Validation Study to Determine the Accuracy of Widespread Promoterless EGFP Reporter at Assessing CRISPR/Cas9-Mediated Homology Directed Repair
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
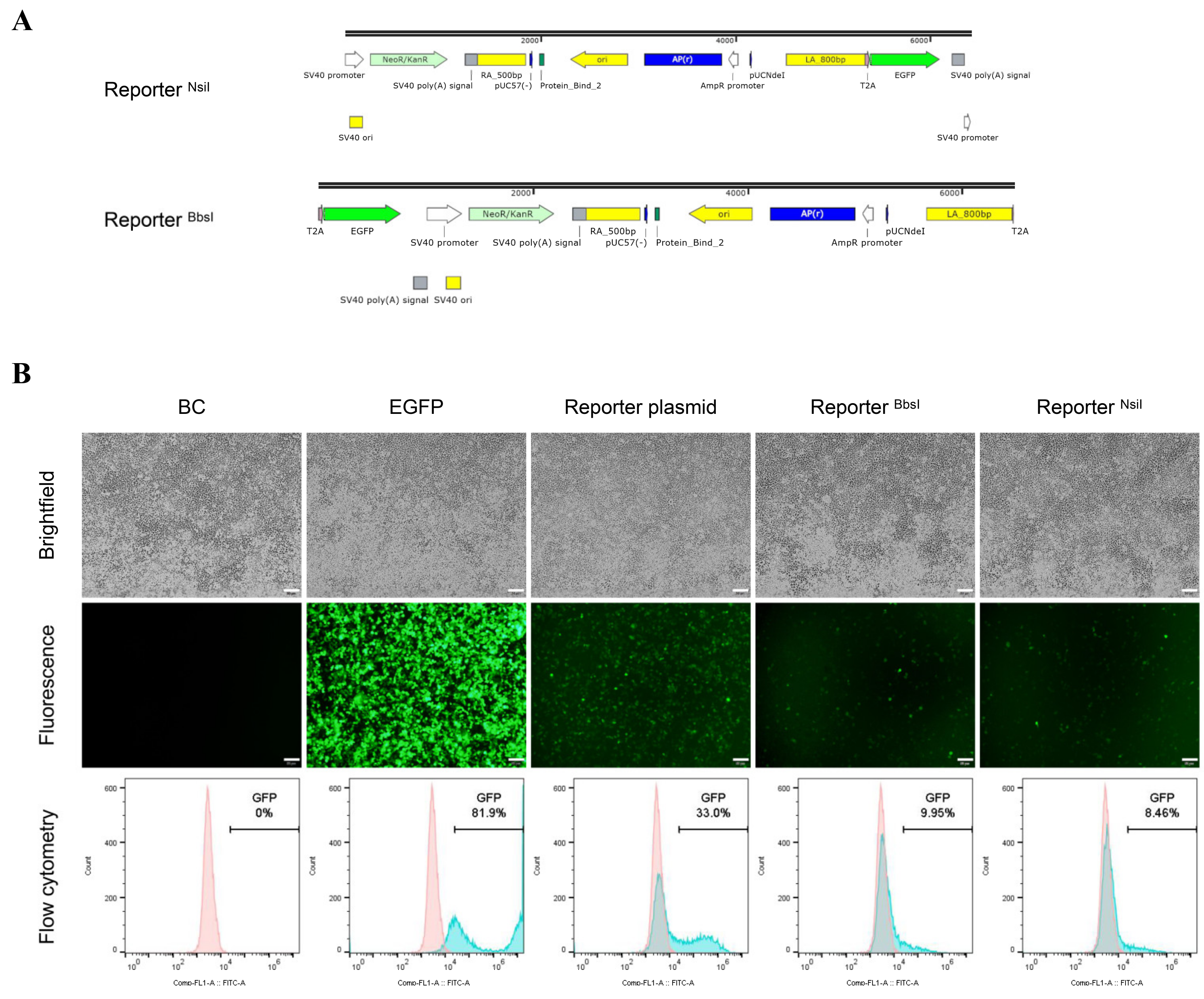
2.2. Design and Construction of the Promoterless EGFP Reporter System to Assess CRISPR/Cas9—Mediated HDR
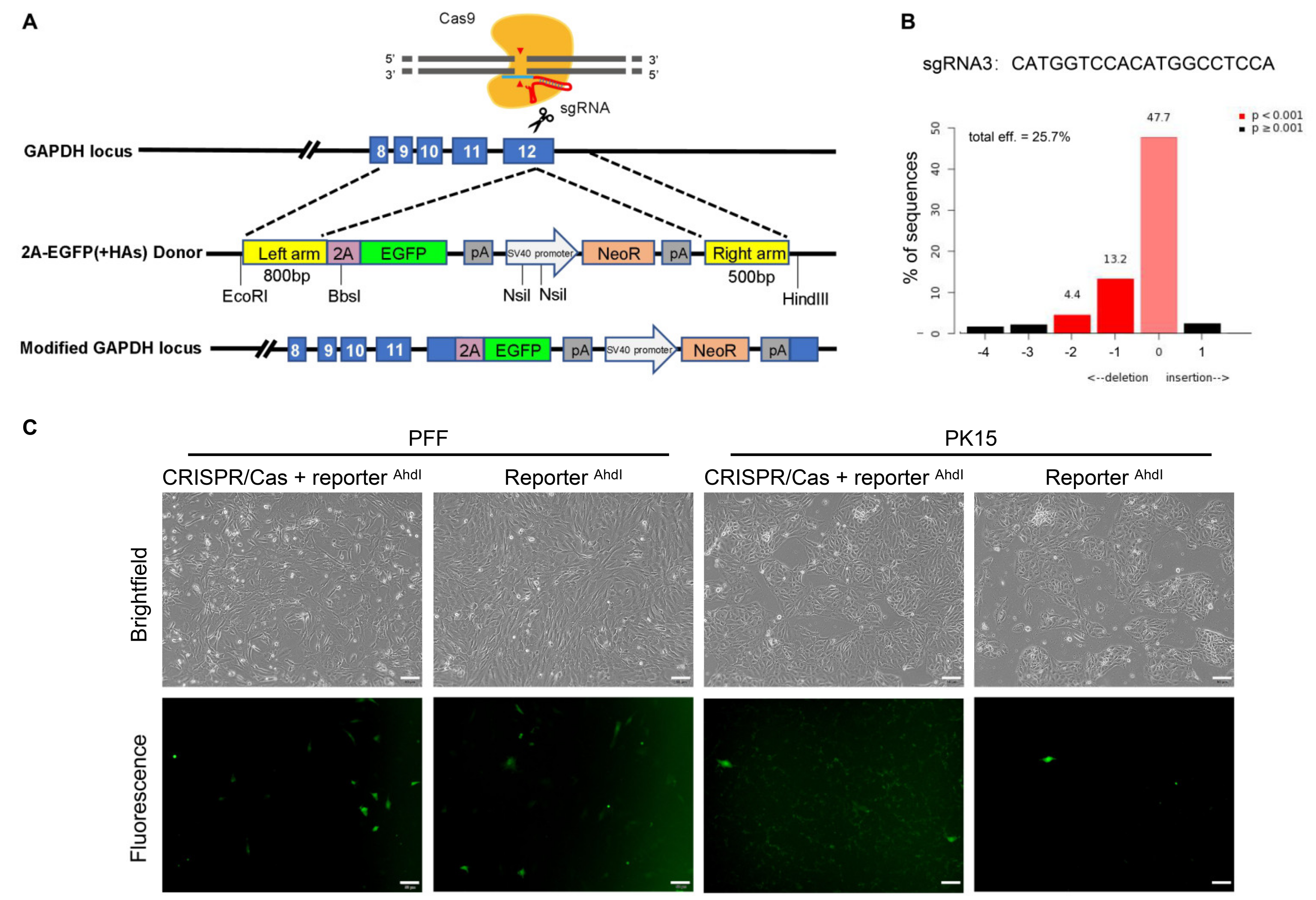
2.3. Assessing the Efficiency of Cas9-Mediated HDR in Porcine Cells Using the Promoterless EGFP Donor
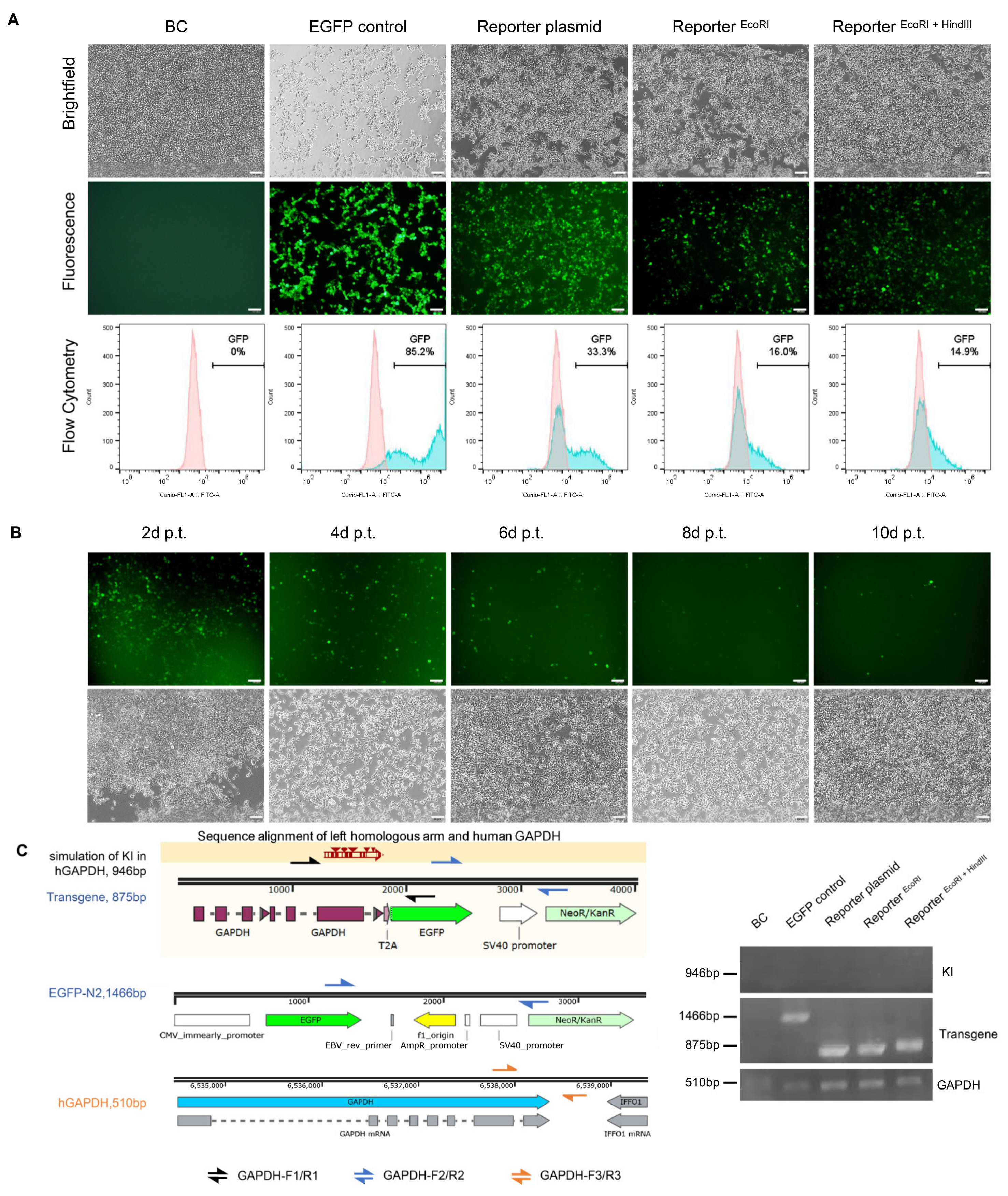
2.4. Evaluating the Promoterless EGFP Reporter in Porcine and Non-Porcine Cells
2.5. Flow Cytometry
2.6. Molecular Analysis of the Targeted Loci
2.7. Predicting Transcription Factor Binding Sites in Different Species
3. Results
3.1. Successful Establishment of the Promoterless EGFP Reporter System and sgRNA Targeting the Porcine GAPDH Locus
3.2. Unexpected EGFP Expression in Porcine Cells Transfected with the Promoterless EGFP Donor and Cas9/sgRNA
3.3. Higher EGFP Expression in Non-Porcine Cells Transfected with the Porcine Promoterless EGFP Donor Targeting GAPDH

3.4. No Precise Insertion but Random Integration in the EGFP Positive Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carlson-Stevermer, J.; Abdeen, A.A.; Kohlenberg, L.; Goedland, M.; Molugu, K.; Lou, M.; Saha, K. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat. Commun. 2017, 8, 1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 2014, 83, 409–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2016, 19, 1–9. [Google Scholar] [CrossRef]
- Xu, K.; Segal, D.J.; Zhang, Z. Editorial: Precise Genome Editing Techniques and Applications. Front. Genet. 2020, 11, 412. [Google Scholar] [CrossRef] [Green Version]
