Yeast Secretes High Amounts of Human Calreticulin without Cellular Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids, Yeast Strains, Media and Growth
2.2. Cell Culture Medium Sample Preparation and SDS-PAGE
2.3. Cell Sample Preparation for 2DE
2.4. Making of NEPHGE Gels
2.5. Casting NEPHGE First-Dimension Gels for 2DE
2.6. Running the First Dimension
2.7. Running the Second Dimension (SDS-PAGE), Fixing and Staining of 2DE Gels
2.8. Image Analysis
2.9. Sample Preparation for LC-MSE
2.10. LC-MSE (Data-Independent Acquisition)-Based Protein Identification
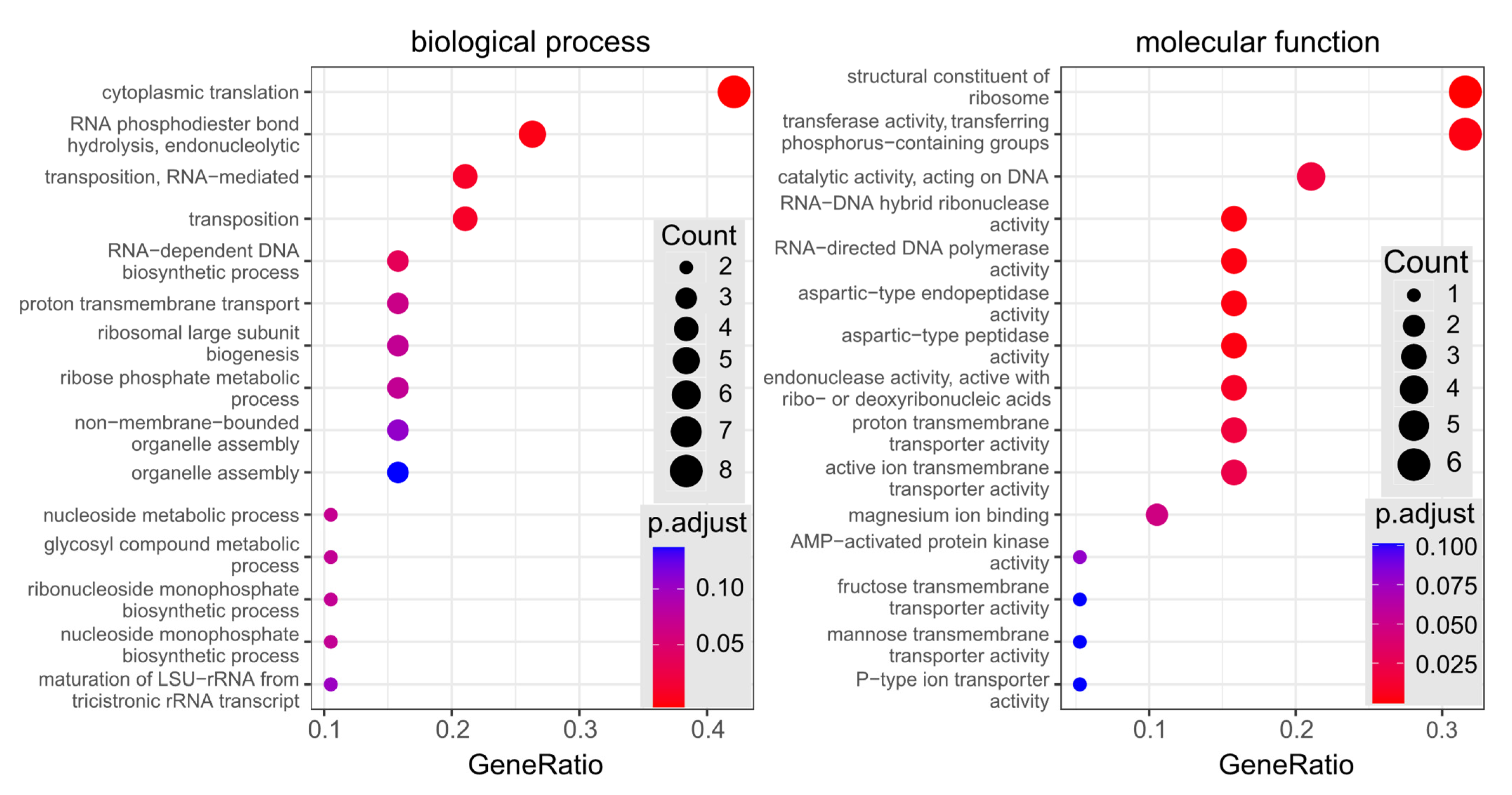
2.11. Bioinformatic Analysis of Differentially Expressed Proteins
3. Results
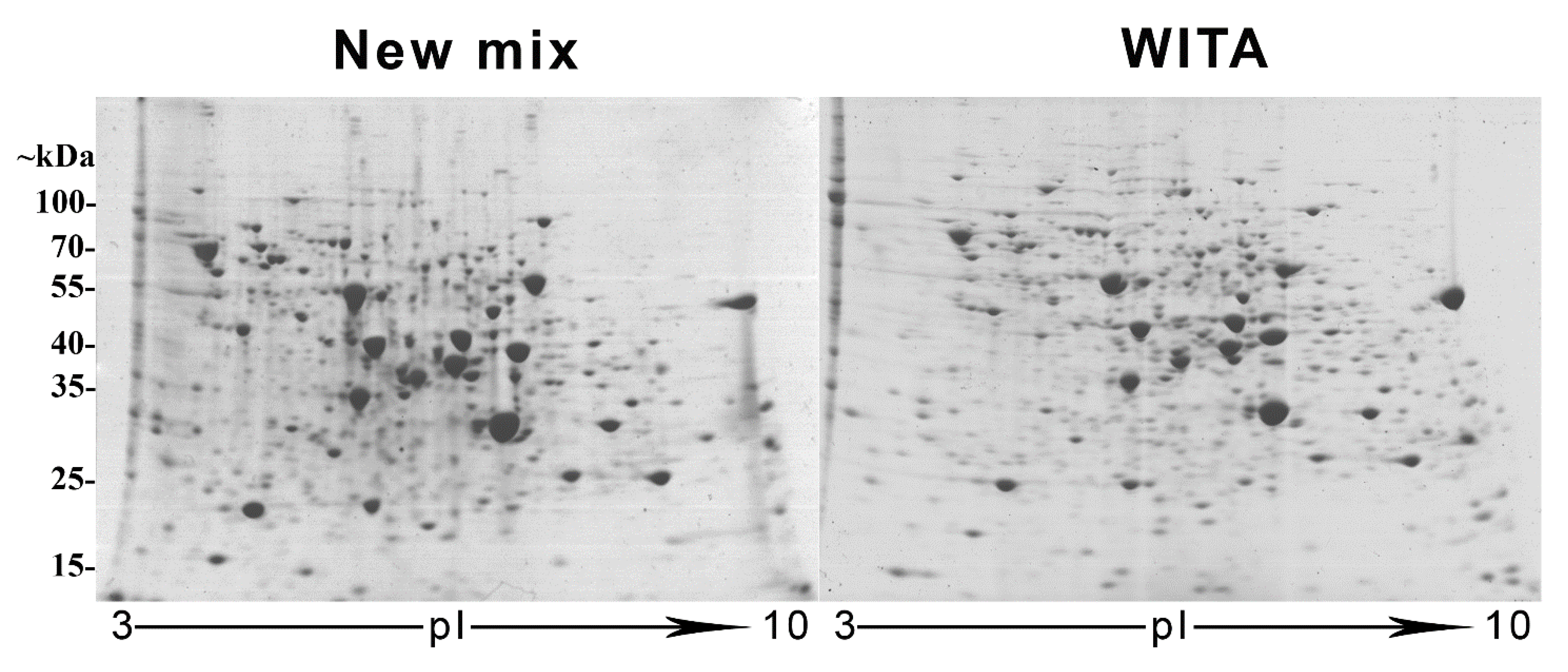
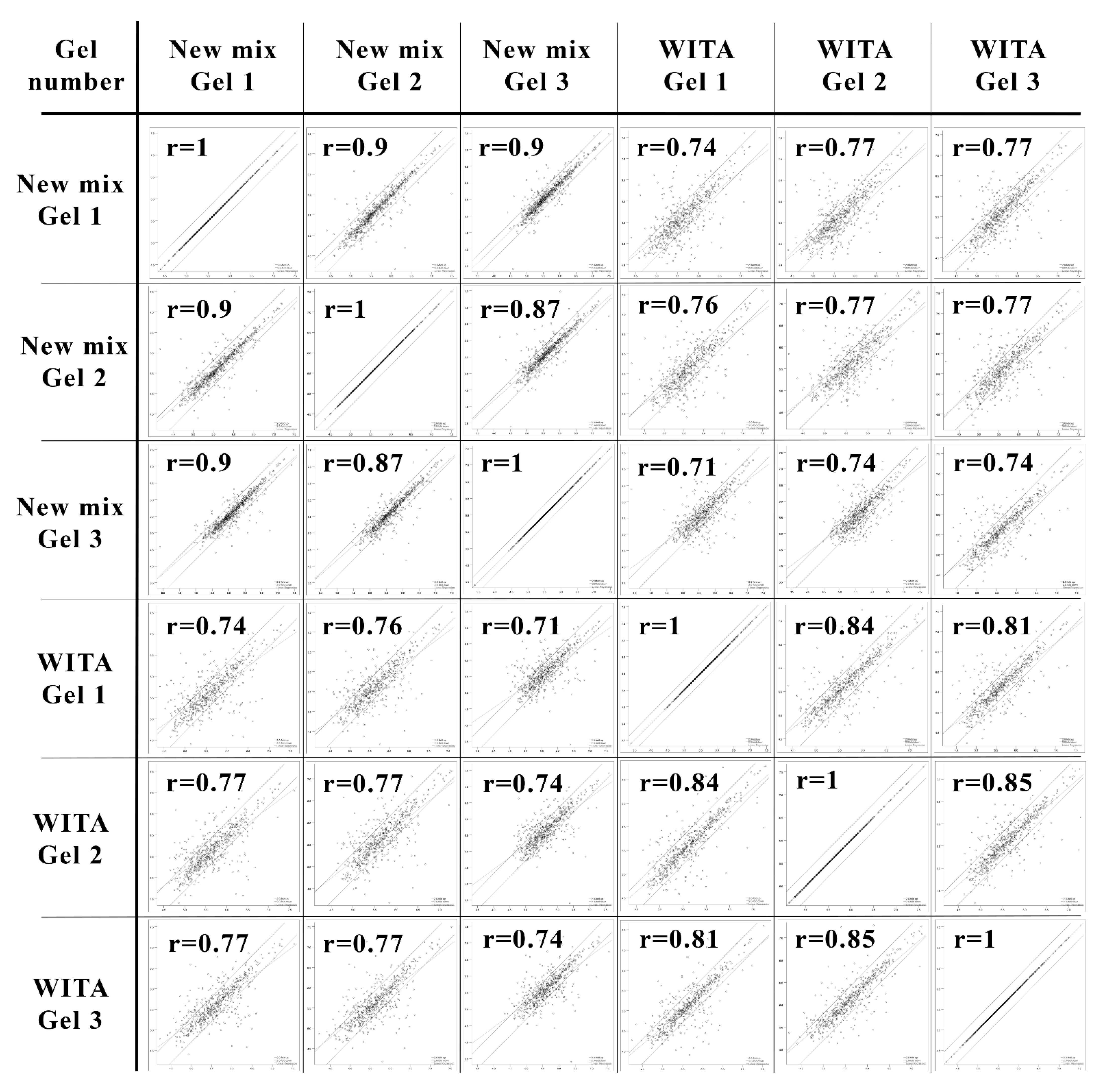
3.1. Restoration of the NEPHGE First-Dimension Gel CA Composition and Comparison to the Commercial “WITA” Gels
3.2. Expression and Secretion of Human Recombinant CALR Protein in Yeast S. cerevisiae
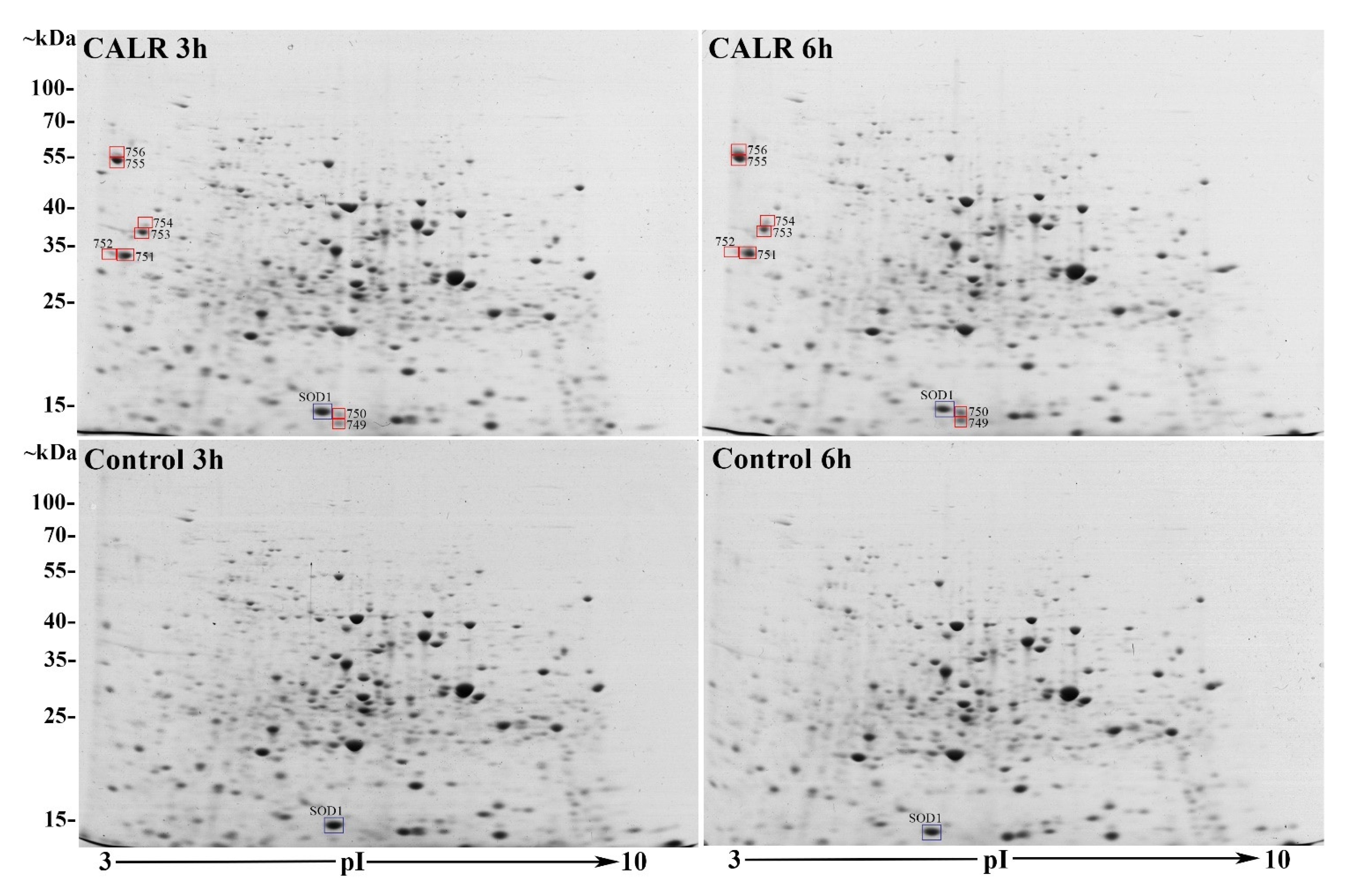
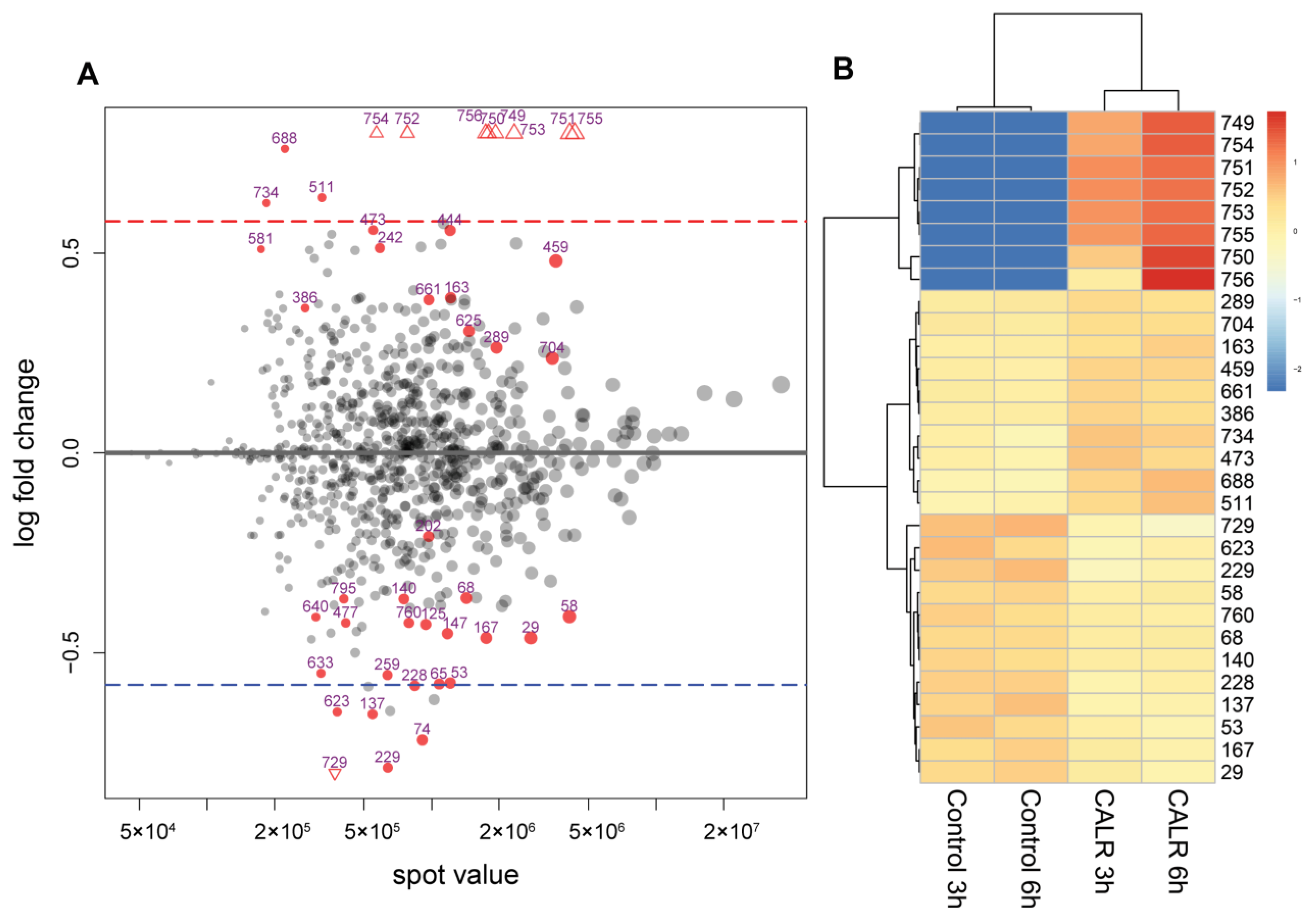
3.3. Comparative 2DE-Based Analysis of CALR-Expressing Yeast Cell Samples vs. Control
3.4. Comparative LC-MSE-Based Analysis of of the Whole Proteome Samples of CALR Expressing and Control Yeast Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Q.; Groenendyk, J.; Michalak, M. Glycoprotein Quality Control and Endoplasmic Reticulum Stress. Molecules 2015, 20, 13689–13704. [Google Scholar] [CrossRef] [PubMed]
- Michalak, M.; Robert Parker, J.M.; Opas, M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium 2002, 32, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Adhikari, R.; Howarth, M.; Nakamura, K.; Gold, M.C.; Hill, A.B.; Knee, R.; Michalak, M.; Elliott, T. Assembly and Antigen-Presenting Function of MHC Class I Molecules in Cells Lacking the ER Chaperone Calreticulin. Immunity 2002, 16, 99–109. [Google Scholar] [CrossRef]
- Kielbik, M.; Szulc-Kielbik, I.; Klink, M. Calreticulin—Multifunctional Chaperone in Immunogenic Cell Death: Potential Significance as a Prognostic Biomarker in Ovarian Cancer Patients. Cells 2021, 10, 130. [Google Scholar] [CrossRef]
- Nanney, L.B.; Woodrell, C.D.; Greives, M.R.; Cardwell, N.L.; Pollins, A.C.; Bancroft, T.A.; Chesser, A.; Michalak, M.; Rahman, M.; Siebert, J.W.; et al. Calreticulin Enhances Porcine Wound Repair by Diverse Biological Effects. Am. J. Pathol. 2008, 173, 610–630. [Google Scholar] [CrossRef]
- Coppolino, M.G.; Woodside, M.J.; Demaurex, N.; Grinstein, S.; St-Arnaud, R.; Dedhar, S. Calreticulin is essential for integrin-mediated calcium signalling and cell adhesion. Nature 1997, 386, 843–847. [Google Scholar] [CrossRef]
- Coppolino, M.G.; Dedhar, S. Bi-directional signal transduction by integrin receptors. Int. J. Biochem. Cell Biol. 2000, 32, 171–188. [Google Scholar] [CrossRef]
- Orr, A.; Elzie, C.A.; Kucik, D.F.; Murphy-Ullrich, J.E. Thrombospondin signaling through the calreticulin/LDL receptor-related protein co-complex stimulates random and directed cell migration. J. Cell Sci. 2003, 116, 2917–2927. [Google Scholar] [CrossRef]
- Gardai, S.J.; McPhillips, K.A.; Frasch, S.C.; Janssen, W.J.; Starefeldt, A.; Murphy-Ullrich, J.E.; Bratton, D.L.; Oldenborg, P.-A.; Michalak, M.; Henson, P.M. Cell-Surface Calreticulin Initiates Clearance of Viable or Apoptotic Cells through trans-Activation of LRP on the Phagocyte. Cell 2005, 123, 321–334. [Google Scholar] [CrossRef]
