unneberg, P.; Bacos, K.; Ling, C. ATAC-seq reveals alterations in open chromatin in pancreatic islets from subjects with type 2 diabetes. Sci. Rep. 2019, 9, 7785. [Google Scholar] [CrossRef]
- Zhu, C.; Yu, M.; Huang, H.; Juric, I.; Abnousi, A.; Hu, R.; Lucero, J.; Behrens, M.M.; Hu, M.; Ren, B. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol. 2019, 26, 1063–1070. [Google Scholar] [CrossRef]
- Sun, Y.; Miao, N.; Sun, T. Detect accessible chromatin using ATAC-sequencing, from principle to applications. Hereditas 2019, 156, 29. [Google Scholar] [CrossRef]
- Boutelle, A.M.; Attardi, L.D. p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol. 2021, 31, 298–310. [Google Scholar] [CrossRef]
- Gallon, J.; Loomis, E.; Curry, E.; Martin, N.; Brody, L.; Garner, I.; Brown, R.; Flanagan, J.M. Chromatin accessibility changes at intergenic regions are associated with ovarian cancer drug resistance. Clin. Epigenetics 2021, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Panda, M.; Tripathi, S.K.; Biswal, B.K. SOX9: An emerging driving factor from cancer progression to drug resistance. Biochim Biophys Acta. Rev. Cancer 2021, 1875, 188517. [Google Scholar] [CrossRef] [PubMed]
- Davies, L.C.; Jenkins, S.J.; Allen, J.E.; Taylor, P.R. Tissue-resident macrophages. Nat. Immunol. 2013, 14, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Casanova-Acebes, M.; Menendez-Gutierrez, M.P.; Porcuna, J.; Alvarez-Errico, D.; Lavin, Y.; Garcia, A.; Kobayashi, S.; Le Berichel, J.; Nunez, V.; Were, F.; et al. RXRs control serous macrophage neonatal expansion and identity and contribute to ovarian cancer progression. Nat. Commun. 2020, 11, 1655. [Google Scholar] [CrossRef]
- Pongor, L.; Harami-Papp, H.; Mehes, E.; Czirok, A.; Gyorffy, B. Cell Dispersal Influences Tumor Heterogeneity and Introduces a Bias in NGS Data Interpretation. Sci. Rep. 2017, 7, 7358. [Google Scholar] [CrossRef]
- Hooda, J.; Novak, M.; Salomon, M.P.; Matsuba, C.; Ramos, R.I.; MacDuffie, E.; Song, M.; Hirsch, M.S.; Lester, J.; Parkash, V.; et al. Early Loss of Histone H2B Monoubiquitylation Alters Chromatin Accessibility and Activates Key Immune Pathways That Facilitate Progression of Ovarian Cancer. Cancer Res. 2019, 79, 760–772. [Google Scholar] [CrossRef]
- Kumari, S.; Devi, G.t.; Badana, A.; Dasari, V.R.; Malla, R.R. CD151-A Striking Marker for Cancer Therapy. Biomark Cancer 2015, 7, 7–11. [Google Scholar] [CrossRef]
- Zhu, J.; Cai, T.; Zhou, J.; Du, W.; Zeng, Y.; Liu, T.; Fu, Y.; Li, Y.; Qian, Q.; Yang, X.H.; et al. CD151 drives cancer progression depending on integrin alpha3beta1 through EGFR signaling in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2021, 40, 192. [Google Scholar] [CrossRef]
- Huang, J.; Zhou, N.; Watabe, K.; Lu, Z.; Wu, F.; Xu, M.; Mo, Y.Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014, 5, e1008. [Google Scholar] [CrossRef]
- Nath, S.; Mukherjee, P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 2014, 20, 332–342. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhan, H.; Zhang, Y.; Yang, J.; Liu, M.; Xu, C.; Fan, X.; Zhang, J.; Zhou, Z.; Shi, X.; et al. ZIP4 promotes non-small cell lung cancer metastasis by activating snail-N-cadherin signaling axis. Cancer Lett. 2021, 521, 71–81. [Google Scholar] [CrossRef]
- Ersahin, T.; Tuncbag, N.; Cetin-Atalay, R. The PI3K/AKT/mTOR interactive pathway. Mol. Biosyst. 2015, 11, 1946–1954. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Janssens, R.; Struyf, S.; Proost, P. The unique structural and functional features of CXCL12. Cell Mol. Immunol. 2018, 15, 299–311. [Google Scholar] [CrossRef]
