MicroRNA-22-3p and MicroRNA-149-5p Inhibit Human Hepatocellular Carcinoma Cell Growth and Metastasis Properties by Regulating Methylenetetrahydrofolate Reductase
Abstract
:1. Introduction
2. Methods
2.1. Cell Culture
2.2. MiRNA Mimic Transfection
2.3. CCK-8 Detects Cell Proliferation
2.4. Real-Time miRNA/mRNA Expression Analyses
2.5. Western Blotting Analyses
2.6. Cell Invasion and Migration Assays
2.7. Statistical Analysis
3. Results
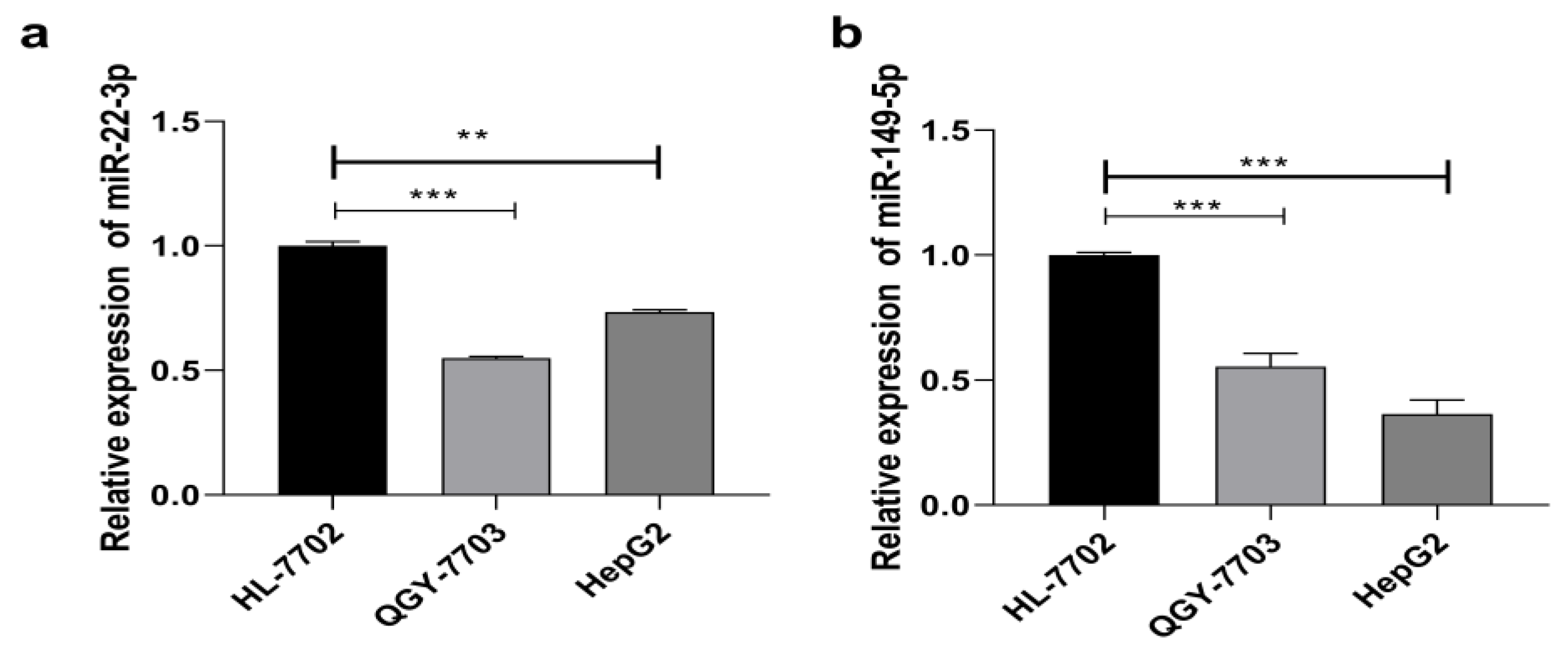
3.1. MiRNA-22-3p/miR-149-5p Are Downregulated in HCC Cell Lines
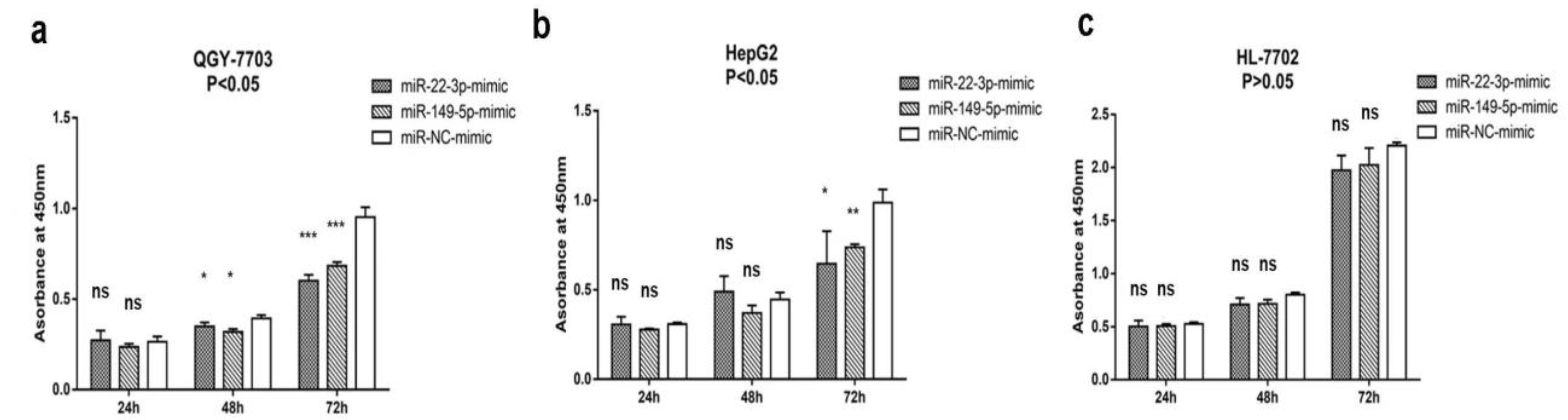
3.2. MiR-22-3p/miR-149-5p Inhibit Hepatocellular Carcinoma Cells Proliferation
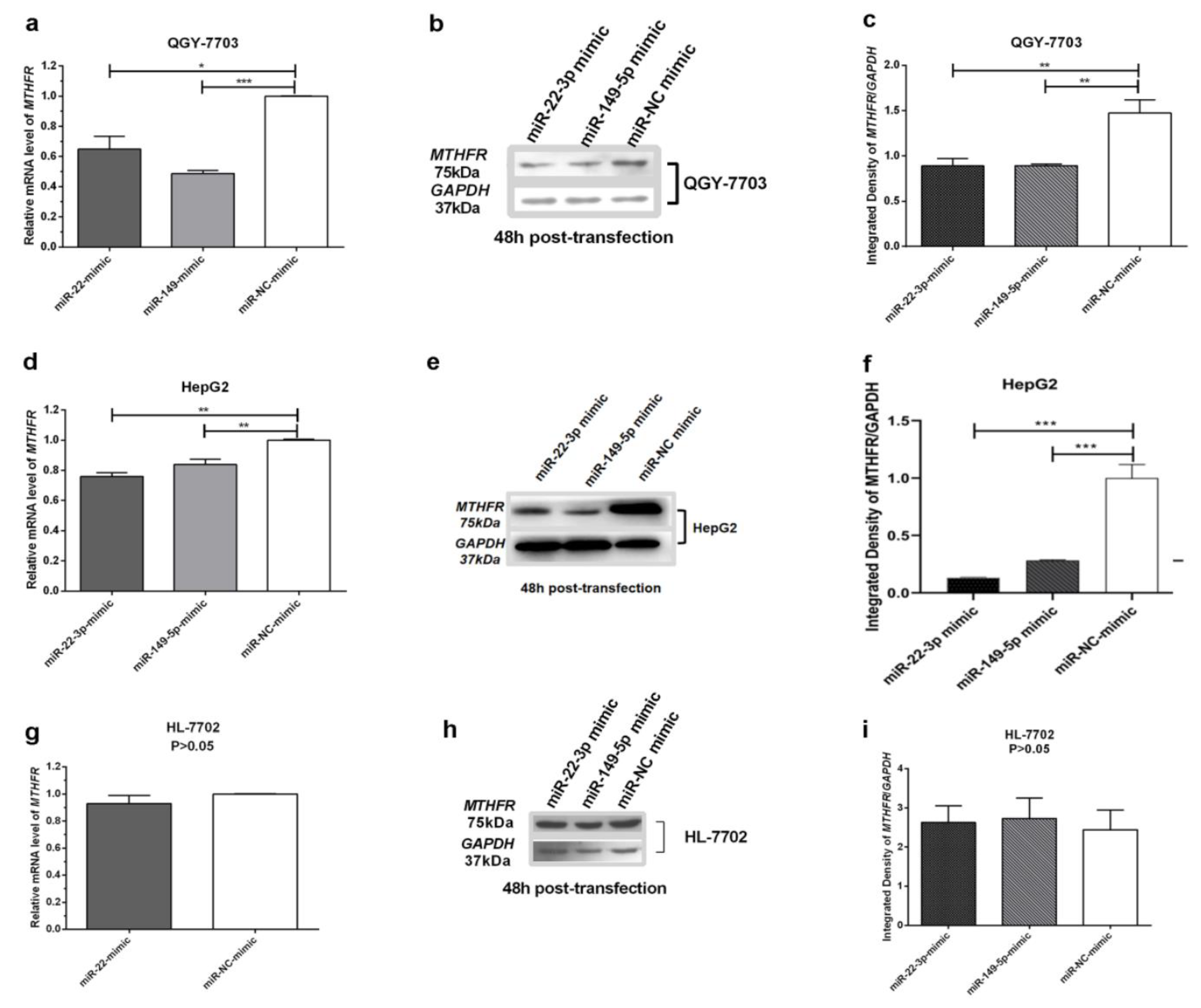
3.3. MiR-22-3p/miR-149-5p Mimic Inhibit the Expression of MTHFR in HCC Cell Lines
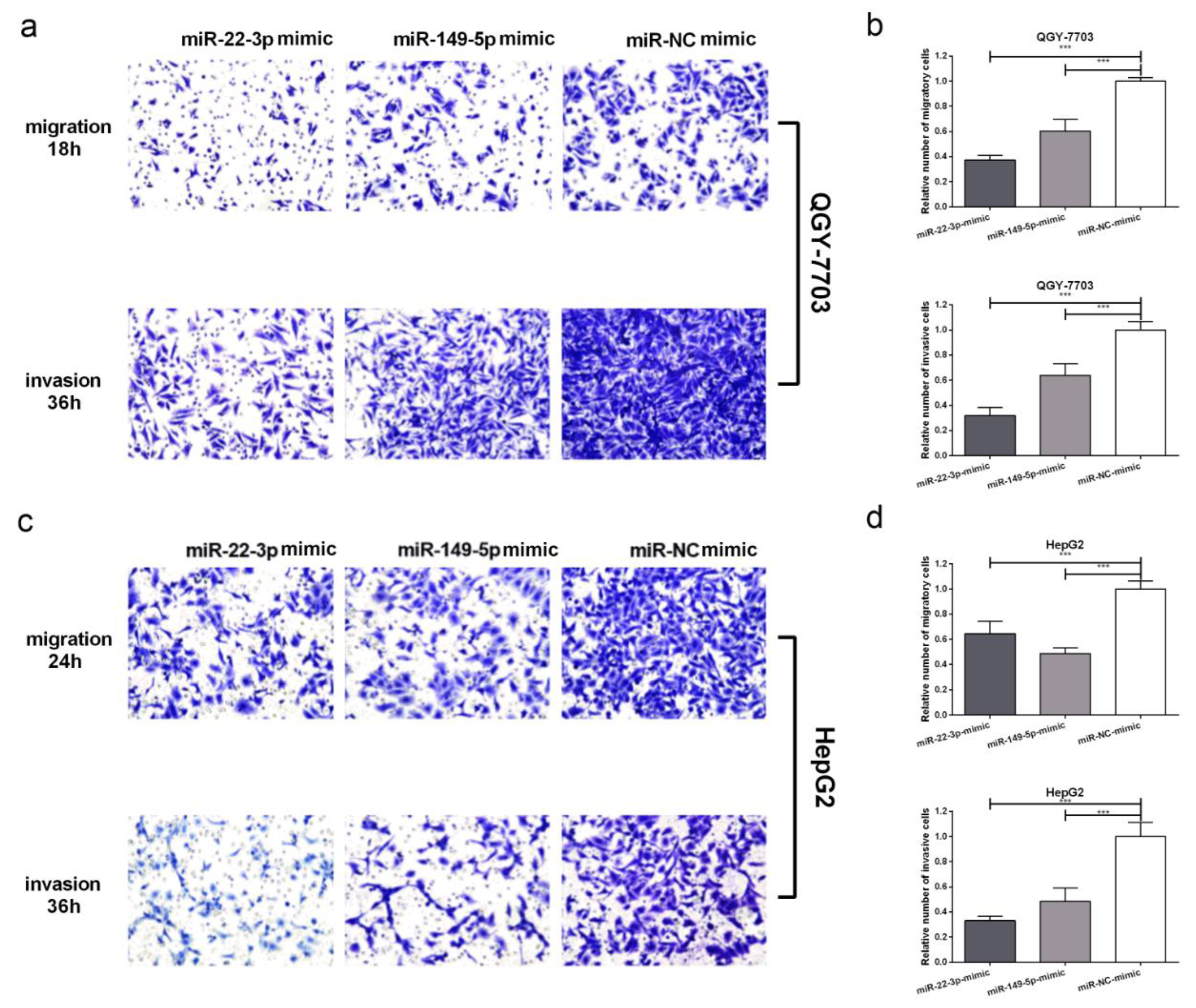
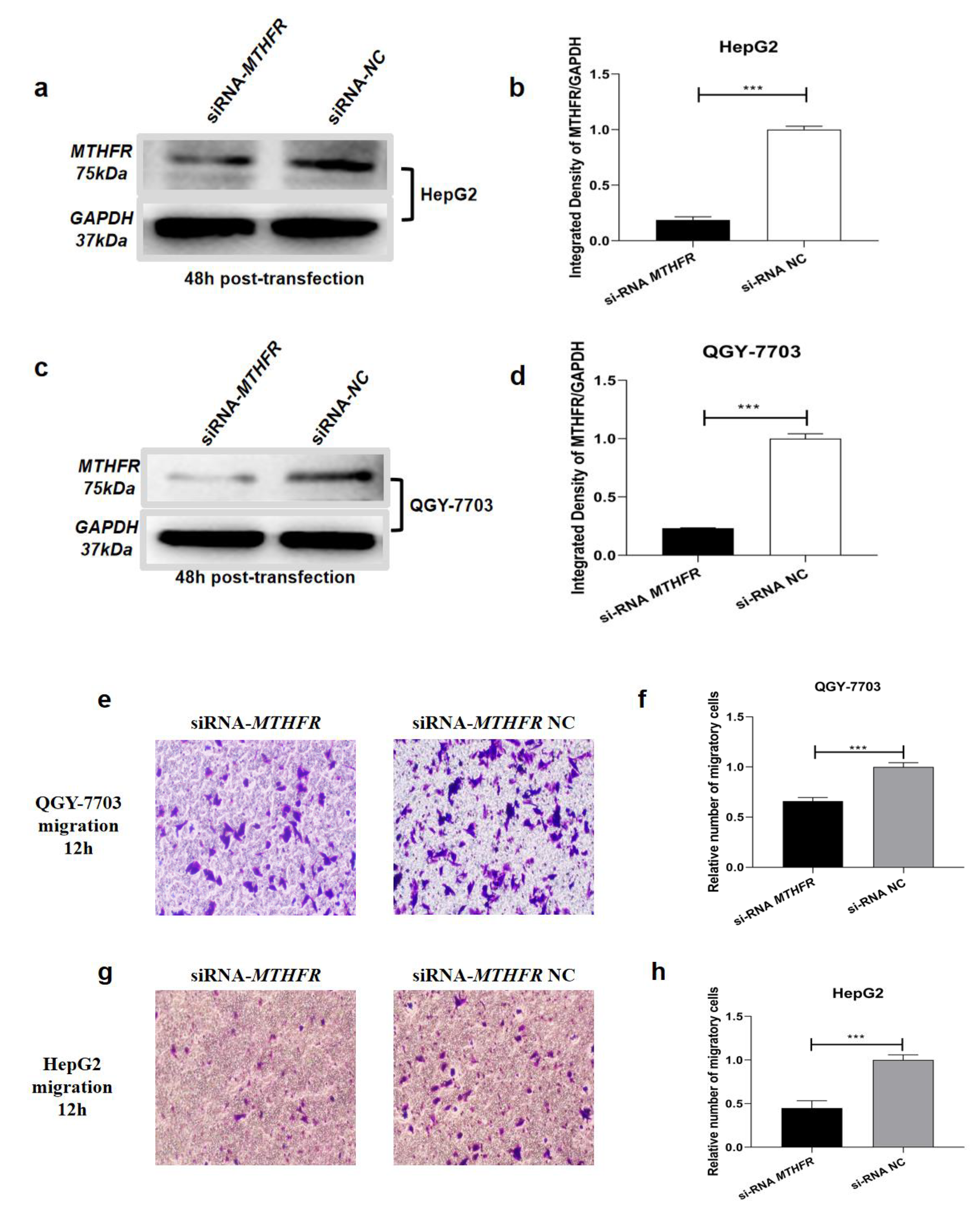
3.4. Transfection of miR-22-3p/miR-149-5p Mimic Suppresses Migration and Invasion in HCC Cells In Vitro
3.5. Transfection of miR-22-3p/miR-149-5p Inhibitor Promote Migration in HCC Cells In Vitro
3.6. Transfection of miR-22-3p/miR-149-5p Mimic Upregulation the Expression of PDCD4/TP53INP1 in QGY-7703