- Tesniere, A.; Apetoh, L.; Ghiringhelli, F.; Joza, N.; Panaretakis, T.; Kepp, O.; Schlemmer, F.; Zitvogel, L.; Kroemer, G. Immunogenic cancer cell death: A key-lock paradigm. Curr. Opin. Immunol. 2008, 20, 504–511. [Google Scholar] [CrossRef]
- Greives, M.R.; Samra, F.; Pavlides, S.C.; Blechman, K.M.; Naylor, S.-M.; Woodrell, C.D.; Cadacio, C.; Levine, J.P.; Bancroft, T.A.; Michalak, M.; et al. Exogenous calreticulin improves diabetic wound healing. Wound Repair Regen. 2012, 20, 715–730. [Google Scholar] [CrossRef]
- Gold, L.I.; Eggleton, P.; Sweetwyne, M.T.; Van Duyn, L.B.; Greives, M.R.; Naylor, S.M.; Michalak, M.; Murphy-Ullrich, J.E. Calreticulin: Non-endoplasmic reticulum functions in physiology and disease. FASEB J. 2010, 24, 665–683. [Google Scholar] [CrossRef]
- Zhang, K.; Kaufman, R.J. Chapter Twenty Identification and Characterization of Endoplasmic Reticulum Stress-Induced Apoptosis In Vivo. Methods Enzymol. 2008, 442, 395–419. [Google Scholar] [CrossRef]
- Tufi, R.; Panaretakis, T.; Bianchi, K.; Criollo, A.; Fazi, B.; Di Sano, F.; Tesniere, A.; Kepp, O.; Brechot, P.P.-; Zitvogel, L.; et al. Reduction of endoplasmic reticulum Ca2+ levels favors plasma membrane surface exposure of calreticulin. Cell Death Differ. 2007, 15, 274–282. [Google Scholar] [CrossRef]
- Liu, P.; Zhao, L.; Kroemer, G.; Kepp, O. Secreted calreticulin mutants subvert anticancer immunosurveillance. OncoImmunology 2020, 9, 1708126. [Google Scholar] [CrossRef]
- Wiersma, V.R.; Michalak, M.; Abdullah, T.M.; Bremer, E.; Eggleton, P. Mechanisms of Translocation of ER Chaperones to the Cell Surface and Immunomodulatory Roles in Cancer and Autoimmunity. Front. Oncol. 2015, 5, 7. [Google Scholar] [CrossRef]
- Ciplys, E.; Zitkus, E.; Gold, L.I.; Daubriac, J.; Pavlides, S.C.; Hojrup, P.; Houen, G.; Wang, W.A.; Michalak, M.; Slibinskas, R. High-level secretion of native recombinant human calreticulin in yeast. Microb. Cell Factories 2015, 14, 165. [Google Scholar] [CrossRef]
- Slibinskas, R.; Ražanskas, R.; Zinkevičiūtė, R.; Čiplys, E. Comparison of first dimension IPG and NEPHGE techniques in two-dimensional gel electrophoresis experiment with cytosolic unfolded protein response in Saccharomyces cerevisiae. Proteome Sci. 2013, 11, 36. [Google Scholar] [CrossRef]
- Zinkevičiūtė, R.; Bakūnaitė, E.; Čiplys, E.; Ražanskas, R.; Raškevičiūtė, J.; Slibinskas, R. Heat shock at higher cell densities improves measles hemagglutinin translocation and human GRP78/BiP secretion in Saccharomyces cerevisiae. New Biotechnol. 2015, 32, 690–700. [Google Scholar] [CrossRef]
- Bjarnadóttir, S.G.; Flengsrud, R. Affinity chromatography, two-dimensional electrophoresis, adapted immunodepletion and mass spectrometry used for detection of porcine and piscine heparin-binding human plasma proteins. J. Chromatogr. B 2014, 944, 107–113. [Google Scholar] [CrossRef]
- Kreipke, C.W.; Morgan, R.; Roberts, G.; Bagchi, M.; Rafols, J.A. Calponin phosphorylation in cerebral cortex microvessels mediates sustained vasoconstriction after brain trauma. Neurol. Res. 2007, 29, 369–374. [Google Scholar] [CrossRef]
