Molecular Mechanisms Underlying CRISPR/Cas-Based Assays for Nucleic Acid Detection
Abstract
1. Introduction
2. Sample Preparation Methods for the Downstream Nucleic Acid Detection
3. Methods of Isothermal Amplification
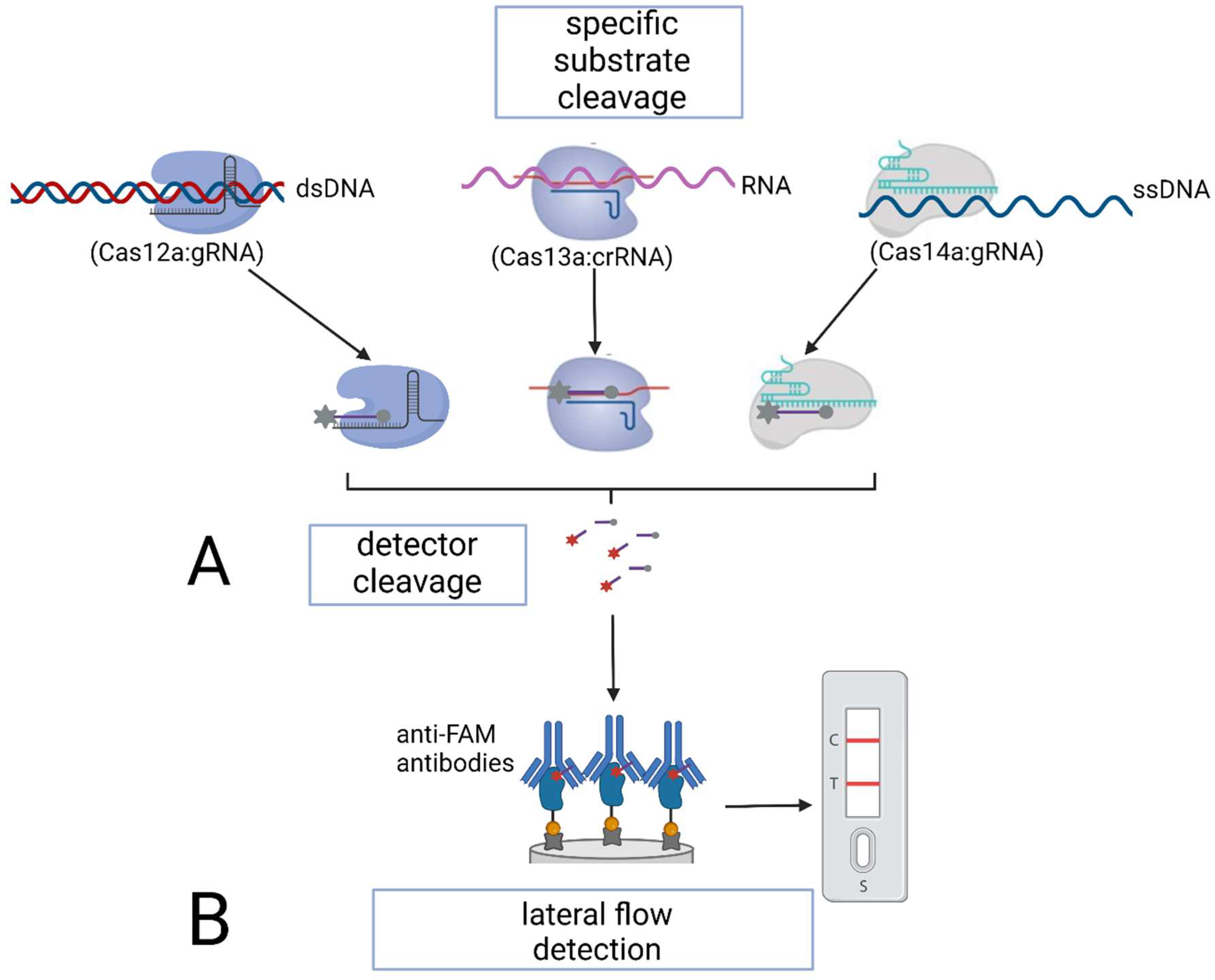
4. Application of Genome-Editing Proteins in Nucleic Acid Detection Assays
5. Other Detection Methods Involving Cas Proteins
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saiki, R.K.; Scharf, S.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985, 230, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90, 2193–2200. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Zhou, W.; Hu, L.; Ying, L.; Zhao, Z.; Chu, P.K.; Yu, X.F. A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018, 9, 5012. [Google Scholar] [CrossRef]
- Wang, X.; Xiong, E.; Tian, T.; Cheng, M.; Lin, W.; Wang, H.; Zhang, G.; Sun, J.; Zhou, X. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9-Mediated Lateral Flow Nucleic Acid Assay. ACS Nano 2020, 14, 2497–2508. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, X.; Chen, X.; Qiu, X.; Qing, G.; Zhang, H.; Zhang, L.; Hu, X.; He, Z.; Zhong, D.; et al. Rolling Circular Amplification (RCA)-Assisted CRISPR/Cas9 Cleavage (RACE) for Highly Specific Detection of Multiple Extracellular Vesicle MicroRNAs. Anal. Chem. 2020, 92, 2176–2185. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef]
- Iwasaki, R.S.; Batey, R.T. SPRINT: A Cas13a-based platform for detection of small molecules. Nucleic Acids Res. 2020, 48, 101. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.-S.F.; Kemball, M.E.; et al. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 1, 5921. [Google Scholar] [CrossRef]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.; Paul, L.; Parham, L.