- Ling, X.; Xie, B.; Gao, X.; Chang, L.; Zheng, W. Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates. Sci. Adv. 2020, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Wang, H.; Zhang, X.; Wu, Z.; Yang, H. A Cas9-transcription factor fusion protein enhances homology-directed repair efficiency. J. Biol. Chem. 2021, 296, 100525. [Google Scholar] [CrossRef]
- Cai, Y.; Cheng, T.; Yao, Y.; Li, X.; Ma, Y. In vivo genome editing rescues photoreceptor degeneration via a Cas9/RecA-mediated homology-directed repair pathway. Sci. Adv. 2019, 5, 3335. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Cheng, J.K.W.; Zhang, Q.; Hughes, N.W.; Xia, Q.; Winslow, M.M.; Cong, L. Microbial single-strand annealing proteins enable CRISPR gene-editing tools with improved knock-in efficiencies and reduced off-target effects. Nucleic Acids Res. 2021, 49, e36. [Google Scholar] [CrossRef]
- Ding, X.; Seebeck, T.; Feng, Y.; Jiang, Y.; Davis, G.D.; Chen, F. Improving CRISPR-Cas9 Genome Editing Efficiency by Fusion with Chromatin-Modulating Peptides. CRISPR J. 2019, 2, 51–63. [Google Scholar] [CrossRef]
- Roche, P.J.R.; Gytz, H.; Hussain, F.; Cameron, C.J.F.; Paquette, D.; Blanchette, M.; Dostie, J.; Nagar, B.; Akavia, U.D. Double-Stranded Biotinylated Donor Enhances Homology-Directed Repair in Combination with Cas9 Monoavidin in Mammalian Cells. CRISPR J. 2018, 1, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Guo, Y.; Tian, Q.; Lan, Y.; Yeh, H.; Zhang, M.; Tasan, I.; Jain, S.; Zhao, H. An efficient gene knock-in strategy using 5′-modified double-stranded DNA donors with