- Klose, J.; Kobalz, U. Two-dimensional electrophoresis of proteins: An updated protocol and implications for a functional analysis of the genome. Electrophor. 1995, 16, 1034–1059. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Russel, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; ISBN 978-087969577-4. [Google Scholar]
- Slibinskas, R.; Samuel, D.; Gedvilaite, A.; Staniulis, J.; Sasnauskas, K. Synthesis of the measles virus nucleoprotein in yeast Pichia pastoris and Saccharomyces cerevisiae. J. Biotechnol. 2004, 107, 115–124. [Google Scholar] [CrossRef]
- O’Farrell, P.Z.; Goodman, H.M.; O’Farrell, P. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 1977, 12, 1133–1142. [Google Scholar] [CrossRef]
- Kirch, W. (Ed.) Pearson’s Correlation Coefficient. In Encyclopedia of Public Health; Springer: Dordrecht, The Netherlands, 2008; pp. 1090–1091. ISBN 978-1-4020-5614-7. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.; Tenzer, S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 2013, 11, 167–170. [Google Scholar] [CrossRef]
- Kuharev, J.; Navarro, P.; Distler, U.; Jahn, O.; Tenzer, S. In-depth evaluation of software tools for data-independent acquisition based label-free quantification. Proteomics 2014, 15, 3140–3151. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Galdieri, L.; Mehrotra, S.; Yu, S.; Vancura, A. Transcriptional Regulation in Yeast during Diauxic Shift and Stationary Phase. OMICS A J. Integr. Biol. 2010, 14, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Escalante-Chong, R.; Savir, Y.; Carroll, S.M.; Ingraham, J.B.; Wang, J.; Marx, C.J.; Springer, M. Galactose metabolic genes in yeast respond to a ratio of galactose and glucose. Proc. Natl. Acad. Sci. USA 2015, 112, 1636–1641. [Google Scholar] [CrossRef] [PubMed]
- Echan, L.A.; Speicher, D.W. Protein Detection in Gels Using Fixation. Curr. Protoc. Protein Sci. 2002, 29, 10.5.1–10.5.18. [Google Scholar] [CrossRef]
- Bonander, N.; Darby, R.A.; Grgic, L.; Bora, N.; Wen, J.; Brogna, S.; Poyner, D.R.; O’Neill, M.A.; Bill, R.M. Altering the ribosomal subunit ratio in yeast maximizes recombinant protein yield. Microb. Cell Factories 2009, 8, 10. [Google Scholar] [CrossRef]
- Li, X.; Chen, G.; Fongang, B.; Homouz, D.; Rowicka, M.; Kudlicki, A. The High-Resolution Timeline of Expression of Ribosomal Protein Genes in Yeast. bioRxiv 2017, 170399. [Google Scholar] [CrossRef]
- Slavov, N.; Semrau, S.; Airoldi, E.; Budnik, B.; van Oudenaarden, A. Differential Stoichiometry among Core Ribosomal Proteins. Cell Rep. 2015, 13, 865–873. [Google Scholar] [CrossRef]
- Moffatt, B.A.; Ashihara, H. Purine and pyrimidine nucleotide synthesis and metabolism. In The Arabidopsis Book; The American Society of Plant Biologists: Rockville, MD, USA, 2002; Volume 1, p. e0018. [Google Scholar]
- Yu, H.; Zhang, Y.; Zhang, D.; Lu, Y.; He, H.; Zheng, F.; Wang, M. Identification of a Ribose-Phosphate Pyrophosphokinase that Can Interact In Vivo with the Anaphase Promoting Complex/Cyclosome. Int. J. Mol. Sci. 2017, 18, 617. [Google Scholar] [CrossRef]
- Allen, L.A.; Zhao, X.-J.; Caughey, W.; Poyton, R.O. Isoforms of Yeast Cytochrome c Oxidase Subunit V Affect the Binuclear Reaction Center and Alter the Kinetics of Interaction with the Isoforms of Yeast Cytochrome c. J. Biol. Chem. 1995, 270, 110–118. [Google Scholar] [CrossRef]