- Obri, A.; Ouararhni, K.; Papin, C.; Diebold, M.L.; Padmanabhan, K.; Marek, M.; Stoll, I.; Roy, L.; Reilly, P.T.; Mak, T.W.; et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 2014, 505, 648–653. [Google Scholar] [CrossRef]
- Meissl, K.; Macho-Maschler, S.; Muller, M.; Strobl, B. The good and the bad faces of STAT1 in solid tumours. Cytokine 2017, 89, 12–20. [Google Scholar] [CrossRef]
- Donehower, L.A.; Soussi, T.; Korkut, A.; Liu, Y.; Schultz, A.; Cardenas, M.; Li, X.; Babur, O.; Hsu, T.K.; Lichtarge, O.; et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep. 2019, 28, 3010. [Google Scholar] [CrossRef]
- Jo, A.; Denduluri, S.; Zhang, B.; Wang, Z.; Yin, L.; Yan, Z.; Kang, R.; Shi, L.L.; Mok, J.; Lee, M.J.; et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis 2014, 1, 149–161. [Google Scholar] [CrossRef]
- Krezel, W.; Ruhl, R.; de Lera, A.R. Alternative retinoid X receptor (RXR) ligands. Mol. Cell Endocrinol. 2019, 491, 110436. [Google Scholar] [CrossRef]
- Wang, F.; El-Saafin, F.; Ye, T.; Stierle, M.; Negroni, L.; Durik, M.; Fischer, V.; Devys, D.; Vincent, S.D.; Tora, L. Histone H2Bub1 deubiquitylation is essential for mouse development, but does not regulate global RNA polymerase II transcription. Cell Death Differ. 2021, 28, 2385–2403. [Google Scholar] [CrossRef]
- Levinson, K.; Dorigo, O.; Rubin, K.; Moore, K. Immunotherapy in Gynecologic Cancers: What We Know Now and Where We Are Headed. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, e126–e140. [Google Scholar] [CrossRef] [PubMed]
- Lobenwein, D.; Kocher, F.; Dobner, S.; Gollmann-Tepekoylu, C.; Holfeld, J. Cardiotoxic mechanisms of cancer immunotherapy—A systematic review. Int. J. Cardiol. 2021, 323, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Palaia, I.; Tomao, F.; Sassu, C.M.; Musacchio, L.; Benedetti Panici, P. Immunotherapy For Ovarian Cancer: Recent Advances And Combination Therapeutic Approaches. Onco. Targets Ther. 2020, 13, 6109–6129. [Google Scholar] [CrossRef] [PubMed]
- Seledtsov, V.I.; von Delwig, A. Clinically feasible and prospective immunotherapeutic interventions in multidirectional comprehensive treatment of cancer. Expert Opin. Biol. Ther. 2021, 21, 323–342. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Xu, Z.; Miao, L.; Huang, L. mRNA Vaccine with Antigen-Specific Checkpoint Blockade Induces an Enhanced Immune Response against Established Melanoma. Mol. Ther. 2018, 26, 420–434. [Google Scholar] [CrossRef]
- Yang, X.; Fan, J.; Wu, Y.; Ma, Z.; Huang, J.; Zhang, Y.; Zhou, Z.; Mo, F.; Liu, X.; Yuan, H.; et al. Synthetic multiepitope neoantigen DNA vaccine for personalized cancer immunotherapy. Nanomedicine 2021, 37, 102443. [Google Scholar] [CrossRef]
- Zamani, P.; Teymouri, M.; Nikpoor, A.R.; Navashenaq, J.G.; Gholizadeh, Z.; Darban, S.A.; Jaafari, M.R. Nanoliposomal vaccine containing long multi-epitope peptide E75-AE36 pulsed PADRE-induced effective immune response in mice TUBO model of breast cancer. Eur. J. Cancer 2020, 129, 80–96. [Google Scholar] [CrossRef]
- Marks, J.R.; Davidoff, A.M.; Kerns, B.J.; Humphrey, P.A.; Pence, J.C.; Dodge, R.K.; Clarke-Pearson, D.L.; Iglehart, J.D.; Bast, R.C., Jr.; Berchuck, A. Overexpression and mutation of p53 in epithelial ovarian cancer. Cancer Res. 1991, 51, 2979–2984. [Google Scholar]
- Havrilesky, L.; Darcy, K.M.; Hamdan, H.; Priore, R.L.; Leon, J.; Bell, J.; Berchuck, A.; Gynecologic Oncology Group, S. Prognostic significance of p53 mutation and p53 overexpression in advanced epithelial ovarian cancer: A Gynecologic Oncology Group Study. J. Clin. Oncol. 2003, 21, 3814–3825. [Google Scholar] [CrossRef]
