Identification of Seven Key Structural Genes in the Anthocyanin Biosynthesis Pathway in Sepals of Hydrangea macrophylla
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sampling
2.2. Determination of Total Anthocyanin Contents
2.3. Full-Length Transcriptome Sequencing and Gene Annotation
2.4. Selection and Validation of Genes
2.4.1. Gene Selection
2.4.2. qRT–PCR analysis
3. Results
3.1. Anthocyanin Contents
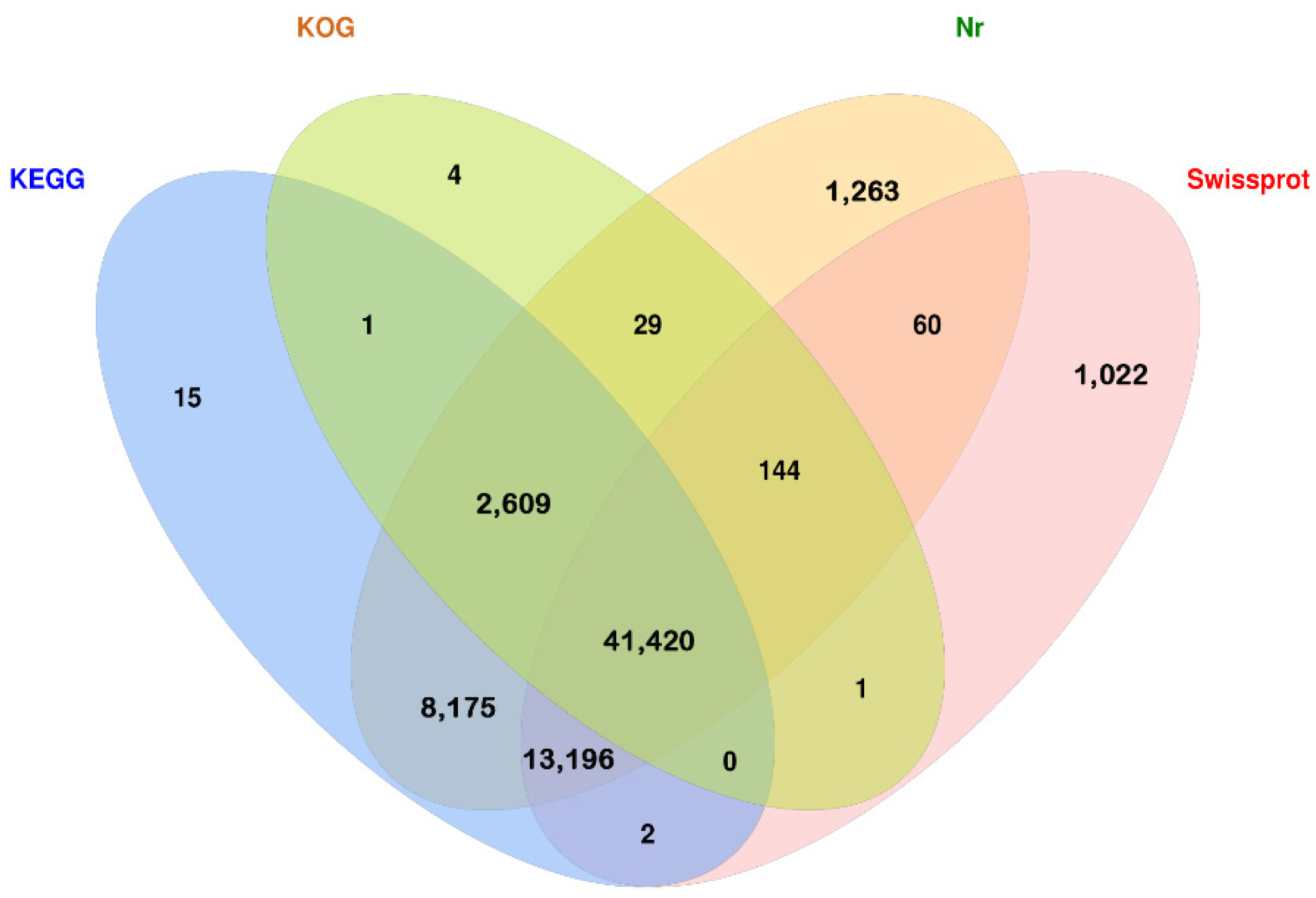
3.2. Analysis of Full-Length Transcriptome Sequencing Data
3.3. Screening of Key Structural Genes Involved in Anthocyanin Biosynthesis
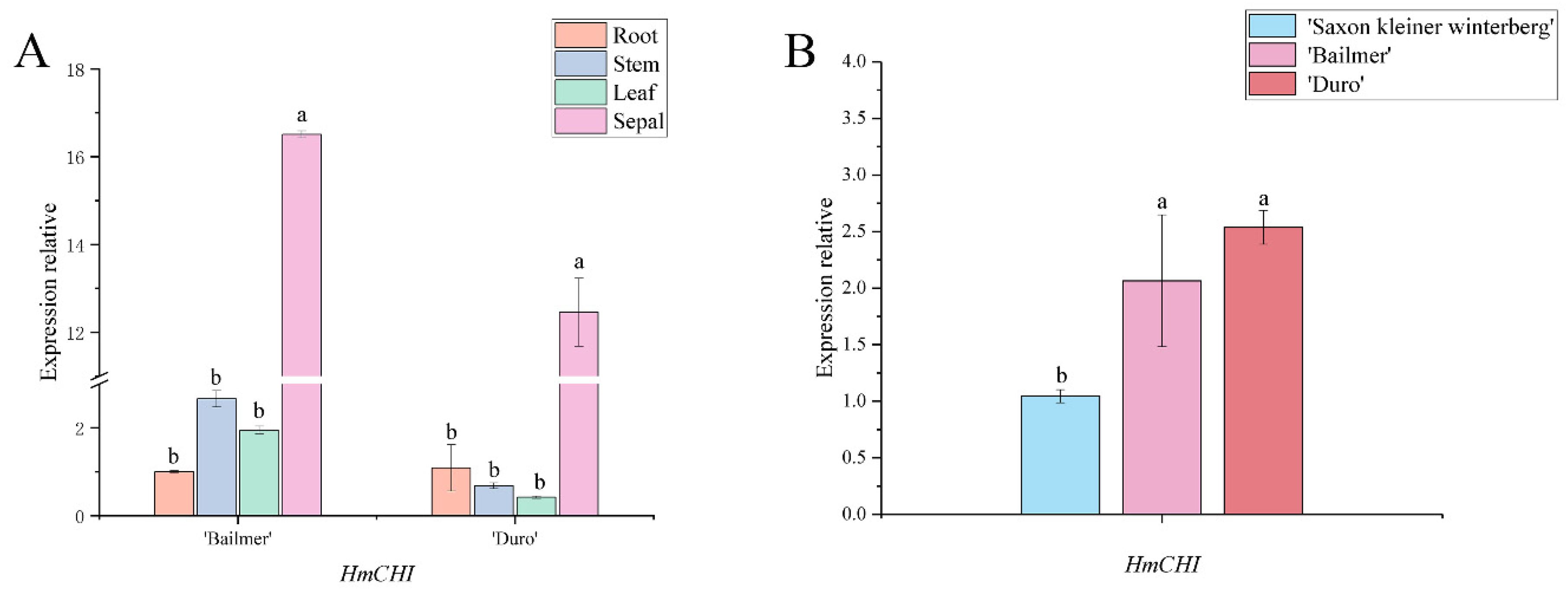
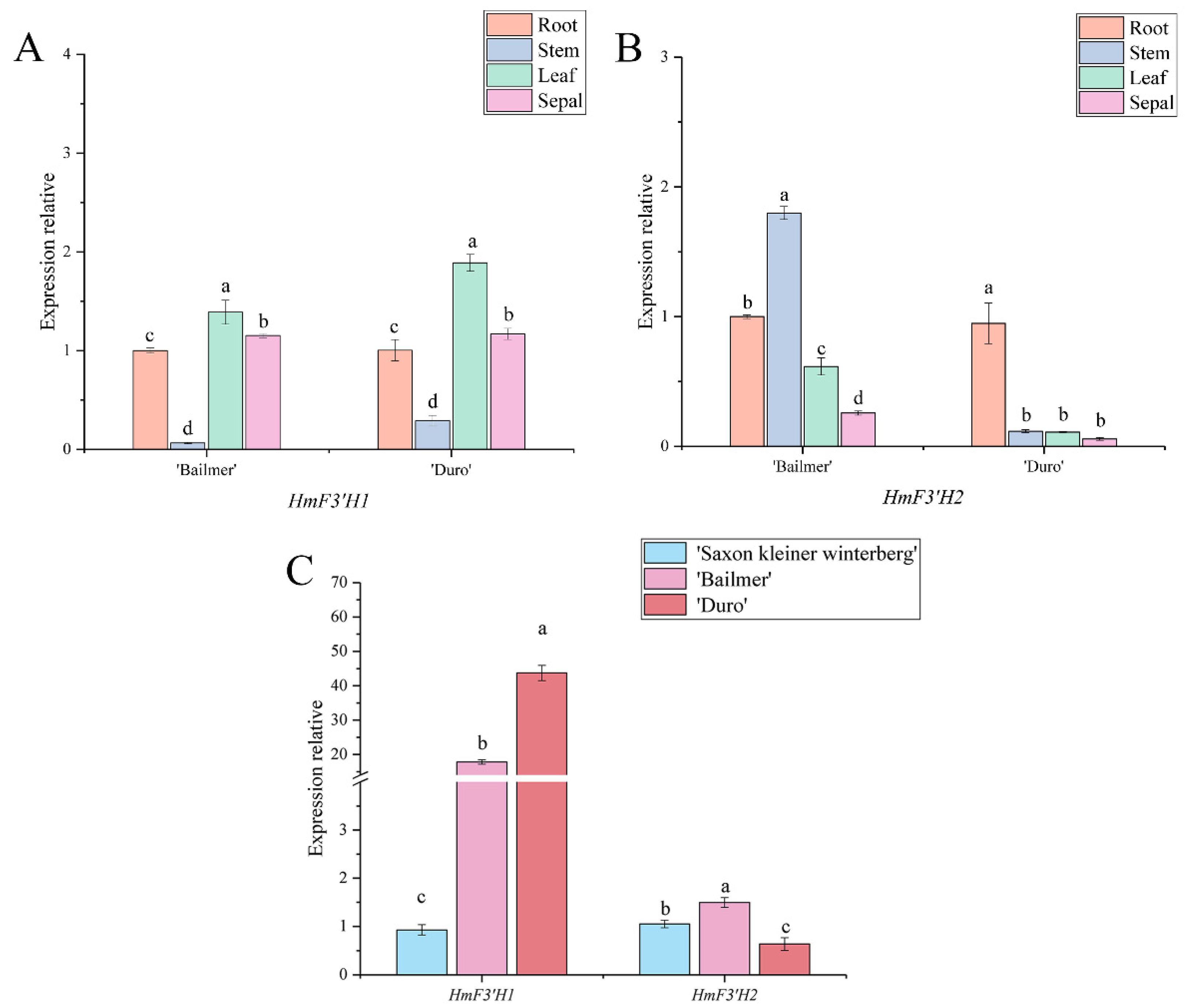
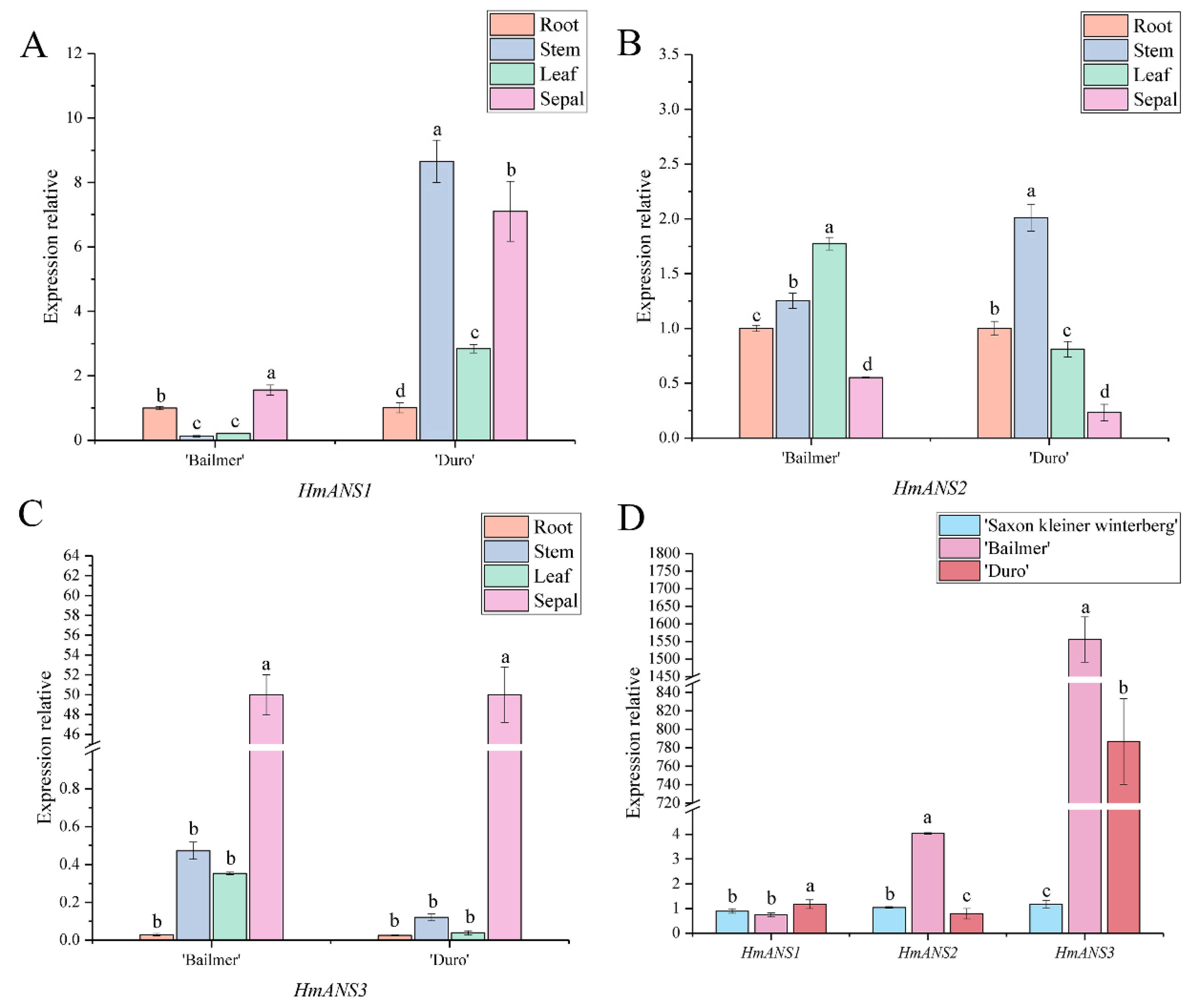
3.4. Expression Pattern Analysis of Key Structural Genes Involved in Anthocyanin Biosynthesis
3.5. Expression Patterns of the Seven Structural Genes Involved in Anthocyanin Biosynthesis during the Development of Sepals
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galopin, G.; Codarin, S.; Viemont, J.D.; Morel, P. Architectural development of inflorescence in Hydrangea macrophylla cv. Hermann Dienemann. HortScience 2008, 43, 361–365. [Google Scholar] [CrossRef]
- Atkins, W.R.G. Hydrogen-ion concentration of the soil in relation to the flower color of Hydrangea hortensis W. and the availability of iron. Sci. Proc. R. Dublin Soc. 1923, 17, 201–210. [Google Scholar] [CrossRef]
- Asen, S.; Siegelman, H.W. Effect of aluminum on absorption spectra of the anthocyanin and flavonols from sepals of Hydrangea macrophylla var. Merveille 1957.