- Burke, P.V.; Raitt, D.C.; Allen, L.A.; Kellogg, E.A.; Poyton, R.O. Effects of Oxygen Concentration on the Expression of Cytochrome c and Cytochrome c Oxidase Genes in Yeast. J. Biol. Chem. 1997, 272, 14705–14712. [Google Scholar] [CrossRef]
- Siso, M.I.G.; Becerra, M.; Maceiras, M.L.; Vázquez, Á.V.; Cerdán, M.E. The yeast hypoxic responses, resources for new biotechnological opportunities. Biotechnol. Lett. 2012, 34, 2161–2173. [Google Scholar] [CrossRef]
- Curcio, M.J.; Lutz, S.; Lesage, P. The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae. Microbiol. Spectr. 2015, 3, 927–964. [Google Scholar] [CrossRef]
- de Godoy, L.M.F.; Olsen, J.V.; Cox, J.; Nielsen, M.L.; Hubner, N.C.; Fröhlich, F.; Walther, T.C.; Mann, M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 2008, 455, 1251–1254. [Google Scholar] [CrossRef]
- Zaratiegui, M. Cross-Regulation between Transposable Elements and Host DNA Replication. Viruses 2017, 9, 57. [Google Scholar] [CrossRef]
- Broach, J. RAS genes in Saccharomyces cerevisiae: Signal transduction in search of a pathway. Trends Genet. 1991, 7, 28–33. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Lorsch, J.R. The Mechanism of Eukaryotic Translation Initiation: New Insights and Challenges. Cold Spring Harb. Perspect. Biol. 2012, 4, a011544. [Google Scholar] [CrossRef]
- Kunkel, J.; Luo, X.; Capaldi, A.P. Integrated TORC1 and PKA signaling control the temporal activation of glucose-induced gene expression in yeast. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Lui, J.; Campbell, S.G.; Ashe, M.P. Inhibition of translation initiation following glucose depletion in yeast facilitates a rationalization of mRNA content. Biochem. Soc. Trans. 2010, 38, 1131–1136. [Google Scholar] [CrossRef]
- Rolland, F.; Winderickx, J.; Thevelein, J.M. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002, 2, 183–201. [Google Scholar] [CrossRef]
- Tudisca, V.; Simpson, C.; Castelli, L.; Lui, J.; Hoyle, N.; Moreno, S.; Ashe, M.; Portela, P. PKA isoforms coordinate mRNA fate during nutrient starvation. J. Cell Sci. 2012, 125, 5221–5232. [Google Scholar] [CrossRef]
- Greatrix, B.W.; van Vuuren, H.J.J. Expression of the HXT13, HXT15 and HXT17 genes in Saccharomyces cerevisiae and stabilization of the HXT1 gene transcript by sugar-induced osmotic stress. Curr. Genet. 2006, 49, 205–217. [Google Scholar] [CrossRef]
- Jordan, P.; Choe, J.-Y.; Boles, E.; Oreb, M. Hxt13, Hxt15, Hxt16 and Hxt17 from Saccharomyces cerevisiae represent a novel type of polyol transporters. Sci. Rep. 2016, 6, 23502. [Google Scholar] [CrossRef]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef]
- Clarkson, B.K.; Gilbert, W.V.; Doudna, J.A. Functional Overlap between eIF4G Isoforms in Saccharomyces cerevisiae. PLoS ONE 2010, 5, e9114. [Google Scholar] [CrossRef]
- Costello, J.L.; Kershaw, C.; Castelli, L.M.; Talavera, D.; Rowe, W.; Sims, P.F.G.; Ashe, M.P.; Grant, C.M.; Hubbard, S.J.; Pavitt, G.D. Dynamic changes in eIF4F-mRNA interactions revealed by global analyses of environmental stress responses. Genome Biol. 2017, 18, 201. [Google Scholar] [CrossRef]
- Gallie, D.R.; Browning, K.S. eIF4G Functionally Differs from eIFiso4G in Promoting Internal Initiation, Cap-independent Translation, and Translation of Structured mRNAs. J. Biol. Chem. 2001, 276, 36951–36960. [Google Scholar] [CrossRef] [PubMed]