short homology arms. Nat. Chem. Biol. 2019, 16, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Chu, V.T.; Weber, T.; Wefers, B.; Wurst, W.; Sander, S.; Rajewsky, K.; Kuhn, R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015, 33, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 2014, 3, e04766. [Google Scholar] [CrossRef] [PubMed]
- Shahryari, A.; Moya, N.; Siehler, J.; Wang, X.; Burtscher, I.; Lickert, H. Increasing Gene Editing Efficiency for CRISPR-Cas9 by Small RNAs in Pluripotent Stem Cells. CRISPR J. 2021, 4, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Han, J.P.; Chang, Y.J.; Song, D.W.; Choi, B.S.; Koo, O.J.; Yi, S.Y.; Park, T.S.; Yeom, S.C. High Homology-Directed Repair Using Mitosis Phase and Nucleus Localizing Signal. Int. J. Mol. Sci. 2020, 21, 13747. [Google Scholar] [CrossRef]
- Yan, N.; Sun, Y.; Fang, Y.; Deng, J.; Mu, L.; Xu, K.; Mymryk, J.S.; Zhang, Z. A Universal Surrogate Reporter for Efficient Enrichment of CRISPR/Cas9-Mediated Homology-Directed Repair in Mammalian Cells. Mol. Ther. Nucleic Acids 2020, 19, 775–789. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, H.; Xiong, T.; Wang, J.; Yang, D.; Zhu, D.; Li, J.; Yang, Y.; Sun, C.; Zhao, Y.; et al. Suppression of SHROOM1 Improves In Vitro and In Vivo Gene Integration by Promoting Homology-Directed Repair. Int. J. Mol. Sci. 2020, 21, 65821. [Google Scholar] [CrossRef]
- Xie, Z.; Pang, D.; Wang, K.; Li, M.; Guo, N.; Yuan, H.; Tang, X. Optimization of a CRISPR/Cas9-mediated Knock-in Strategy at the Porcine Rosa26 Locus in Porcine Foetal Fibroblasts. Sci. Rep. 2017, 7, 3036. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Inoue, K.I.; Furuta, Y.; Kiyonari, H. Pronuclear Microinjection during S-Phase Increases the Efficiency of CRISPR-Cas9-Assisted Knockin of Large DNA Donors in Mouse Zygotes. Cell Rep. 2020, 31, 107653. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Becerra, G.; Kadonaga, J.T. Enhancement of homology-directed repair with chromatin donor templates in cells. eLife 2020, 9, e55780. [Google Scholar] [CrossRef] [PubMed]
- Paix, A.; Folkmann, A.; Goldman, D.H.; Kulaga, H.; Grzelak, M.J.; Rasoloson, D.; Seydoux, G. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl. Acad. Sci. USA 2017, 114, e10745–e10754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Qu, Y.; Cheng, J.K.W.; Hughes, N.W.; Zhang, Q.; Wang, M.; Cong, L. dCas9-based gene editing for cleavage-free genomic knock-in of long sequences. Nat. Cell Biol. 2022, 24, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Concordet, J.P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Haeussler, M.; Schonig, K.; Eckert, H.; Eschstruth, A.; Mianne, J.; Renaud, J.B.; Schneider-Maunoury, S.; Shkumatava, A.; Teboul, L.; Kent, J.; et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016, 17, 148. [Google Scholar] [CrossRef]
- Allen, F.; Crepaldi, L.; Alsinet, C.; Strong, A.J.; Kleshchevnikov, V.; De Angeli, P.; Palenikova, P.; Khodak, A.; Kiselev, V.; Kosicki, M.; et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2019, 37, 64–72. [Google Scholar] [CrossRef]
- Shen, M.W.; Arbab, M.; Hsu, J.Y.; Worstell, D.; Culbertson, S.J.; Krabbe, O.; Cassa, C.A.; Liu, D.R.; Gifford, D.K.; Sherwood, R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 2018, 563, 646–651. [Google Scholar] [CrossRef]