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El-Serag, H.B.; Mason, A.C. Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med. 1999, 340, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Chekulaeva, M.; Filipowicz, W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009, 21, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.A.; Liu, C.G.; Franssila, K.; Suster, S.; et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef]
- Sun, H.; Han, B.; Zhai, H.; Cheng, X.; Ma, K. Significant association between MTHFR C677T polymorphism and hepatocellular carcinoma risk: A meta-analysis. Tumour Biol. 2014, 35, 189–193. [Google Scholar] [CrossRef]
- Mu, L.N.; Cao, W.; Zhang, Z.F.; Cai, L.; Jiang, Q.W.; You, N.C.; Goldstein, B.Y.; Wei, G.R.; Chen, C.W.; Lu, Q.Y.; et al. Methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms and the risk of primary hepatocellular carcinoma (HCC) in a Chinese population. Cancer Causes Control. 2007, 18, 665–675. [Google Scholar] [CrossRef]
- Qiao, K.; Zhang, S.; Trieu, C.; Dai, Q.; Huo, Z.; Du, Y.; Lu, W.; Hou, W. Genetic Polymorphism of MTHFR C677T Influences Susceptibility to HBV-Related Hepatocellular Carcinoma in a Chinese Population: A Case-Control Study. Clin Lab. 2017, 63, 787–795. [Google Scholar] [CrossRef]