- Leffers, N.; Lambeck, A.J.; Gooden, M.J.; Hoogeboom, B.N.; Wolf, R.; Hamming, I.E.; Hepkema, B.G.; Willemse, P.H.; Molmans, B.H.; Hollema, H.; et al. Immunization with a P53 synthetic long peptide vaccine induces P53-specific immune responses in ovarian cancer patients, a phase II trial. Int. J. Cancer 2009, 125, 2104–2113. [Google Scholar] [CrossRef]
- Vermeij, R.; Leffers, N.; Hoogeboom, B.N.; Hamming, I.L.; Wolf, R.; Reyners, A.K.; Molmans, B.H.; Hollema, H.; Bart, J.; Drijfhout, J.W.; et al. Potentiation of a p53-SLP vaccine by cyclophosphamide in ovarian cancer: A single-arm phase II study. Int. J. Cancer 2012, 131, E670–E680. [Google Scholar] [CrossRef]
- Hardwick, N.R.; Frankel, P.; Ruel, C.; Kilpatrick, J.; Tsai, W.; Kos, F.; Kaltcheva, T.; Leong, L.; Morgan, R.; Chung, V.; et al. p53-Reactive T Cells Are Associated with Clinical Benefit in Patients with Platinum-Resistant Epithelial Ovarian Cancer After Treatment with a p53 Vaccine and Gemcitabine Chemotherapy. Clin. Cancer Res. 2018, 24, 1315–1325. [Google Scholar] [CrossRef]
- Davodabadi, F.; Sarhadi, M.; Arabpour, J.; Sargazi, S.; Rahdar, A.; Diez-Pascual, A.M. Breast cancer vaccines: New insights into immunomodulatory and nano-therapeutic approaches. J. Control Release 2022, 349, 844–875. [Google Scholar] [CrossRef]
- Advani, S.; Kopetz, S. Ongoing and future directions in the management of metastatic colorectal cancer: Update on clinical trials. J. Surg. Oncol. 2019, 119, 642–652. [Google Scholar] [CrossRef]
- Gaiger, A.; Reese, V.; Disis, M.L.; Cheever, M.A. Immunity to WT1 in the animal model and in patients with acute myeloid leukemia. Blood 2000, 96, 1480–1489. [Google Scholar] [CrossRef]
- Nishida, S.; Morimoto, S.; Oji, Y.; Morita, S.; Shirakata, T.; Enomoto, T.; Tsuboi, A.; Ueda, Y.; Yoshino, K.; Shouq, A.; et al. Cellular and Humoral Immune Responses Induced by an HLA Class I-restricted Peptide Cancer Vaccine Targeting WT1 Are Associated With Favorable Clinical Outcomes in Advanced Ovarian Cancer. J. Immunother. 2022, 45, 56–66. [Google Scholar] [CrossRef]
- Odunsi, K.; Jungbluth, A.A.; Stockert, E.; Qian, F.; Gnjatic, S.; Tammela, J.; Intengan, M.; Beck, A.; Keitz, B.; Santiago, D.; et al. NY-ESO-1 and LAGE-1 cancer-testis antigens are potential targets for immunotherapy in epithelial ovarian cancer. Cancer Res. 2003, 63, 6076–6083. [Google Scholar]
- Odunsi, K.; Qian, F.; Matsuzaki, J.; Mhawech-Fauceglia, P.; Andrews, C.; Hoffman, E.W.; Pan, L.; Ritter, G.; Villella, J.; Thomas, B.; et al. Vaccination with an NY-ESO-1 peptide of HLA class I/II specificities induces integrated humoral and T cell responses in ovarian cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 12837–12842. [Google Scholar] [CrossRef]
- Odunsi, K.; Matsuzaki, J.; Karbach, J.; Neumann, A.; Mhawech-Fauceglia, P.; Miller, A.; Beck, A.; Morrison, C.D.; Ritter, G.; Godoy, H.; et al. Efficacy of vaccination with recombinant vaccinia and fowlpox vectors expressing NY-ESO-1 antigen in ovarian cancer and melanoma patients. Proc. Natl. Acad. Sci. USA 2012, 109, 5797–5802. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, X.; Hu, B.; He, P.; Jiang, X.; Wang, Z.; Zhu, H.; Hu, L.; Yu, M.; Feng, M. Generated SecPen_NY-ESO-1_ubiquitin-pulsed dendritic cell cancer vaccine elicits stronger and specific T cell immune responses. Acta. Pharm. Sin. B 2021, 11, 476–487. [Google Scholar] [CrossRef]
- Yu, S.; Yi, M.; Qin, S.; Wu, K. Next generation chimeric antigen receptor T cells: Safety strategies to overcome toxicity. Mol. Cancer 2019, 18, 125. [Google Scholar] [CrossRef] [PubMed]