- Chen, W.; McKenna, A.; Schreiber, J.; Haeussler, M.; Yin, Y.; Agarwal, V.; Noble, W.S.; Shendure, J. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 2019, 47, 7989–8003. [Google Scholar] [CrossRef] [Green Version]
- Zhong, G.; Li, H.; Bai, J.; Pang, S.; He, C.; Du, X.; Wang, H.; Zhang, Q.; Xie, S.; Du, H.; et al. Advancing the predictivity of skin sensitization by applying a novel HMOX1 reporter system. Arch. Toxicol. 2018, 92, 3103–3115. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef] [PubMed]
- Paquet, D.; Kwart, D.; Chen, A.; Sproul, A.; Jacob, S.; Teo, S.; Olsen, K.M.; Gregg, A.; Noggle, S.; Tessier-Lavigne, M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 2016, 533, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Tatiossian, K.J.; Clark, R.D.E.; Huang, C.; Thornton, M.E.; Grubbs, B.H.; Cannon, P.M. Rational Selection of CRISPR-Cas9 Guide RNAs for Homology-Directed Genome Editing. Mol. Ther. 2021, 29, 1057–1069. [Google Scholar] [CrossRef]
- Liu, R.; Liang, L.; Freed, E.F.; Gill, R.T. Directed Evolution of CRISPR/Cas Systems for Precise Gene Editing. Trends Biotechnol. 2021, 39, 262–273. [Google Scholar] [CrossRef]
- He, X.; Tan, C.; Wang, F.; Wang, Y.; Zhou, R.; Cui, D.; You, W.; Zhao, H.; Ren, J.; Feng, B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016, 44, e85. [Google Scholar] [CrossRef] [Green Version]
- Mou, H.; Ozata, D.M.; Smith, J.L.; Sheel, A.; Kwan, S.Y.; Hough, S.; Kucukural, A.; Kennedy, Z.; Cao, Y.; Xue, W. CRISPR-SONIC: Targeted somatic oncogene knock-in enables rapid in vivo cancer modeling. Genome Med. 2019, 11, 21. [Google Scholar] [CrossRef]