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Scheler, O.; Kaplinski, L.; Glynn, B.; Palta, P.; Parkel, S.; Toome, K.; Maher, M.; Barry, T.; Remm, M.; Kurg, A. Detection of NASBA amplified bacterial tmRNA molecules on SLICSel designed microarray probe. BMC Biotechnol. 2011, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Zhang, J.Z.; Zhang, M.J. Application of NASBA and RPA in detection of pathogenic bacteria. Zhonghua Liu Xing Bing Xue Za Zhi 2019, 40, 1018–1022. [Google Scholar] [PubMed]
- Zhai, L.; Liu, H.; Li, J.; Lu, Z.; Bie, X. A duplex real-time NASBA assay targeting a serotype-specific gene for rapid detection of viable Salmonella Paratyphi C in retail foods of animal origin. Can. J. Microbiol. 2022, 68, 259–268. [Google Scholar] [CrossRef]
- Cook, N. The use of NASBA for the detection of microbial pathogens in food and environmental samples. J. Microbiol. Methods 2003, 68, 165–174. [Google Scholar] [CrossRef]
- Romano, J.W.; van Gemen, B.; Kievits, T. NASBA: A novel, isothermal detection technology for qualitative and quantitative HIV-1 RNA measurements. Clin. Lab. Med. 1996, 16, 89–103. [Google Scholar] [CrossRef]
- Kia, V.; Tafti, A.; Paryan, M.; Mohammadi-Yeganeh, S. Evaluation of real-time NASBA assay for the detection of SARS-CoV-2 compared with real-time PCR. Ir. J. Med. Sci. 2022, 6, 1–7. [Google Scholar] [CrossRef]
- Lau, L.T.; Fung, Y.W.; Yu, A.C. Detection of animal viruses using nucleic acid sequence-based amplification (NASBA). Dev. Biol. 2006, 126, 7–15. [Google Scholar]
- Lau, L.T.; Reid, S.M.; King, D.P.; Lau, A.M.; Shaw, A.E.; Ferris, N.P.; Yu, A.C. Detection of foot-and-mouth disease virus by nucleic acid sequence-based amplification (NASBA). Vet. Microbiol. 2008, 126, 101–110. [Google Scholar] [CrossRef]