- Nakatsura, T.; Nishimura, Y. Usefulness of the novel oncofetal antigen glypican-3 for diagnosis of hepatocellular carcinoma and melanoma. BioDrugs 2005, 19, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Sakata, J.; Utsumi, F.; Sekiya, R.; Kajiyama, H.; Shibata, K.; Kikkawa, F.; Nakatsura, T. Efficacy of glypican-3-derived peptide vaccine therapy on the survival of patients with refractory ovarian clear cell carcinoma. Oncoimmunology 2016, 5, e1238542. [Google Scholar] [CrossRef] [PubMed]
- Ukai, M.; Yokoi, A.; Yoshida, K.; Suzuki, S.; Shibata, K.; Kikkawa, F.; Nakatsura, T.; Kajiyama, H. Extracellular miRNAs as Predictive Biomarkers for Glypican-3-Derived Peptide Vaccine Therapy Response in Ovarian Clear Cell Carcinoma. Cancers 2021, 13, 550. [Google Scholar] [CrossRef]
- Pang, N.; Shi, J.; Qin, L.; Chen, A.; Tang, Y.; Yang, H.; Huang, Y.; Wu, Q.; Li, X.; He, B.; et al. IL-7 and CCL19-secreting CAR-T cell therapy for tumors with positive glypican-3 or mesothelin. J. Hematol. Oncol. 2021, 14, 118. [Google Scholar] [CrossRef]
- Hubbe, M.L.; Jaehger, D.E.; Andresen, T.L.; Andersen, M.H. Leveraging Endogenous Dendritic Cells to Enhance the Therapeutic Efficacy of Adoptive T-Cell Therapy and Checkpoint Blockade. Front. Immunol. 2020, 11, 578349. [Google Scholar] [CrossRef]
- Correll, A.; Tuettenberg, A.; Becker, C.; Jonuleit, H. Increased regulatory T-cell frequencies in patients with advanced melanoma correlate with a generally impaired T-cell responsiveness and are restored after dendritic cell-based vaccination. Exp. Dermatol. 2010, 19, e213–e221. [Google Scholar] [CrossRef]
- Tanyi, J.L.; Bobisse, S.; Ophir, E.; Tuyaerts, S.; Roberti, A.; Genolet, R.; Baumgartner, P.; Stevenson, B.J.; Iseli, C.; Dangaj, D.; et al. Personalized cancer vaccine effectively mobilizes antitumor T cell immunity in ovarian cancer. Sci. Transl. Med 2018, 10, eaao5931. [Google Scholar] [CrossRef]
- Morisaki, T.; Hikichi, T.; Onishi, H.; Morisaki, T.; Kubo, M.; Hirano, T.; Yoshimura, S.; Kiyotani, K.; Nakamura, Y. Intranodal Administration of Neoantigen Peptide-loaded Dendritic Cell Vaccine Elicits Epitope-specific T Cell Responses and Clinical Effects in a Patient with Chemorefractory Ovarian Cancer with Malignant Ascites. Immunol. Investig. 2021, 50, 562–579. [Google Scholar] [CrossRef]
- Caro, A.A.; Deschoemaeker, S.; Allonsius, L.; Coosemans, A.; Laoui, D. Dendritic Cell Vaccines: A Promising Approach in the Fight against Ovarian Cancer. Cancers 2022, 14, 4037. [Google Scholar] [CrossRef]
- Fares, C.M.; Van Allen, E.M.; Drake, C.G.; Allison, J.P.; Hu-Lieskovan, S. Mechanisms of Resistance to Immune Checkpoint Blockade: Why Does Checkpoint Inhibitor Immunotherapy Not Work for All Patients? Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 147–164. [Google Scholar] [CrossRef]
- Gonzalez, N.L.; Puwanant, A.; Lu, A.; Marks, S.M.; Zivkovic, S.A. Myasthenia triggered by immune checkpoint inhibitors: New case and literature review. Neuromuscul. Disord. 2017, 27, 266–268. [Google Scholar] [CrossRef]
- Kreileder, M.; Barrett, I.; Bendtsen, C.; Brennan, D.; Kolch, W. Signaling Dynamics Regulating Crosstalks between T-Cell Activation and Immune Checkpoints. Trends Cell Biol. 2021, 31, 224–235. [Google Scholar] [CrossRef]