- Mohammadi, Z.; Karamzadeh, A.; Tabatabaiefar, M.A.; Khanahmad, H. Evidence for expression of promoterless GFP cassette: Is GFP an ideal reporter gene in biotechnology science? Res. Pharm. Sci. 2019, 14, 351–358. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Zuo, Q.; Feng, D.; He, C.; Lin, C.; Huang, D.; Wan, Y.; Chen, F.; Mo, G.; Sun, Q.; et al. Validation Study to Determine the Accuracy of Widespread Promoterless EGFP Reporter at Assessing CRISPR/Cas9-Mediated Homology Directed Repair. Curr. Issues Mol. Biol. 2022, 44, 1688-1700. https://doi.org/10.3390/cimb44040116
Xu W, Zuo Q, Feng D, He C, Lin C, Huang D, Wan Y, Chen F, Mo G, Sun Q, et al. Validation Study to Determine the Accuracy of Widespread Promoterless EGFP Reporter at Assessing CRISPR/Cas9-Mediated Homology Directed Repair. Current Issues in Molecular Biology. 2022; 44(4):1688-1700. https://doi.org/10.3390/cimb44040116
Chicago/Turabian StyleXu, Wanqing, Qingxia Zuo, Dongyan Feng, Changsheng He, Cailing Lin, Dongchao Huang, Yanbin Wan, Feng Chen, Guosheng Mo, Qi Sun, and et al. 2022. "Validation Study to Determine the Accuracy of Widespread Promoterless EGFP Reporter at Assessing CRISPR/Cas9-Mediated Homology Directed Repair" Current Issues in Molecular Biology 44, no. 4: 1688-1700. https://doi.org/10.3390/cimb44040116
APA StyleXu, W., Zuo, Q., Feng, D., He, C., Lin, C., Huang, D., Wan, Y., Chen, F., Mo, G., Sun, Q., Du, H., & Huang, L. (2022). Validation Study to Determine the Accuracy of Widespread Promoterless EGFP Reporter at Assessing CRISPR/Cas9-Mediated Homology Directed Repair. Current Issues in Molecular Biology, 44(4), 1688-1700. https://doi.org/10.3390/cimb44040116