- Li, C.; Ni, J.; Liu, Y.X.; Wang, H.; Liang, Z.Q.; Wang, X. Response of MiRNA-22-3p and MiRNA-149-5p to Folate Deficiency and the Differential Regulation of MTHFR Expression in Normal and Cancerous Human Hepatocytes. PLoS ONE 2017, 12, e0168049. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Qiao, Z.G.; Li, J.; Liu, J.J.; Song, S.L.; Zhao, X.P.; Miao, P.; Tang, T.T.; Wang, L.; Liu, W.C.; et al. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: Evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget 2016, 7, 44252–44265. [Google Scholar] [CrossRef]
- Koufaris, C.; Valbuena, G.N.; Pomyen, Y.; Tredwell, G.D.; Nevedomskaya, E.; Lau, C.H.; Yang, T.; Benito, A.; Ellis, J.K.; Keun, H.C. Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene 2016, 35, 2766–2776. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.H.; Tang, J.L.; Li, C.; Kong, J.L.; Wang, J.; Wu, Y.H.; Xu, E.P.; Lai, M. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015, 356, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.X.; Wang, M.; Xu, D.; Yang, J.H.; Kang, H.F.; Wang, X.J.; Lin, S.; Yang, P.T.; Liu, X.H.; Dai, Z.J. Quantitative Assessment of the Association between Genetic Variants in MicroRNAs and Colorectal Cancer Risk. Biomed. Res. Int. 2015, 2015, 276410. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, J.W.; Yuan, Y. Comprehensive assessment of the association between miRNA polymorphisms and gastric cancer risk. Mutat. Res. Rev. Mutat. Res. 2015, 763, 148–160. [Google Scholar] [CrossRef]
- Yang, F.; Hu, Y.; Liu, H.X.; Wan, Y.J. MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J. Bio. Chem. 2015, 290, 6507–6515. [Google Scholar] [CrossRef]
- Chen, J.; Wu, F.X.; Luo, H.L.; Liu, J.J.; Luo, T.; Bai, T.; Li, L.Q.; Fan, X.H. Berberine upregulates miR-22-3p to suppress hepatocellular carcinoma cell proliferation by targeting Sp1. Am. J. Transl. Res. 2016, 8, 4932–4941. [Google Scholar]
- Chen, M.; Hu, W.; Xiong, C.L.; Qu, Z.; Yin, C.Q.; Wang, Y.H.; Luo, C.L.; Guan, Q.; Yuan, C.H.; Wang, F.B. miR-22 targets YWHAZ to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Oncotarget 2016, 7, 80751–80764. [Google Scholar] [CrossRef]
- Luo, L.J.; Zhang, L.P.; Duan, C.Y.; Wang, B.; He, N.N.; Abulimiti, P.; Lin, Yan. The inhibition role of miR-22 in hepatocellular carcinoma cell migration and invasion via targeting CD147. Cancer cell Int. 2017, 17, 17. [Google Scholar] [CrossRef]
- Luo, G.; Chao, Y.L.; Tang, B.; Li, B.S.; Xiao, Y.F.; Xie, R.; Wang, S.M.; Wu, Y.Y.; Dong, H.; Liu, X.D.; et al. miR-149 represses metastasis of hepatocellular carcinoma by targeting actin-regulatory proteins PPM1F. Oncotarget 2015, 6, 37808–37823. [Google Scholar] [CrossRef] [PubMed]
- Marsit, C.J.; Eddy, K.; Kelsey, K.T. MicroRNA responses to cellular stress. Cancer Res. 2006, 66, 10843–10848. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, H.; Li, N.; Li, X.P.; Zhang, H.B.; Song, Q.B.; Peng, M. MicroRNA-641 inhibits lung cancer cells proliferation, metastasis but promotes apoptosis in cells by targeting PDCD4. Int J Clin Exp Pathol. 2017, 10, 8211–8221. [Google Scholar] [PubMed]
- Li, N.; Cui, T.; Guo, W.; Wang, D.; Mao, L. MiR-155-5p accelerates the metastasis of cervical cancer cell via targeting TP53INP1. Onco Targets Ther. 2019, 12, 3181–3196. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Li, X.; Wang, H.; Guo, X.; Xue, J.; Wang, X.; Ni, J. MicroRNA-22-3p and MicroRNA-149-5p Inhibit Human Hepatocellular Carcinoma Cell Growth and Metastasis Properties by Regulating Methylenetetrahydrofolate Reductase. Curr. Issues Mol. Biol. 2022, 44, 952-962. https://doi.org/10.3390/cimb44020063