- Goyer, C.; Altmann, M.; Lee, H.S.; Blanc, A.; Deshmukh, M.; Woolford, J.L.; Trachsel, H.; Sonenberg, N. TIF4631 and TIF4632: Two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 1993, 13, 4860–4874. [Google Scholar] [CrossRef] [PubMed][Green Version]
- von der Haar, T.; McCarthy, J.E.G. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 2002, 46, 531–544. [Google Scholar] [CrossRef] [PubMed]
- E Basson, M.; Thorsness, M.; Rine, J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc. Natl. Acad. Sci. USA 1986, 83, 5563–5567. [Google Scholar] [CrossRef]
- Hampton, R.Y.; Garza, R.M. Protein Quality Control as a Strategy for Cellular Regulation: Lessons from Ubiquitin-Mediated Regulation of the Sterol Pathway. Chem. Rev. 2009, 109, 1561–1574. [Google Scholar] [CrossRef]
- Wangeline, M.A.; Hampton, R.Y. “Mallostery”—Ligand-dependent protein misfolding enables physiological regulation by ERAD. J. Biol. Chem. 2018, 293, 14937–14950. [Google Scholar] [CrossRef]
- Yamada, M.; Hayatsu, N.; Matsuura, A.; Ishikawa, F. Y′-Help1, a DNA Helicase Encoded by the Yeast Subtelomeric Y′ Element, Is Induced in Survivors Defective for Telomerase. J. Biol. Chem. 1998, 273, 33360–33366. [Google Scholar] [CrossRef]
- Fernandes, A.R.; Sá-Correia, I. Transcription patterns ofPMA1andPMA2genes and activity of plasma membrane H+-ATPase in Saccharomyces cerevisiae during diauxic growth and stationary phase. Yeast 2003, 20, 207–219. [Google Scholar] [CrossRef]
- Martin-Perez, M.; Villén, J. Determinants and Regulation of Protein Turnover in Yeast. Cell Syst. 2017, 5, 283–294.e5. [Google Scholar] [CrossRef]
- Schlesser, A.; Ulaszewski, S.; Ghislain, M.; Goffeau, A. A second transport ATPase gene in Saccharomyces cerevisiae. J. Biol. Chem. 1988, 263, 19480–19487. [Google Scholar] [CrossRef]
- Supply, P.; Wach, A.; Goffeau, A. Enzymatic properties of the PMA2 plasma membrane-bound H(+)-ATPase of Saccharomyces cerevisiae. J. Biol. Chem. 1993, 268, 19753–19759. [Google Scholar] [CrossRef]
- Rabilloud, T.; Chevallet, M.; Luche, S.; Lelong, C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J. Proteom. 2010, 73, 2064–2077. [Google Scholar] [CrossRef]
- Čiplys, E.; Paškevičius, T.; Žitkus, E.; Bielskis, J.; Ražanskas, R.; Šneideris, T.; Smirnovas, V.; Kaupinis, A.; Tester, D.J.; Ackerman, M.J.; et al. Mapping human calreticulin regions important for structural stability. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140710. [Google Scholar] [CrossRef]
- Weinhandl, K.; Winkler, M.; Glieder, A.; Camattari, A. Carbon source dependent promoters in yeasts. Microb. Cell Factories 2014, 13, 5. [Google Scholar] [CrossRef]
- Čiplys, E.; Žitkus, E.; Slibinskas, R. Native signal peptide of human ERp57 disulfide isomerase mediates secretion of active native recombinant ERp57 protein in yeast Saccharomyces cerevisiae. Protein Expr. Purif. 2013, 89, 131–135. [Google Scholar] [CrossRef]
- Čiplys, E.; Aučynaitė, A.; Slibinskas, R. Generation of human ER chaperone BiP in yeast Saccharomyces cerevisiae. Microb. Cell Factories 2014, 13, 22. [Google Scholar] [CrossRef]
- Peters, L.R.; Raghavan, M. Endoplasmic Reticulum Calcium Depletion Impacts Chaperone Secretion, Innate Immunity, and Phagocytic Uptake of Cells. J. Immunol. 2011, 187, 919–931. [Google Scholar] [CrossRef]