- Park, S.J.; Chang, S.J.; Suh, D.H.; Kong, T.W.; Song, H.; Kim, T.H.; Kim, J.W.; Kim, H.S.; Lee, S.J. A phase IA dose-escalation study of PHI-101, a new checkpoint kinase 2 inhibitor, for platinum-resistant recurrent ovarian cancer. BMC Cancer 2022, 22, 28. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Chang, H.W.; Frey, G.; Liu, H.; Xing, C.; Steinman, L.; Boyle, W.J.; Short, J.M. Generating tumor-selective conditionally active biologic anti-CTLA4 antibodies via protein-associated chemical switches. Proc. Natl. Acad. Sci. USA 2021, 118, e2020606118. [Google Scholar] [CrossRef]
- Loke, P.; Allison, J.P. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc. Natl. Acad. Sci. USA 2003, 100, 5336–5341. [Google Scholar] [CrossRef]
- Hamanishi, J.; Mandai, M.; Iwasaki, M.; Okazaki, T.; Tanaka, Y.; Yamaguchi, K.; Higuchi, T.; Yagi, H.; Takakura, K.; Minato, N.; et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 3360–3365. [Google Scholar] [CrossRef]
- Hino, R.; Kabashima, K.; Kato, Y.; Yagi, H.; Nakamura, M.; Honjo, T.; Okazaki, T.; Tokura, Y. Tumor cell expression of programmed cell death-1 ligand 1 is a prognostic factor for malignant melanoma. Cancer 2010, 116, 1757–1766. [Google Scholar] [CrossRef]
- Tseng, S.Y.; Otsuji, M.; Gorski, K.; Huang, X.; Slansky, J.E.; Pai, S.I.; Shalabi, A.; Shin, T.; Pardoll, D.M.; Tsuchiya, H. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J. Exp. Med. 2001, 193, 839–846. [Google Scholar] [CrossRef]
- Fecher, L.A.; Agarwala, S.S.; Hodi, F.S.; Weber, J.S. Ipilimumab and its toxicities: A multidisciplinary approach. Oncologist 2013, 18, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A.; Shapira-Frommer, R.; Santin, A.D.; Lisyanskaya, A.S.; Pignata, S.; Vergote, I.; Raspagliesi, F.; Sonke, G.S.; Birrer, M.; Provencher, D.M.; et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: Results from the phase II KEYNOTE-100 study. Ann. Oncol. 2019, 30, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Mehnert, J.M.; Varga, A.; Brose, M.S.; Aggarwal, R.R.; Lin, C.C.; Prawira, A.; de Braud, F.; Tamura, K.; Doi, T.; Piha-Paul, S.A.; et al. Safety and antitumor activity of the anti-PD-1 antibody pembrolizumab in patients with advanced, PD-L1-positive papillary or follicular thyroid cancer. BMC Cancer 2019, 19, 196. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Xiong, N.; Cheng, S.C.; Barry, W.T.; Penson, R.T.; Konstantinopoulos, P.A.; Hoffman, M.A.; Horowitz, N.; Dizon, D.S.; Stover, E.H.; et al. Combined pembrolizumab and pegylated liposomal doxorubicin in platinum resistant ovarian cancer: A phase 2 clinical trial. Gynecol. Oncol. 2020, 159, 72–78. [Google Scholar] [CrossRef]
- Lin, Y.C.; Wen, K.C.; Sung, P.L.; Chou, Y.T.; Liew, P.L.; Chen, L.Y.; Huang, R.L.; Lai, H.C.; Chang, L.T. Complete remission of heavily treated ovarian clear cell carcinoma with ARID1A mutations after pembrolizumab and bevacizumab combination therapy: A case report. J. Ovarian. Res. 2020, 13, 143. [Google Scholar] [CrossRef]
- Hamanishi, J.; Mandai, M.; Ikeda, T.; Minami, M.; Kawaguchi, A.; Murayama, T.; Kanai, M.; Mori, Y.; Matsumoto, S.; Chikuma, S.; et al. Safety and Antitumor Activity of Anti-PD-1 Antibody, Nivolumab, in Patients With Platinum-Resistant Ovarian Cancer. J. Clin. Oncol. 2015, 33, 4015–4022. [Google Scholar] [CrossRef]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef]
- Zamarin, D.; Burger, R.A.; Sill, M.W.; Powell, D.J., Jr.; Lankes, H.A.; Feldman, M.D.; Zivanovic, O.; Gunderson, C.; Ko, E.; Mathews, C.; et al. Randomized Phase II Trial of Nivolumab Versus Nivolumab and Ipilimumab for Recurrent or Persistent Ovarian Cancer: An NRG Oncology Study. J. Clin. Oncol. 2020, 38, 1814–1823. [Google Scholar] [CrossRef]
- Herbst, R.S.; Soria, J.C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef]