- Wu, Q.; Suo, C.; Brown, T.; Wang, T.; Teichmann, S.A.; Bassett, A.R. INSIGHT: A population-scale COVID-19 testing strategy combining point-of-care diagnosis with centralized high-throughput sequencing. Sci. Adv. 2021, 7, eabe5054. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, N.; Reed, A.; Gerasimova, Y.V.; Kolpashchikov, D.M. Split Dapoxyl Aptamer for Sequence-Selective Analysis of Nucleic Acid Sequence Based Amplification Amplicons. Anal. Chem. 2019, 91, 2667–2671. [Google Scholar] [CrossRef]
- Lamhoujeb, S.; Charest, H.; Fliss, I.; Ngazoa, S.; Jean, J. Real-time molecular beacon NASBA for rapid and sensitive detection of norovirus GII in clinical samples. Can. J. Microbiol. 2009, 55, 1375–1380. [Google Scholar] [CrossRef]
- Morabito, K.; Wiske, C.; Tripathi, A. Engineering Insights for Multiplexed Real-Time Nucleic Acid Sequence-Based Amplification (NASBA): Implications for Design of Point-of-Care Diagnostics. Mol. Diagn. Ther. 2013, 17, 185–192. [Google Scholar] [CrossRef]
- Ju, Y.; Kim, J.; Park, Y.; Lee, C.Y.; Kim, K.; Hong, K.H.; Lee, H.; Yong, D.; Park, H.G. Rapid and accurate clinical testing for COVID-19 by nicking and extension chain reaction system-based amplification (NESBA). Biosens. Bioelectron. 2022, 196, 113689. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2020, 145, 1950–1960. [Google Scholar] [CrossRef]
- Mayboroda, O.; Gonzalez Benito, A.; Sabaté del Rio, J.; Svobodova, M.; Julich, S.; Tomaso, H.; O’Sullivan, C.K.; Katakis, I. Isothermal solid-phase amplification system for detection of Yersinia pestis. Anal. Bioanal. Chem. 2016, 408, 671–676. [Google Scholar] [CrossRef]