- Strayle, J.; Pozzan, T.; Rudolph, H.K. Steady-state free Ca2+ in the yeast endoplasmic reticulum reaches only 10 microM and is mainly controlled by the secretory pathway pump Pmr1. EMBO J. 1999, 18, 4733–4743. [Google Scholar] [CrossRef]
- Twyman, R.; George, A. Principles of Proteomics; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Martínez, J.L.; Meza, E.; Petranovic, D.; Nielsen, J. The impact of respiration and oxidative stress response on recombinant α-amylase production by Saccharomyces cerevisiae. Metab. Eng. Commun. 2016, 3, 205–210. [Google Scholar] [CrossRef]
- Tyo, K.E.; Liu, Z.; Petranovic, D.; Nielsen, J. Imbalance of heterologous protein folding and disulfide bond formation rates yields runaway oxidative stress. BMC Biol. 2012, 10, 16. [Google Scholar] [CrossRef]
- Haynes, C.M.; Titus, E.A.; Cooper, A.A. Degradation of Misfolded Proteins Prevents ER-Derived Oxidative Stress and Cell Death. Mol. Cell 2004, 15, 767–776. [Google Scholar] [CrossRef]
- Brodsky, J.L. Cleaning Up: ER-Associated Degradation to the Rescue. Cell 2012, 151, 1163–1167. [Google Scholar] [CrossRef]
- Hou, J.; Tyo, K.E.; Liu, Z.; Petranovic, D.; Nielsen, J. Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 491–510. [Google Scholar] [CrossRef]
- Pelham, H.R. Insights from yeast endosomes. Curr. Opin. Cell Biol. 2002, 14, 454–462. [Google Scholar] [CrossRef]
- Ho, B.; Baryshnikova, A.; Brown, G.W. Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Syst. 2018, 6, 192–205.e3. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.-Q.; Liang, S.-L.; Han, S.-Y.; Zheng, S.-P.; Ye, Y.-R.; Lin, Y. Quantitative iTRAQ LC-MS/MS proteomics reveals the cellular response to heterologous protein overexpression and the regulation of HAC1 in Pichia pastoris. J. Proteom. 2013, 91, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-W.; Klein, T.; Cassidy, L.; Linke, D.; Lange, S.; Anders, U.; Bureik, M.; Heinzle, E.; Schneider, K.; Tholey, A. Comparative Proteome Analysis in Schizosaccharomyces pombe Identifies Metabolic Targets to Improve Protein Production and Secretion. Mol. Cell. Proteom. 2016, 15, 3090–3106. [Google Scholar] [CrossRef] [PubMed]
- Mattanovich, D.; Branduardi, P.; Dato, L.; Gasser, B.; Sauer, M.; Porro, D. Recombinant Protein Production in Yeasts. Tumor Prev. Genet. 2011, 824, 329–358. [Google Scholar] [CrossRef]
- Huang, M.; Bao, J.; Hallström, B.M.; Petranovic, D.; Nielsen, J. Efficient protein production by yeast requires global tuning of metabolism. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Zahrl, R.J.; Gasser, B.; Mattanovich, D.; Ferrer, P. Detection and Elimination of Cellular Bottlenecks in Protein-Producing Yeasts. In Methods in Pharmacology and Toxicology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1923, pp. 75–95. [Google Scholar]
- Kastberg, L.L.B.; Ard, R.; Jensen, M.K.; Workman, C.T. Burden Imposed by Heterologous Protein Production in Two Major Industrial Yeast Cell Factories: Identifying Sources and Mitigation Strategies. Front. Fungal Biol. 2022, 3, 3. [Google Scholar] [CrossRef]
- Gasser, B.; Saloheimo, M.; Rinas, U.; Dragosits, M.; Rodríguez-Carmona, E.; Baumann, K.; Giuliani, M.; Parrilli, E.; Branduardi, P.; Lang, C.; et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: A host comparative overview. Microb. Cell Factories 2008, 7, 11. [Google Scholar] [CrossRef]