- Liu, J.F.; Gordon, M.; Veneris, J.; Braiteh, F.; Balmanoukian, A.; Eder, J.P.; Oaknin, A.; Hamilton, E.; Wang, Y.; Sarkar, I.; et al. Safety, clinical activity and biomarker assessments of atezolizumab from a Phase I study in advanced/recurrent ovarian and uterine cancers. Gynecol. Oncol. 2019, 154, 314–322. [Google Scholar] [CrossRef]
- Moroney, J.W.; Powderly, J.; Lieu, C.H.; Bendell, J.C.; Eckhardt, S.G.; Chang, C.W.; Molinero, L.; Spahn, J.; Williams, P.; Lin, Y.G.; et al. Safety and Clinical Activity of Atezolizumab Plus Bevacizumab in Patients with Ovarian Cancer: A Phase Ib Study. Clin. Cancer Res. 2020, 26, 5631–5637. [Google Scholar] [CrossRef]
- Boyerinas, B.; Jochems, C.; Fantini, M.; Heery, C.R.; Gulley, J.L.; Tsang, K.Y.; Schlom, J. Antibody-Dependent Cellular Cytotoxicity Activity of a Novel Anti-PD-L1 Antibody Avelumab (MSB0010718C) on Human Tumor Cells. Cancer Immunol. Res. 2015, 3, 1148–1157. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Russell, J.; Hamid, O.; Bhatia, S.; Terheyden, P.; D’Angelo, S.P.; Shih, K.C.; Lebbe, C.; Linette, G.P.; Milella, M.; et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: A multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1374–1385. [Google Scholar] [CrossRef]
- Hamilton, G. Avelumab: Search for combinations of immune checkpoint inhibition with chemotherapy. Expert Opin. Biol. Ther. 2021, 21, 311–322. [Google Scholar] [CrossRef]
- Disis, M.L.; Taylor, M.H.; Kelly, K.; Beck, J.T.; Gordon, M.; Moore, K.M.; Patel, M.R.; Chaves, J.; Park, H.; Mita, A.C.; et al. Efficacy and Safety of Avelumab for Patients With Recurrent or Refractory Ovarian Cancer: Phase 1b Results From the JAVELIN Solid Tumor Trial. JAMA Oncol. 2019, 5, 393–401. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Fujiwara, K.; Ledermann, J.A.; Oza, A.M.; Kristeleit, R.; Ray-Coquard, I.L.; Richardson, G.E.; Sessa, C.; Yonemori, K.; Banerjee, S.; et al. Avelumab alone or in combination with chemotherapy versus chemotherapy alone in platinum-resistant or platinum-refractory ovarian cancer (JAVELIN Ovarian 200): An open-label, three-arm, randomised, phase 3 study. Lancet Oncol. 2021, 22, 1034–1046. [Google Scholar] [CrossRef]
- Monk, B.J.; Colombo, N.; Oza, A.M.; Fujiwara, K.; Birrer, M.J.; Randall, L.; Poddubskaya, E.V.; Scambia, G.; Shparyk, Y.V.; Lim, M.C.; et al. Chemotherapy with or without avelumab followed by avelumab maintenance versus chemotherapy alone in patients with previously untreated epithelial ovarian cancer (JAVELIN Ovarian 100): An open-label, randomised, phase 3 trial. Lancet Oncol. 2021, 22, 1275–1289. [Google Scholar] [CrossRef]
- Song, D.G.; Ye, Q.; Santoro, S.; Fang, C.; Best, A.; Powell, D.J., Jr. Chimeric NKG2D CAR-expressing T cell-mediated attack of human ovarian cancer is enhanced by histone deacetylase inhibition. Hum. Gene Ther. 2013, 24, 295–305. [Google Scholar] [CrossRef]
- Yan, W.; Hu, H.; Tang, B. Advances Of Chimeric Antigen Receptor T Cell Therapy In Ovarian Cancer. Onco. Targets Ther. 2019, 12, 8015–8022. [Google Scholar] [CrossRef]
- Morello, A.; Sadelain, M.; Adusumilli, P.S. Mesothelin-Targeted CARs: Driving T Cells to Solid Tumors. Cancer Discov. 2016, 6, 133–146. [Google Scholar] [CrossRef]
- Schoutrop, E.; El-Serafi, I.; Poiret, T.; Zhao, Y.; Gultekin, O.; He, R.; Moyano-Galceran, L.; Carlson, J.W.; Lehti, K.; Hassan, M.; et al. Mesothelin-Specific CAR T Cells Target Ovarian Cancer. Cancer Res. 2021, 81, 3022–3035. [Google Scholar] [CrossRef] [PubMed]