- Martorell, S.; Palanca, S.; Maquieira, Á.; Tortajada-Genaro, L.A. Blocked recombinase polymerase amplification for mutation analysis of PIK3CA gene. Anal. Biochem. 2018, 544, 49–56. [Google Scholar] [CrossRef]
- Kersting, S.; Rausch, V.; Bier, F.F.; von Nickisch-Rosenegk, M. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malar. J. 2014, 13, 99. [Google Scholar] [CrossRef]
- Rosser, A.; Rollinson, D.; Forrest, M.; Webster, B. Isothermal Recombinase Polymerase amplification (RPA) of Schistosoma haematobium DNA and oligochromatographic lateral flow detection. Parasites Vectors 2015, 8, 446. [Google Scholar] [CrossRef]
- Krõlov, K.; Frolova, J.; Tudoran, O.; Suhorutsenko, J.; Lehto, T.; Sibul, H.; Mäger, I.; Laanpere, M.; Tulp, I.; Langel, Ü. Sensitive and rapid detection of Chlamydia trachomatis by recombinase polymerase amplification directly from urine samples. J. Mol. Diagn. 2015, 16, 127–135. [Google Scholar] [CrossRef]
- Lai, M.Y.; Ooi, C.H.; Lau, Y.L. Validation of SYBR green I based closed-tube loop-mediated isothermal amplification (LAMP) assay for diagnosis of knowlesi malaria. Malar. J. 2021, 20, 166. [Google Scholar] [CrossRef]
- Singh, R.; Singh, D.P.; Savargaonkar, D.; Singh, O.P.; Bhatt, R.M.; Valecha, N. Evaluation of SYBR green I based visual loop-mediated isothermal amplification (LAMP) assay for genus and species-specific diagnosis of malaria in P. vivax and P. falciparum endemic regions. J. Vector Borne Dis. 2017, 54, 54–56. [Google Scholar]
- Rolando, J.C.; Jue, E.; Barlow, J.T.; Ismagilov, R. Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification. Nucleic Acids Res. 2020, 48, 42. [Google Scholar] [CrossRef]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Hu, L.; Zhong, H.; Wang, H.; Yusa, S.; Weiss, T.C.; Romaniuk, P.J.; Pickerill, S.; You, Q. Cross priming amplification: Mechanism and optimization for isothermal DNA amplification. Sci. Rep. 2012, 2, 246. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Xu, X.; Wang, X.; Zuo, K.; Li, Z.; Leng, C.; Kan, Y.; Yao, L.; Bi, Y. Novel polymerase spiral reaction assay for the visible molecular detection of porcine circovirus type 3. BMC Vet. Res. 2019, 15, 322. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Dong, D.; Yang, Z.; Zou, D.; Chen, Z.; Yuan, J.; Huang, L. Polymerase Spiral Reaction (PSR): A novel isothermal nucleic acid amplification method. Sci. Rep. 2015, 29, 123–127. [Google Scholar] [CrossRef]
- Gupta, V.; Chakravarti, S.; Chander, V.; Majumder, S.; Bhat, S.A.; Gupta, V.K.; Nandi, S. Polymerase spiral reaction (PSR): A novel, visual isothermal amplification method for detection of canine parvovirus 2 genomic DNA. Arch. Virol. 2017, 162, 1995–2001. [Google Scholar] [CrossRef]
- Wharam, S.D.; Marsh, P.; Lloyd, J.S.; Ray, T.D.; Mock, G.A.; Assenberg, R.; McPhee, J.E.; Brown, P.; Weston, A.; Cardy, D.L.N. Specific detection of DNA and RNA targets using a novel isothermal nucleic acid amplification assay based on the formation of a three-way junction structure. Nucleic Acids Res. 2001, 29, e54. [Google Scholar] [CrossRef]
- Hall, M.J.; Wharam, S.D.; Weston, A.; Cardy, D.L.; Wilson, W.H. Use of signal-mediated amplification of RNA technology (SMART) to detect marine cyanophage DNA. Biotechniques 2002, 32, 604–606, 608–611. [Google Scholar] [CrossRef]
- Wang, G.; Ding, X.; Hu, J.; Wu, W.; Sun, J.; Mu, Y. Unusual isothermal multimerization and amplification by the strand-displacing DNA polymerases with reverse transcription activities. Sci. Rep. 2017, 7, 13928. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.G.; Woolley, A.E.; Segel, M.; Barretto, R.; Ranu, A.; Macrae, R.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef]