- Mattanovich, D.; Gasser, B.; Hohenblum, H.; Sauer, M. Stress in recombinant protein producing yeasts. J. Biotechnol. 2004, 113, 121–135. [Google Scholar] [CrossRef]
- Čiplys, E.; Samuel, D.; Juozapaitis, M.; Sasnauskas, K.; Slibinskas, R. Overexpression of human virus surface glycoprotein precursors induces cytosolic unfolded protein response in Saccharomyces cerevisiae. Microb. Cell Factories 2011, 10, 37. [Google Scholar] [CrossRef]
- Kauffman, K.; Pridgen, E.; Doyle, F.; Dhurjati, P.; Robinson, A. Decreased Protein Expression and Intermittent Recoveries in BiP Levels Result from Cellular Stress during Heterologous Protein Expression in Saccharomyces cerevisiae. Biotechnol. Prog. 2002, 18, 942–950. [Google Scholar] [CrossRef]
- Hiller, M.M.; Finger, A.; Schweiger, M.; Wolf, D.H. ER Degradation of a Misfolded Luminal Protein by the Cytosolic Ubiquitin-Proteasome Pathway. Science 1996, 273, 1725–1728. [Google Scholar] [CrossRef]
- Nishikawa, S.-I.; Fewell, S.W.; Kato, Y.; Brodsky, J.L.; Endo, T. Molecular Chaperones in the Yeast Endoplasmic Reticulum Maintain the Solubility of Proteins for Retrotranslocation and Degradation. J. Cell Biol. 2001, 153, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Knop, M.; Finger, A.; Braun, T.; Hellmuth, K.; Wolf, D.H. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996, 15, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Bordallo, J.; Plemper, R.K.; Finger, A.; Wolf, D.H. Der3p/Hrd1p Is Required for Endoplasmic Reticulum-associated Degradation of Misfolded Lumenal and Integral Membrane Proteins. Mol. Biol. Cell 1998, 9, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Hampton, R.Y.; Gardner, R.; Rine, J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 1996, 7, 2029–2044. [Google Scholar] [CrossRef]
- Pilon, M.; Schekman, R.; Römisch, K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997, 16, 4540–4548. [Google Scholar] [CrossRef]
- Chouquet, A.; Paidassi, H.; Ling, W.L.; Frachet, P.; Houen, G.; Arlaud, G.J.; Gaboriaud, C. X-Ray Structure of the Human Calreticulin Globular Domain Reveals a Peptide-Binding Area and Suggests a Multi-Molecular Mechanism. PLoS ONE 2011, 6, e17886. [Google Scholar] [CrossRef]
| Cellular Proteins with Upregulated Expression in CALR Samples | |||||
|---|---|---|---|---|---|
| Protein Name | Description | Functional Group | Log2FC | p-Value (<0.01) | FDR (<0.05) |
| RL17B | 60S ribosomal protein | Ribosome biogenesis [37,38,39] | 13.56115 | 2.37783 × 10−11 | 4.1 × 10−8 |
| RL26A | 60S ribosomal protein | Ribosome biogenesis [37,38,39] | 12.45301 | 5.09354 × 10−11 | 4.39 × 10−8 |
| RL15B | 60S ribosomal protein | Ribosome biogenesis [37,38,39] | 11.49495 | 2.22033 × 10−10 | 1.28 × 10−7 |
| KPR | Ribose-phosphate pyrophosphokinase 2 necessary for de novo and salvage synthesis of nucleotides | Nucleotide metabolism [40,41] | 13.14576 | 7.28579 × 10−10 | 2.51 × 10−7 |
| COX5B | Hypoxia-induced Cytochrome c oxidase polypeptide 5B is a terminal oxidase of the mitochondrial respiratory chain | Energy [42,43,44] | 11.0593 | 9.82351 × 10−9 | 2.82 × 10−6 |
| YC21B | Transposon Ty2-C Gag-Pol polyprotein | Unknown function in the cellular processes [45,46,47] | 14.18216 | 2.47468 × 10−6 | 0.000328 |