- Felder, M.; Kapur, A.; Gonzalez-Bosquet, J.; Horibata, S.; Heintz, J.; Albrecht, R.; Fass, L.; Kaur, J.; Hu, K.; Shojaei, H.; et al. MUC16 (CA125): Tumor biomarker to cancer therapy, a work in progress. Mol. Cancer 2014, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Cheng, Z.; Luo, L.; Yang, Y.; Zhang, Z.; Ma, H.; Chen, T.; Huang, X.; Lin, S.Y.; Jin, M.; et al. C-terminus of MUC16 activates Wnt signaling pathway through its interaction with beta-catenin to promote tumorigenesis and metastasis. Oncotarget 2016, 7, 36800–36813. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, J. Therapeutic effect of dual CAR-T targeting PDL1 and MUC16 antigens on ovarian cancer cells in mice. BMC Cancer 2020, 20, 678. [Google Scholar] [CrossRef] [PubMed]
- Coelho, R.; Marcos-Silva, L.; Ricardo, S.; Ponte, F.; Costa, A.; Lopes, J.M.; David, L. Peritoneal dissemination of ovarian cancer: Role of MUC16-mesothelin interaction and implications for treatment. Expert. Rev. Anticancer Ther. 2018, 18, 177–186. [Google Scholar] [CrossRef]
- Coney, L.R.; Tomassetti, A.; Carayannopoulos, L.; Frasca, V.; Kamen, B.A.; Colnaghi, M.I.; Zurawski, V.R., Jr. Cloning of a tumor-associated antigen: MOv18 and MOv19 antibodies recognize a folate-binding protein. Cancer Res. 1991, 51, 6125–6132. [Google Scholar]
- Kandalaft, L.E.; Powell, D.J., Jr.; Coukos, G. A phase I clinical trial of adoptive transfer of folate receptor-alpha redirected autologous T cells for recurrent ovarian cancer. J. Transl. Med. 2012, 10, 157. [Google Scholar] [CrossRef]
- Zuo, S.; Wen, Y.; Panha, H.; Dai, G.; Wang, L.; Ren, X.; Fu, K. Modification of cytokine-induced killer cells with folate receptor alpha (FRalpha)-specific chimeric antigen receptors enhances their antitumor immunity toward FRalpha-positive ovarian cancers. Mol. Immunol. 2017, 85, 293–304. [Google Scholar] [CrossRef]
- Berchuck, A.; Kamel, A.; Whitaker, R.; Kerns, B.; Olt, G.; Kinney, R.; Soper, J.T.; Dodge, R.; Clarke-Pearson, D.L.; Marks, P.; et al. Overexpression of HER-2/neu is associated with poor survival in advanced epithelial ovarian cancer. Cancer Res. 1990, 50, 4087–4091. [Google Scholar]
- Jiang, D.; Im, H.J.; Sun, H.; Valdovinos, H.F.; England, C.G.; Ehlerding, E.B.; Nickles, R.J.; Lee, D.S.; Cho, S.Y.; Huang, P.; et al. Radiolabeled pertuzumab for imaging of human epidermal growth factor receptor 2 expression in ovarian cancer. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 1296–1305. [Google Scholar] [CrossRef]
- Song, X.; Sun, K.; Hu, J.; Zhou, J. Suppression of human epidermal growth factor receptor 2 via interference increases the chemosensitivity of ovarian carcinoma. Oncol. Lett. 2016, 11, 3028–3032. [Google Scholar] [CrossRef]
- Hyrenius-Wittsten, A.; Su, Y.; Park, M.; Garcia, J.M.; Alavi, J.; Perry, N.; Montgomery, G.; Liu, B.; Roybal, K.T. SynNotch CAR circuits enhance solid tumor recognition and promote persistent antitumor activity in mouse models. Sci. Transl. Med. 2021, 13, 591. [Google Scholar] [CrossRef]
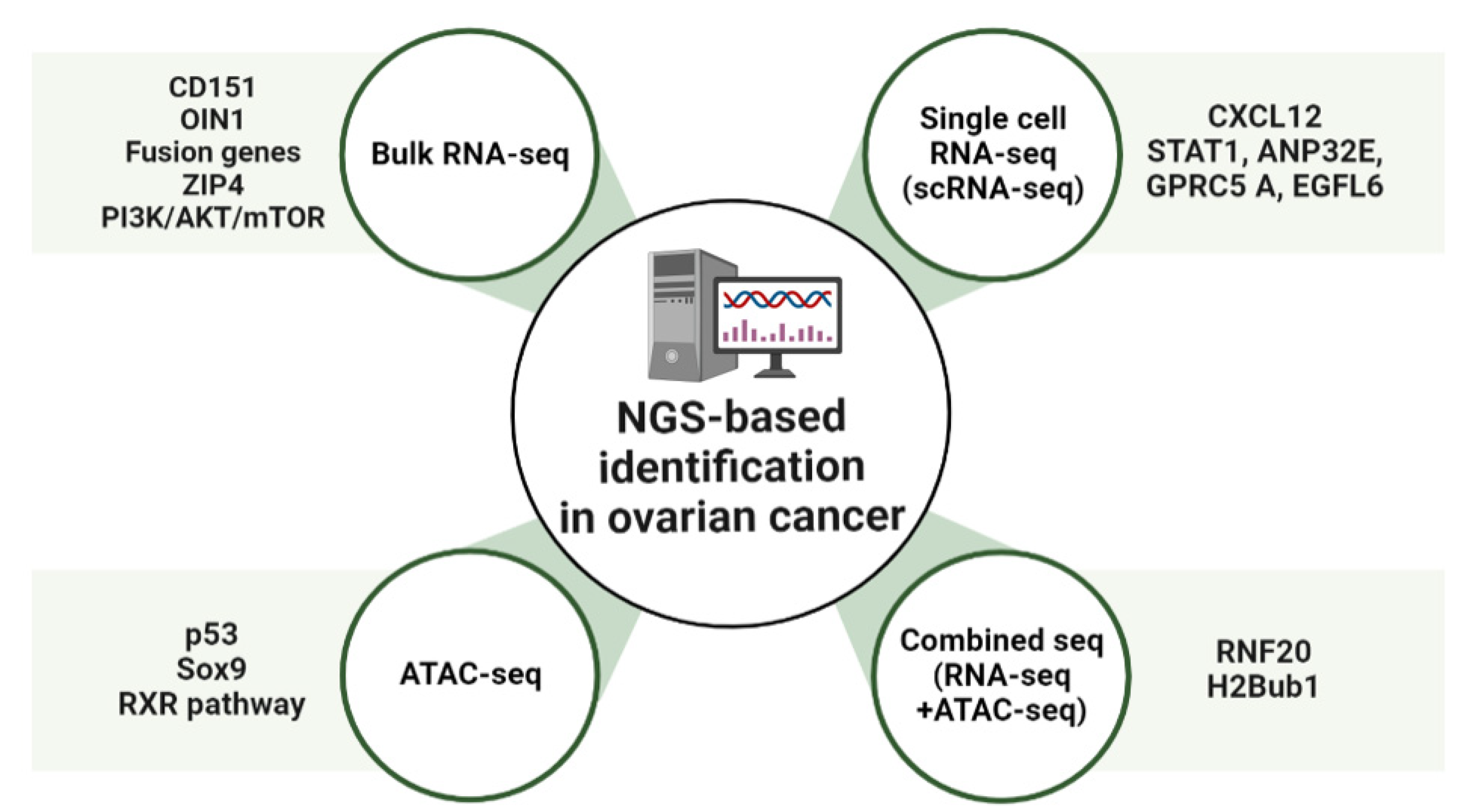
| Seq. Type | Molecule | Function | Reference |
|---|---|---|---|
| Bulk RNA-seq | OIN1(ovarian cancer long intergenic noncoding RNA1) | Unannotated lincRNA originated from chromosome 10q21; Contribute to ovarian cancer progression and lead to suppression of apoptosis | [37] |
| CD151 | Maintaining normal cellular integrity, cell-to-cell communication, wound healing, platelet aggregation, trafficking, cell motility and angiogenesis; Association with various stages of cancer, including metastatic cascade and primary tumor growth | [67,68] | |