- Yuan, C.; Tian, T.; Sun, J.; Hu, M.; Wang, X.; Xiong, E.; Cheng, M.; Bao, Y.; Lin, W.; Jiang, J.; et al. Universal and Naked-Eye Gene Detection Platform Based on the Clustered Regularly Interspaced Short Palindromic Repeats/Cas12a/13a System. Anal. Chem. 2020, 92, 4029–4037. [Google Scholar] [CrossRef] [PubMed]
- López-Valls, M.; Escalona-Noguero, C.; Rodríguez-Díaz, C.; Pardo, D.; Castellanos, M.; Milán-Rois, P.; Martínez-Garay, C.; Coloma, R.; Abreu, M.; Cantón, R.; et al. CASCADE: Naked eye-detection of SARS-CoV-2 using Cas13a and gold nanoparticles. Anal. Chim. Acta 2022, 1205, 339749. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, T.; Wang, M.; Ledesma-Amaro, R.; Lu, Y.; Hu, X.; Zhang, T.; Yang, M.; Li, Y.; Xiang, J.; et al. CRISPR-Cas13a cascade-based viral RNA assay for detecting SARS-CoV-2 and its mutations in clinical samples. Sens. Actuators B Chem. 2022, 362, 131765. [Google Scholar] [CrossRef]
- Chi, Z.; Wu, Y.; Chen, L.; Yang, H.; Khan, M.R.; Busquets, R.; Huang, N.; Lin, X.; Deng, R.; Yamg, W.; et al. CRISPR-Cas14a-integrated strand displacement amplification for rapid and isothermal detection of cholangiocarcinoma associated circulating microRNAs. Anal. Chim. Acta 2022, 1205, 339763. [Google Scholar] [CrossRef]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.; Kumar, R.; Stephens, S.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333. [Google Scholar] [CrossRef]
- Jia, F.; Li, X.; Zhang, C.; Tang, X. The expanded development and application of CRISPR system for sensitive nucleotide detection. Protein Cell 2017, 11, 624–629. [Google Scholar] [CrossRef]