| YD22B | Transposon Ty2-DR2 Gag-Pol polyprotein | Unknown function in the cellular processes [45,46,47] | 14.18216 | 2.47468 × 10−6 | 0.000328 |
| KAPC | cAMP-dependent protein kinase type 3 essential member of the Ras signalling pathway | Regulation of cell growth, stress resistance and metabolism [48,49,50,51,52,53] | 12.55319 | 4.67411 × 10−6 | 0.000576 |
| HXT15 | Hexose transporter HXT15 promotes growth of non-fermentable carbon sources in case of glucose starvation | Energy/transport [54,55,56] | 9.376681 | 7.99509 × 10−5 | 0.00862 |
| RL7A | 60S ribosomal protein | Ribosome biogenesis [37,38,39] | 0.782346 | 0.000475619 | 0.043181 |
| IF4F2 | Eukaryotic initiation factor 4F subunit p130 limiting factor of translation initiation and ribosome recruitment | Translation initiation [57,58,59,60,61] | 0.574644 | 0.000406732 | 0.038978 |
| YD11A | Transposon Ty1-DR1 Gag polyprotein | Unknown function in the cellular processes [45,46,47] | −16.3721 | 5.51007 × 10−10 | 2.38 × 10−7 |
| YN12B | Transposon Ty1-NL2 Gag-Pol polyprotein | Unknown function in the cellular processes [45,46,47] | −10.17748 | 6.56423 × 10−8 | 1.59 × 10−5 |
| HMDH2 | Hypoxia induced 3-hydroxy–3-methylglutaryl-coenzyme A reductase 2 a rate-limiting member in sterol biosynthesis pathway | Lipid biosynthesis [62,63,64] | −13.22143 | 9.92516 × 10−8 | 1.9 × 10−5 |
| YIR7 | Y′ element ATP-dependent helicase YIL177C | Telomerase-independent telomere maintenance [65] | −10.7232 | 3.52 × 10−7 | 6.064 × 10−5 |
| PMA2 | Plasma membrane ATPase 2 creates proton gradient for secondary nutrient transport, elevated expression during carbon starvation | Nutrient transport and pH homeostasis [66,67,68,69] | −2.233914 | 1.33254 × 10−6 | 0.000209 |
| RSSA1 | 40S ribosomal protein | Ribosome biogenesis [37,38,39] | −1.852668 | 1.46542 × 10−5 | 0.001685 |
| RL22B | 60S ribosomal protein | Ribosome biogenesis [37,38,39] | −2.51894 | 0.000285 | 0.028895 |
| KPR4 | Ribose-phosphate pyrophosphokinase 4, necessary for de novo and salvage synthesis of nucleotides | Nucleotide metabolism [40,41] | −1.568709 | 0.000516 | 0.044519 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zinkevičiūtė, R.; Ražanskas, R.; Kaupinis, A.; Macijauskaitė, N.; Čiplys, E.; Houen, G.; Slibinskas, R. Yeast Secretes High Amounts of Human Calreticulin without Cellular Stress. Curr. Issues Mol. Biol. 2022, 44, 1768-1787. https://doi.org/10.3390/cimb44050122
Zinkevičiūtė R, Ražanskas R, Kaupinis A, Macijauskaitė N, Čiplys E, Houen G, Slibinskas R. Yeast Secretes High Amounts of Human Calreticulin without Cellular Stress. Current Issues in Molecular Biology. 2022; 44(5):1768-1787. https://doi.org/10.3390/cimb44050122
Chicago/Turabian StyleZinkevičiūtė, Rūta, Raimundas Ražanskas, Algirdas Kaupinis, Neringa Macijauskaitė, Evaldas Čiplys, Gunnar Houen, and Rimantas Slibinskas. 2022. "Yeast Secretes High Amounts of Human Calreticulin without Cellular Stress" Current Issues in Molecular Biology 44, no. 5: 1768-1787. https://doi.org/10.3390/cimb44050122
APA StyleZinkevičiūtė, R., Ražanskas, R., Kaupinis, A., Macijauskaitė, N., Čiplys, E., Houen, G., & Slibinskas, R. (2022). Yeast Secretes High Amounts of Human Calreticulin without Cellular Stress. Current Issues in Molecular Biology, 44(5), 1768-1787. https://doi.org/10.3390/cimb44050122