Li C, Li X, Wang H, Guo X, Xue J, Wang X, Ni J. MicroRNA-22-3p and MicroRNA-149-5p Inhibit Human Hepatocellular Carcinoma Cell Growth and Metastasis Properties by Regulating Methylenetetrahydrofolate Reductase. Current Issues in Molecular Biology. 2022; 44(2):952-962. https://doi.org/10.3390/cimb44020063
Chicago/Turabian StyleLi, Chao, Xiang Li, Han Wang, Xihan Guo, Jinglun Xue, Xu Wang, and Juan Ni. 2022. "MicroRNA-22-3p and MicroRNA-149-5p Inhibit Human Hepatocellular Carcinoma Cell Growth and Metastasis Properties by Regulating Methylenetetrahydrofolate Reductase" Current Issues in Molecular Biology 44, no. 2: 952-962. https://doi.org/10.3390/cimb44020063
APA StyleLi, C., Li, X., Wang, H., Guo, X., Xue, J., Wang, X., & Ni, J. (2022). MicroRNA-22-3p and MicroRNA-149-5p Inhibit Human Hepatocellular Carcinoma Cell Growth and Metastasis Properties by Regulating Methylenetetrahydrofolate Reductase. Current Issues in Molecular Biology, 44(2), 952-962. https://doi.org/10.3390/cimb44020063