- Karvelis, T.; Bigelyte, G.; Young, J.K.; Hou, Z.; Zedaveinyte, R.; Budre, K.; Paulraj, S.; Djukanovic, V.; Gasior, S.; Silanskas, A.; et al. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020, 48, 5016–5023. [Google Scholar] [CrossRef] [PubMed]
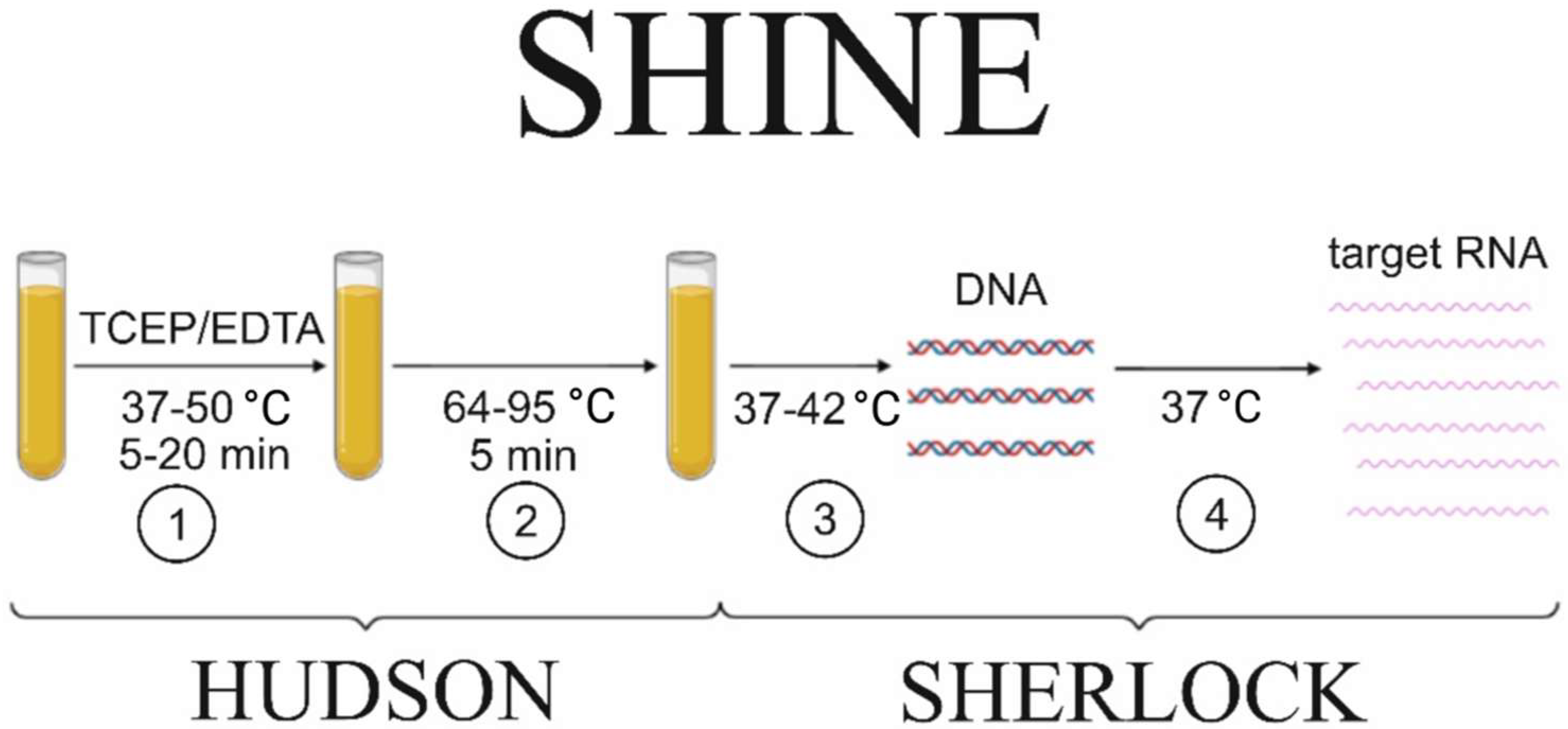
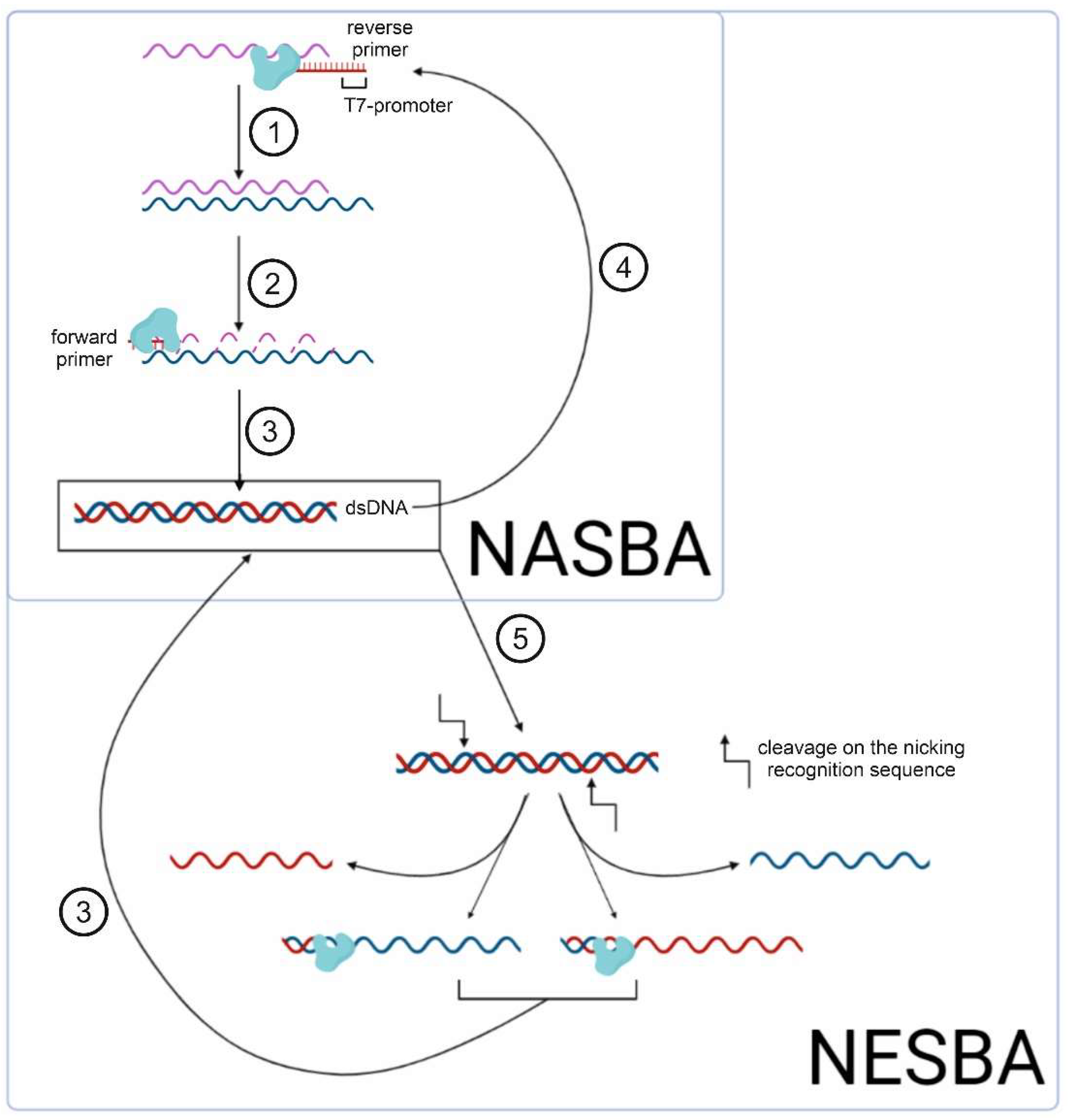
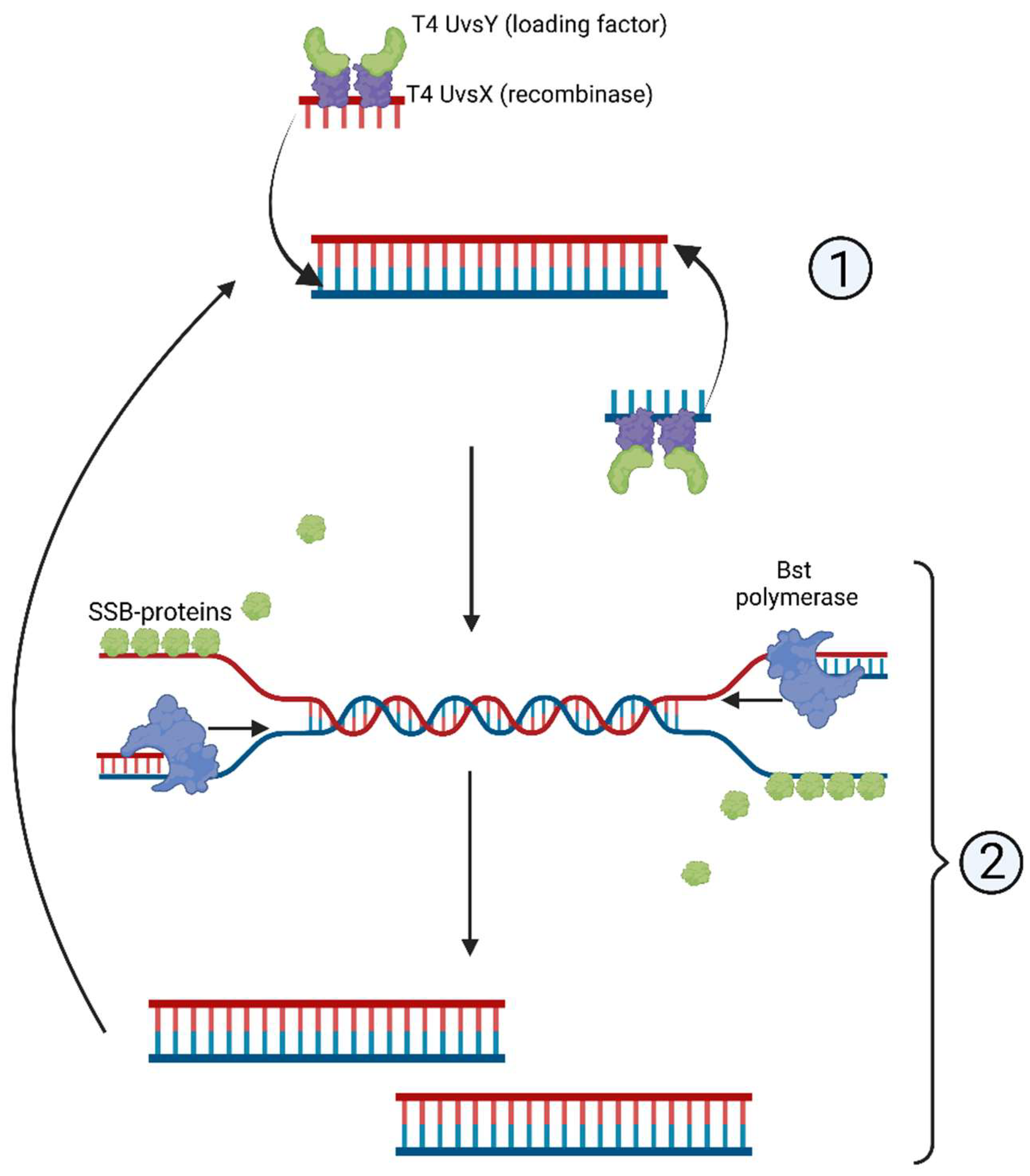

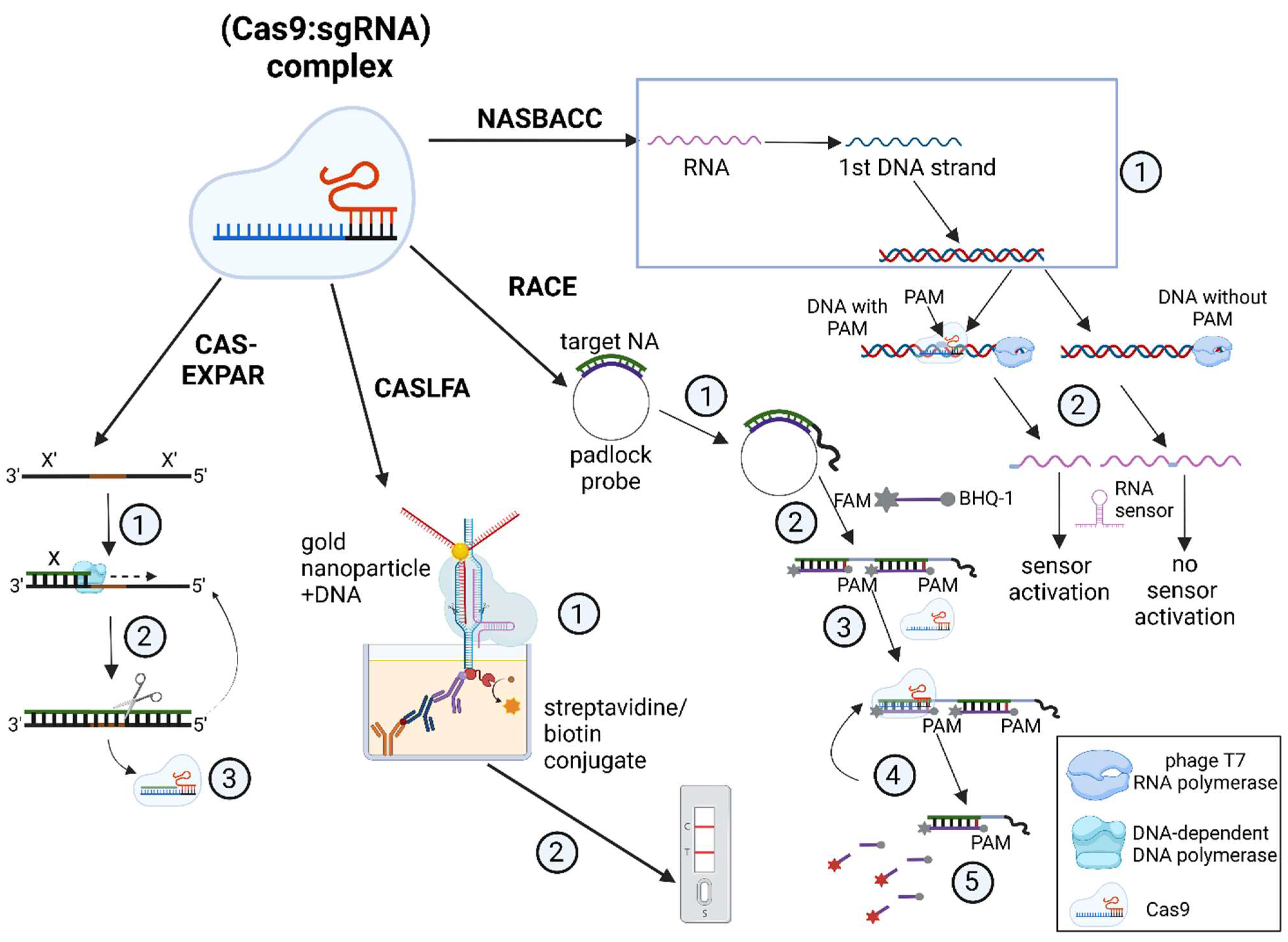
| Detection System | CRISPR/Cas | Amplification | Target (DNA/RNA) | Detection Object | References |
|---|---|---|---|---|---|
| Cas type-II-based detection systems | |||||
| CAS-EXPAR | Cas9 | PCR | dsDNA, miRNA | L. monocytogenes | [5] |
| NASBACC | Cas9 | NASBA | dsDNA | Zika virus | [6] |
| CRISDA | Cas9 | SDA | dsDNA | Human genomic DNA | [7] |
| CASLFA | Cas9 (dCas9) | RPA/PCR | dsDNA | L. monocytogenes, African swine fever virus | [8] |
| RACE | Cas9 | RCA | miRNA | Vesicular miRNAs (miR-121, 122) | [9] |
| Cas type-V- and type-VI-based detection systems | |||||
| DETECTR | Cas12a | RPA | dsDNA | Human papillomavirus | [10] |
| HOLMES | Cas12a | PCR | dsDNA | Pseudorabies virus, Japanese encephalitis virus | [11] |
| HOLMES v.2 | Cas12b | Asymmetric PCR, LAMP, RT–LAMP | dsDNA | Japanese encephalitis virus | [12] |
| SHERLOCK | Cas13a | RPA/RT–RPA | DNA/RNA | Zika virus, Dengue virus | [13] |
| SHERLOCK v.2 | Cas13a, Cas13b, Cas12a | RPA/RT–RPA | DNA/RNA | Zika virus, Dengue virus | [14] |
| CARMEN | Cas13a | RPA/RT–RPA | DNA/RNA | 169 human viruses | [15] |
| SPRINT | Cas13a | RPA/RT–RPA | DNA/RNA | Inorganic compounds, B. subtilis | [16] |
| SHINE | Cas13a | RPA/RT–RPA | DNA/RNA | SARS-CoV-2 | [17] |
| DETECTR–Cas14 | Cas14 | LAMP | ssDNA | human HERC2 gene | [43] |
| STOP | Cas13a | RPA/RT–RPA | DNA/RNA | SARS-CoV-2 | [51] |