| UCA1 (urothelial cancer-associated 1) | Biomarker for diagnosis of bladder cancer; Promote proliferation, migration, and immune response in gastric cancer | [69] | |
| MUC1 | Single pass type Ⅰ transmembrane protein with a heavily glycosylated extracellular domain (oncoprotein); Protection to underlying epithelia; Maintain pluripotency and self-renewal ability in embryonic stem cells | [70] | |
| ZIP4 | A zinc transporter; Related to the process of tumor growth and metastasis of many cancers | [71] | |
| PI3K–AKT–mTOR pathway | A group of plasma membrane-associated lipid kinases; Regulate a broad spectrum of cellular mechanisms involving survival, proliferation, growth, metabolism, angiogenesis and metastasis | [72,73] | |
| scRNA-seq | CXCL12 | Ligand for CXCR4; Important factor in physiological and pathological processes involving embryogenesis, hematopoiesis, angiogenesis and inflammation | [74] |
| ANP32E | Interact with a short region of the docking domain of H2A.Z chaperon | [75,76] | |
| STAT1 | Regulate a variety of cellular processes, such as antimicrobial activities, cell proliferation and cell death | [75,76] | |
| ATAC-seq | TP53 (p53) | Tumor suppressor gene; Inhibit cell division or survival in response to various stresses | [77] |
| SOX9 | Transcription factor; Determine cell fate during embryonic development | [78] | |
| RXR (Retinoid X receptor) | Regulate various functions by virtue of their dimerization with other nuclear hormone receptors, leading to activities of different signaling pathways | [79] | |
| Combined seq | H2Bub1 | weaken DNA–histone interactions and interrupt chromatin compaction | [80] |
| Type of Mechanism | Agents | FDA Approval | Combination with | Ref. |
|---|---|---|---|---|
| Peptide vaccines | ||||
| P53 | P53-SLP vaccine | no | Cyclophosphamide; Gemcitabine | [90,91,92] |
| WT1 | WT1 vaccine | no | [95,96] | |
| NYESO-1 | NYESO-1 vaccine | no | Secretin-penetratin | [97,98,99,100] |
| GPC3 | GPC3 vaccine | no | miR-375, miR-93a, miR-1128; CAR-T cell | [103,104,105] |
| DC | DC protein vaccine | yes | [107] | |
| DC | Neoantigen vaccine | no | [108] | |
| Checkpoint inhibitor | ||||
| CTLA-4 | PHI-101 | yes | [114] | |
| CTLA-4 | Ipilimumab | yes | [115] | |
| PD-1 | Pembrolizumab | yes | Doxorubicin; Bevacizumab | [122,123,124,125] |
| PD-1 | Nivolumab | yes | Ipilimumab | [126,128] |
| PD-L1 | Atezolizumab | yes | Bevacizumab | [130,131] |
| PD-L1 | Avelumab | yes | Axitinib; Docetaxel; Doxolubicin | [134,135,136] |
| PD-L1 | Avelumab | yes | Carboplatin + Paclitaxel | [137] |
| CAR-T | ||||
| Mesothelin | MSTL-CAR-T | no | [141] | |
| MUC16 | PD1-MUC16-CAR-T | no | [144] | |
| FRα | FRα-CAR-T | no | [147] | |
| FRα | FRα-CAR-T | no | Cytokine-inducing killer cell | [148] |
| HER2 | HER2-CAR-T | no | Pertuzumab | [150] |
| HER2 | HER2-CAR-T | no | Synthetic Notch | [152] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, H.; Kim, A.; Jang, H. Immunotherapeutic Approaches in Ovarian Cancer. Curr. Issues Mol. Biol. 2023, 45, 1233-1249. https://doi.org/10.3390/cimb45020081
Yoon H, Kim A, Jang H. Immunotherapeutic Approaches in Ovarian Cancer. Current Issues in Molecular Biology. 2023; 45(2):1233-1249. https://doi.org/10.3390/cimb45020081
Chicago/Turabian StyleYoon, Hyunho, Ayoung Kim, and Hoon Jang. 2023. "Immunotherapeutic Approaches in Ovarian Cancer" Current Issues in Molecular Biology 45, no. 2: 1233-1249. https://doi.org/10.3390/cimb45020081
APA StyleYoon, H., Kim, A., & Jang, H. (2023). Immunotherapeutic Approaches in Ovarian Cancer. Current Issues in Molecular Biology, 45(2), 1233-1249. https://doi.org/10.3390/cimb45020081