- Okada, M.; Okawa, K. The Quantity of Aluminum and Phosphorus in Plants and Its Influence on the Sepal Color of Hydrangea macrophylla DC. Engei Gakkai Zasshi 2007, 42, 361–370. [Google Scholar] [CrossRef]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Takeda, K.; Yamashita, T.; Takahashi, A.; Timberlake, C.F. Stable blue complexes of anthocyanin-aluminium-3-p-coumaroyl- or 3-caffeoyl-quinic acid involved in the blueing of Hydrangea flower. Phytochemistry 1990, 29, 1089–1091. [Google Scholar] [CrossRef]
- Yoshida, K.; Toyama-Kato, Y.; Kameda, K.; Kondo, T. Sepal color variation of Hydrangea macrophylla and vacuolar pH value measured with a proton-selective microelectrode. Plant Cell Physiol. 2003, 44, 262–268. [Google Scholar] [CrossRef]
- Ryan, P.R.; Delhaize, E. The convergent evolution of aluminium resistance in plants exploits a convenient currency. Funct. Plant Biol. 2010, 37, 275–284. [Google Scholar] [CrossRef]
- Takeda, K.; Kubota, R.; Yagioka, C. Copigments in the blueing of sepal colour of Hydrangea macrophylla. Pergamon 1985, 24, 1207–1209. [Google Scholar] [CrossRef]
- Goto, T.; Kondo, T. Structure and molecular stacking of anthocyanins—Flower color variation. Angew. Chem. Int. Ed. 1991, 30, 17–33. [Google Scholar] [CrossRef]
- Kondo, T.; Toyama-Kato, Y.; Yoshida, K. Essential structure of co-pigment for blue sepal-color development of hydrangea. Tetrahedron Lett. 2005, 46, 6645–6649. [Google Scholar] [CrossRef]
- Yoshida, K.; Kitahara, S.; Ito, D.; Kondo, T. Ferric ions involved in the flower color development of the himalayan blue poppy. meconopsis grandis. Phytochemistry 2006, 67, 992–998. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Toyama, K.Y.; Kondo, T. Synthesis of designed acylquinic acid derivatives involved in blue color development of Hydrangea and their co-pigmentation effect. Heterocycles 2007, 72, 239–254. [Google Scholar] [CrossRef]
- Ito, T.; Oyama, K.I.; Yoshida, K. Direct observation of Hydrangea blue-complex composed of 3-O-glucosyldelphinidin, Al3+ and 5-O-acylquinic acid by esi-mass spectrometry. Molecules 2018, 23, 1424. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Aoki, D.; Fukushima, K.; Yoshida, K. Direct mapping of hydrangea blue-complex in sepal tissues of hydrangea macrophylla. Sci. Rep. 2019, 9, 5450. [Google Scholar] [CrossRef] [PubMed]
- Mol, J.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 6, 212–217. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Akiyama, T.; Shibuya, M.; Liu, H.M.; Ebizuka, Y. p-Coumaroyltriacetic acid synthase, a new homologue of chalcone synthase, from Hydrangea macrophylla var. thunbergii. FEBS J. 2010, 263, 834–839. [Google Scholar] [CrossRef]
- Chen, D.D.; Li, M.; Peng, J.Q.; Tian, Y.C.; Chao, S.J. Association between HmDFR gene expression and flower color of Hydrangea macrophylla. Plant Physiol. J. 2020, 56, 1641–1649. [Google Scholar]