| Colorimetric detection | Cas12a/Cas13a | PCR, RPA | DNA/RNA | A. tumefaciens, African swine fever virus, Listeria monocytogenes, Staphylococcus aureus, Neisseria encephalitis, Salmonella typhimurium, Enterobacter sakazakii, Pseudomonas aeruginosa, Vibrio parahemolyticus (Cas12); miRNA, L. monocytogenes, S. aureus, S. typhimurium, E. sakazakii, P. aeruginosa, and V. parahemolyticus (Cas13) | [52] |
| CASCADE | Cas13a | RPA, NASBA | DNA/RNA | SARS-CoV-2 | [53] |
| Cas13C | Cas13a | Combination of T7-transcription and synthesis of dsDNA by the Klenow fragment | RNA | SARS-CoV-2 | [54] |
| Cas14SDA | Cas14a | SDA | miRNA | miR-21 in cholangiocarcinoma cells | [55] |
| Amplification-free detection | Cas13a | -------------- | RNA | SARS-CoV-2, MERS-CoV, Influenza A and B viruses | [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antropov, D.N.; Stepanov, G.A. Molecular Mechanisms Underlying CRISPR/Cas-Based Assays for Nucleic Acid Detection. Curr. Issues Mol. Biol. 2023, 45, 649-662. https://doi.org/10.3390/cimb45010043
Antropov DN, Stepanov GA. Molecular Mechanisms Underlying CRISPR/Cas-Based Assays for Nucleic Acid Detection. Current Issues in Molecular Biology. 2023; 45(1):649-662. https://doi.org/10.3390/cimb45010043
Chicago/Turabian StyleAntropov, Denis N., and Grigory A. Stepanov. 2023. "Molecular Mechanisms Underlying CRISPR/Cas-Based Assays for Nucleic Acid Detection" Current Issues in Molecular Biology 45, no. 1: 649-662. https://doi.org/10.3390/cimb45010043
APA StyleAntropov, D. N., & Stepanov, G. A. (2023). Molecular Mechanisms Underlying CRISPR/Cas-Based Assays for Nucleic Acid Detection. Current Issues in Molecular Biology, 45(1), 649-662. https://doi.org/10.3390/cimb45010043