- Xue, C.; Han, J.Y.; Peng, J.Q.; Yang, Y.H.; Xiao, H.L.; Huang, S.Q.; Dong, X.J. Cloning and Expression Analysis of HmF3H Gene Related to Flower Color of Hydrangea macrophylla. Mol. Plant Breed. 2021, 19, 59–64. [Google Scholar] [CrossRef]
- Gordon, S.P.; Tseng, E.; Salamov, A.; Zhang, J.W.; Meng, X.D.; Zhao, Z.Y.; Kang, D.W.; Underwood, J.; Grigoriev, L.V.; Figueroa, M.; et al. Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS ONE 2015, 10, e0132628. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The challenges of moving chemicals within and out of cells: Insights into the transport of plant natural products. Planta 2004, 219, 906–909. [Google Scholar] [CrossRef] [PubMed]
- Ito, D.; Shinkai, Y.; Kato, Y.; Kondo, T.; Yoshida, K. Chemical studies on different color development in blue- and red-colored sepal cells of hydrangea macrophylla. Biosci. Biotechnol. Biochem. 2009, 73, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.T.; Yuan, J.Z.; Chen, H.X. Effect of Aluminum Stress on Flower Color of Hydrangea Cultivar ‘Meihong Mother’. Mol. Plant Breed. 2021, 19, 6164–6171. [Google Scholar] [CrossRef]
- Yoshida, K.; Oyama, K.I.; Kondo, T. Insight into chemical mechanisms of sepal color development and variation in hydrangea: Reviews. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2021, 97, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T. Evolution of gene families. Gene 2000, 259, 45–52. [Google Scholar] [CrossRef]
- Wang, Y.S.; Xu, Y.J.; Gao, L.P.; Yu, O.; Wang, X.Z.; He, X.J.; Jiang, X.L.; Liu, Y.J.; Xia, T. Functional analysis of flavonoid 3’,5’-hydroxylase from tea plant (Camellia sinensis): Critical role in the accumulation of catechins. BMC Plant Biol. 2014, 14, 347. [Google Scholar] [CrossRef]
- Xu, L.; Yang, P.; Yuan, S.; Feng, Y.; Xu, H.; Cao, Y.; Ming, J. Transcriptome Analysis Identifies Key Candidate Genes Mediating Purple Ovary Coloration in Asiatic Hybrid Lilies. Int. J. Mol. Sci. 2016, 17, 1881. [Google Scholar] [CrossRef]
- Peng, J.; Dong, X.; Xue, C.; Liu, Z.; Cao, F. Exploring the Molecular Mechanism of Blue Flower Color Formation in Hydrangea macrophylla cv. “Forever Summer”. Front. Plant Sci. 2021, 12, 585665. [Google Scholar] [CrossRef]
- Jiang, L.; Fan, Z.; Ran, T.; Yin, H.; Li, J.; Zhou, X. Flavonoid 3′-hydroxylase of Camellia nitidissima Chi. promotes the synthesis of polyphenols better than flavonoids. Mol. Biol. Rep. 2021, 48, 3903–3912. [Google Scholar] [CrossRef] [PubMed]







| Protein Name | Gene Name | Family Member | Isoforms ID | Primer Sequence (5′-3′) | Amplicon Length |
|---|---|---|---|---|---|
| Chalcone synthase | CHS | HmCHS1 | Isoform0051878 | F-AATTTCAGCGCATGTGTGACAATT | 134 |
| R-CCACCACCATGTCTTGTCTAGC | |||||
| HmCHS2 | F-AGGGCGCACGTGTTCTTGTT | 125 | |||
| Isoform0001712 | R-GATCACCGCTGATGCACCGT | ||||
| Isoform0002955 | |||||
| Chalcone isomerase | CHI | HmCHI | Isoform0062450 | F-ATTGTTCCTCGGTGGCGCAG | 124 |
| Isoform0064242 | R-GCCCTTCCACTTAACGGCGA | ||||
| Flavanone 3-hydroxylase | F3H | HmF3H1 | Isoform0002931 | F-AGGGATGGTGGGAACACTTGG | 89 |
| R-TTGCTCAGATAATGACCATGGTCG | |||||
| Isoform0053472 | |||||
| HmF3H2 | Isoform0056122 | F-CCTCTTCTTTCAAGGAGGTGGTG | 211 | ||
| R-CTCTGGTTCAGGACATGGTGG | |||||
| HmF3H3 | Isoform0060184 | F-AACCCTCCTTCTTACAGAGAAGCT | 135 | ||
| R-TTCACCACCACCAGCATCC | |||||
| Flavonoid 3′-hydroxylase | F3′H | HmF3′H1 | Isoform0047984 | F-AAGCCTTATTGATGGACATGGTGG | 178 |
| R-TACGGGAGTTTTTGGATGTGGGAC | |||||
| HmF3′H2 | Isoform0013683 | F-GCAACTCCGATTCGGCTCCT | 119 | ||
| Isoform0014970 | R-ACTTTCCGGCGGCGGTTTTA | ||||
| Isoform0048829 | |||||
| Isoform0034725 | |||||
| Isoform0042732 | |||||
| Flavonoid 3′5′-hydroxylase | F3′5′H | HmF3′5′H | Isoform0001658 | F-AGGGCAAGCCGGACTTTCTT | 109 |
| R-CCGGCAGTGAACAAATTCAAGAGTA | |||||
| Dihydroflavonol-4-reductase | DFR | HmDFR1 | Isoform0047641 | F-TCTCTGGAGCTCGCTTACGG | 151 |
| Isoform0055957 | R-TCAATCCTCCAACATTGACCGAG | ||||
| Isoform0057874 | |||||
| HmDFR2 | F-GCTGCCAAAGGCTGAAAAGAACT | 146 | |||
| Isoform0054800 | R-ACTTCATTCTCAGGGTCCTTGGA | ||||
| Anthocyanidin synthase | ANS | HmANS1 | F-AGCCATCCACGGGAGTGCA | 104 | |
| Isoform0064501 | R-CAAGCACCAATCTCTTCTCCTCCA | ||||
| HmANS2 | Isoform0002842 | F-TGGTGATCAAATACAGGTTCTAAGC | 98 | ||
| Isoform0004318 | R-AGAAGGCAAGCGATACGCG | ||||
| Isoform0048013 | |||||
| HmANS3 | Isoform0054522 | F-CCTCTCGGTGCTATCCCTCG | 134 | ||
| R-CCTAGAGCGAGCTCCGGTTG |
| HmCHS1 | HmCHI | HmF3H1 | HmF3′H1 | HmF3′5′H | HmDFR2 | HmANS3 | |
|---|---|---|---|---|---|---|---|
| Content of anthocyanins | 0.91 1 | 0.97 1 | 0.97 1 | 0.90 1 | 0.91 1 | 1.00 1 | 0.96 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, H.; Zhang, G.; Chu, Z.; Liu, C.; Yuan, S. Identification of Seven Key Structural Genes in the Anthocyanin Biosynthesis Pathway in Sepals of Hydrangea macrophylla. Curr. Issues Mol. Biol. 2022, 44, 4167-4180. https://doi.org/10.3390/cimb44090286
Qi H, Zhang G, Chu Z, Liu C, Yuan S. Identification of Seven Key Structural Genes in the Anthocyanin Biosynthesis Pathway in Sepals of Hydrangea macrophylla. Current Issues in Molecular Biology. 2022; 44(9):4167-4180. https://doi.org/10.3390/cimb44090286
Chicago/Turabian StyleQi, Hui, Gaitian Zhang, Zhiyun Chu, Chun Liu, and Suxia Yuan. 2022. "Identification of Seven Key Structural Genes in the Anthocyanin Biosynthesis Pathway in Sepals of Hydrangea macrophylla" Current Issues in Molecular Biology 44, no. 9: 4167-4180. https://doi.org/10.3390/cimb44090286
APA StyleQi, H., Zhang, G., Chu, Z., Liu, C., & Yuan, S. (2022). Identification of Seven Key Structural Genes in the Anthocyanin Biosynthesis Pathway in Sepals of Hydrangea macrophylla. Current Issues in Molecular Biology, 44(9), 4167-4180. https://doi.org/10.3390/cimb44090286





