Identification and Characterization of Polymorphisms in piRNA Regions
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Statistical Analysis
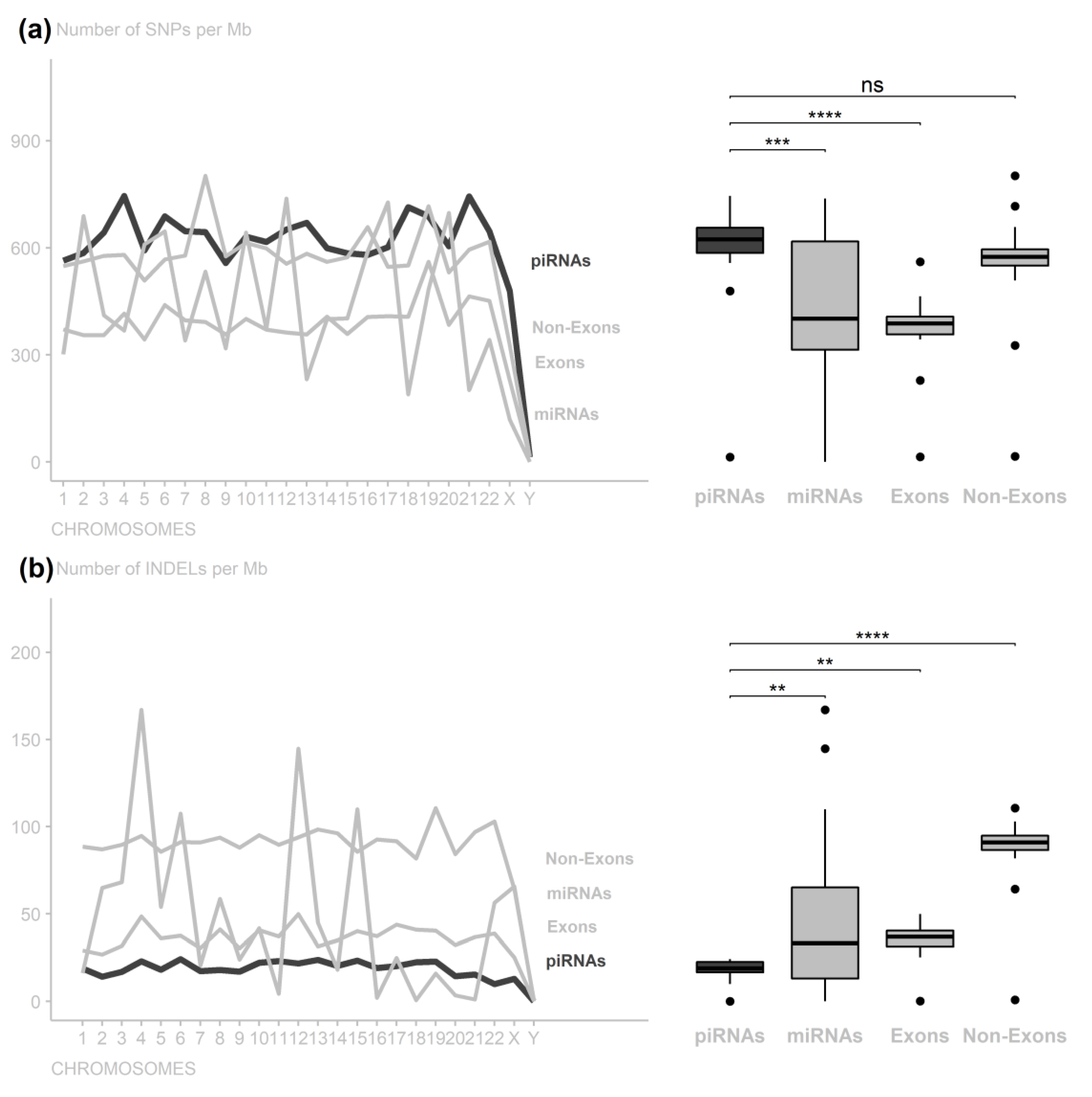
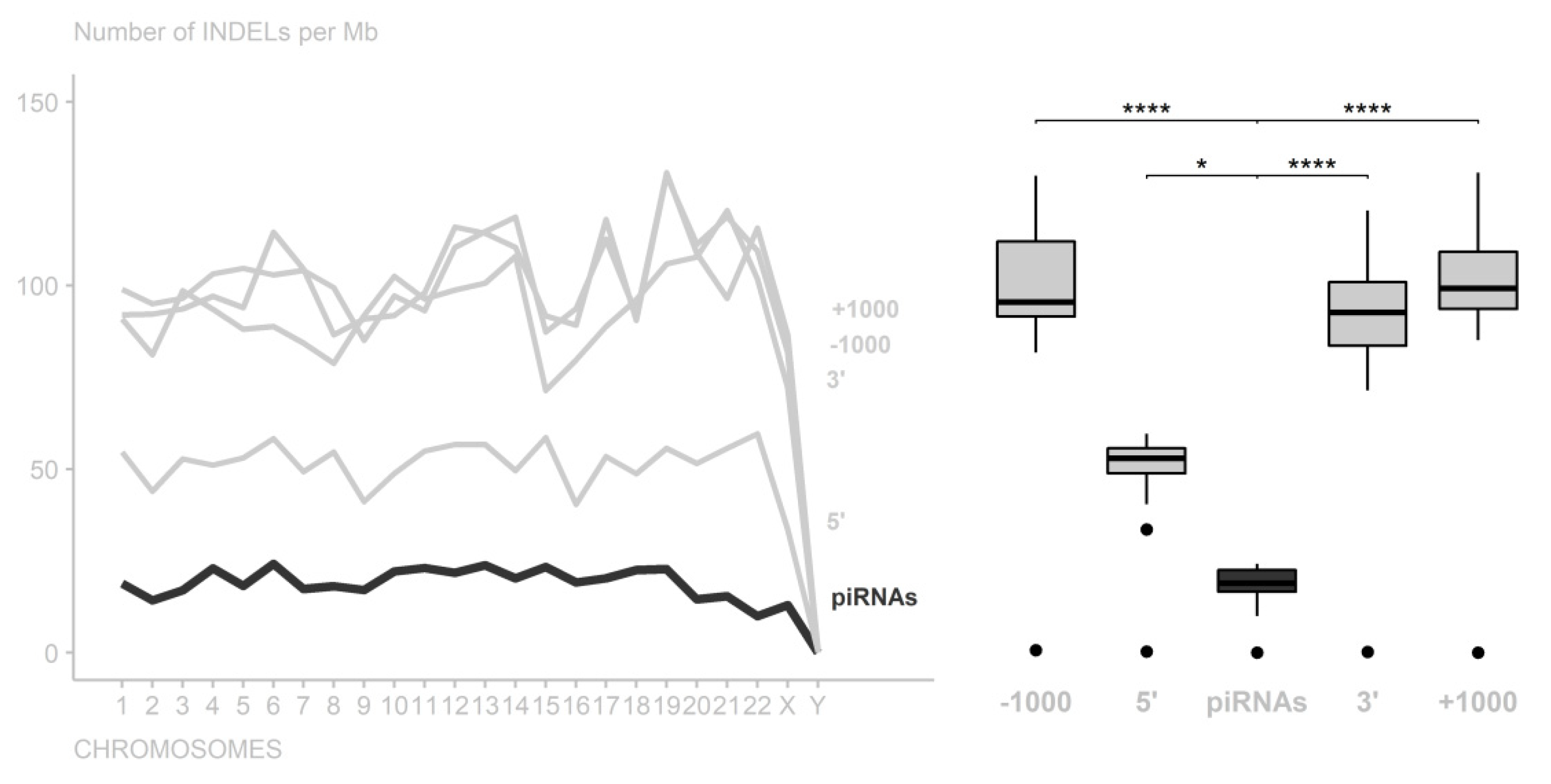
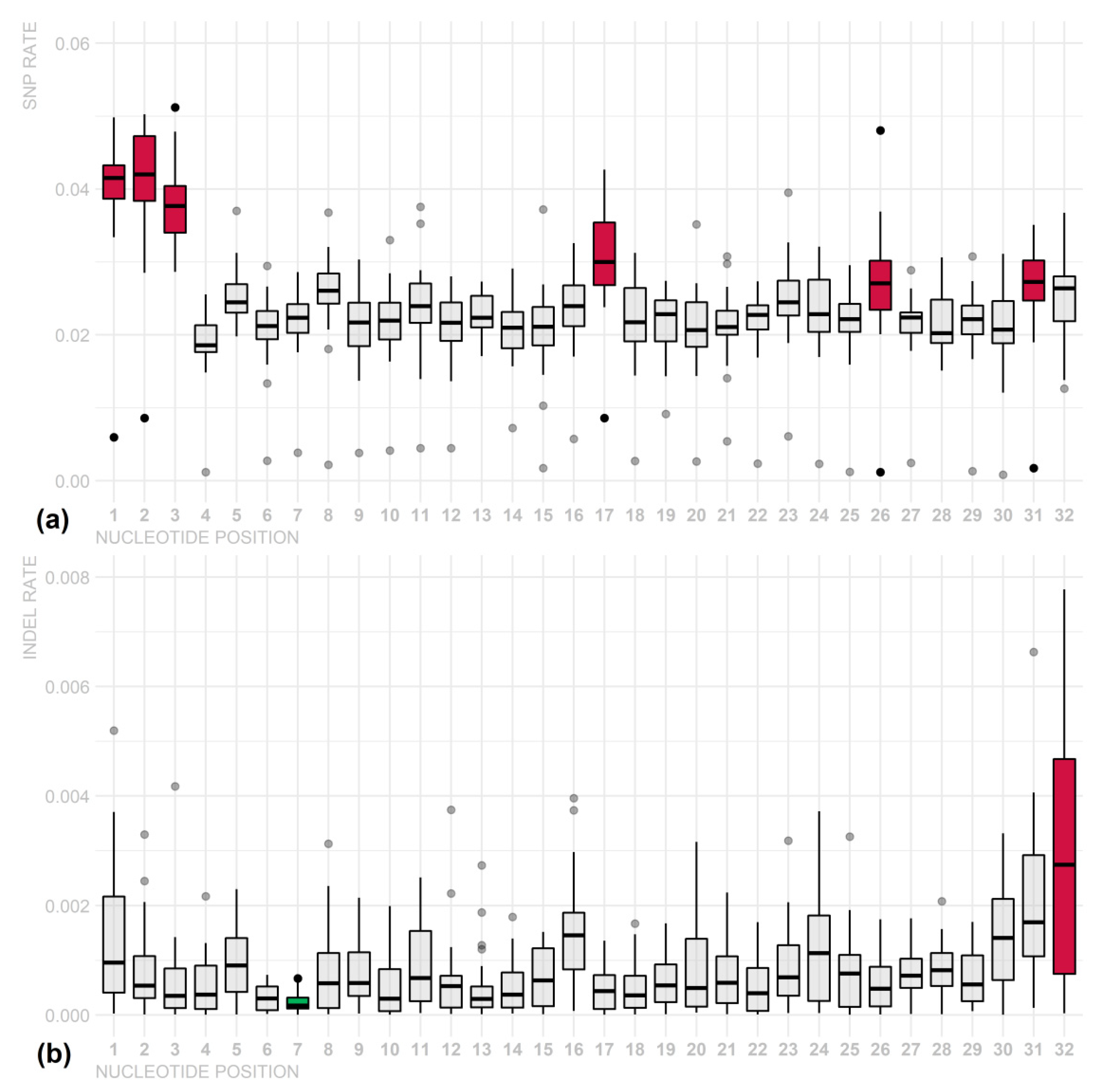
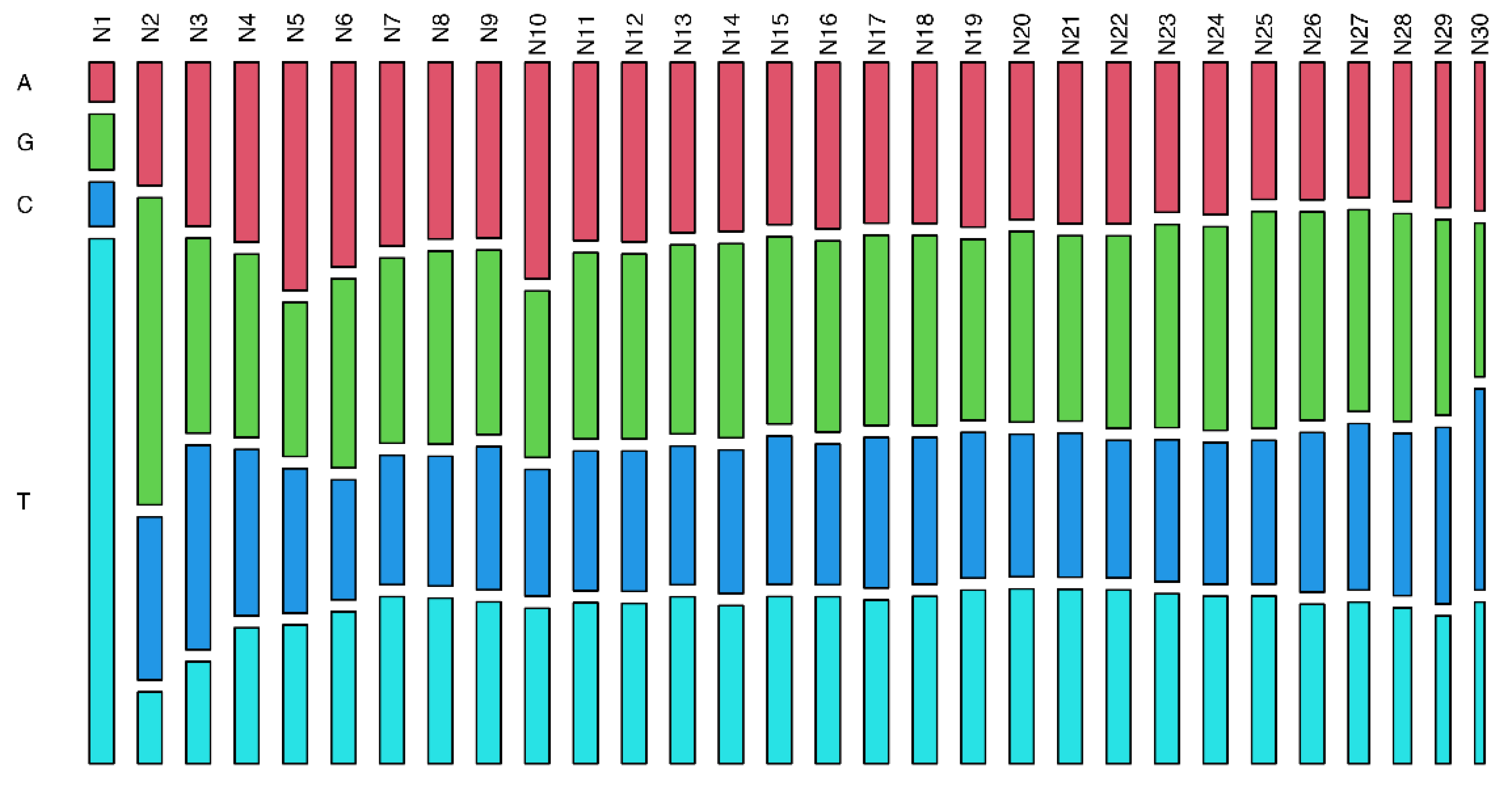
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Eddy, S.R. The ENCODE project: Missteps overshadowing a success. CURBIO 2013, 23, R259–R261. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Samuels, D.; Zhao, S.; Xiang, Y.; Zhao, Y.; Guo, Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes 2017, 8, 366. [Google Scholar] [CrossRef] [PubMed]
- Rands, C.M.; Meader, S.; Ponting, C.P.; Lunter, G. 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage. PLoS Genet. 2014, 10, e1004525. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Clark, D.; Mao, L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013, 336, 46–52. [Google Scholar] [CrossRef]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The Small RNA Profile during Drosophila melanogaster Development. Dev. Cell 2003, 5, 337–350. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef]
- Grivna, S.T.; Beyret, E.; Wang, Z.; Lin, H. (2006)A novel class of small RNAs in the mouse permatogenic cells.pdf. Genes Dev. 2006, 20, 1709–1714. [Google Scholar] [CrossRef]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef]
- Li, P. Non-coding RNAs and gastric cancer. World J. Gastroenterol. 2014, 20, 5411. [Google Scholar] [CrossRef]
- Weick, E.-M.; Miska, E.A. piRNAs: From biogenesis to function. Development 2014, 141, 3458–3471. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Wang, P.J. Mammalian piRNAs. Spermatogenesis 2014, 4, e27889. [Google Scholar] [CrossRef]
- Ponnusamy, M.; Yan, K.; Liu, C.; Li, P.; Wang, K. European Journal of Cell Biology PIWI family emerging as a decisive factor of cell fate: An overview. Eur. J. Cell Biol. 2017, 96, 746–757. [Google Scholar] [CrossRef]
- Gou, L.-T.; Dai, P.; Yang, J.-H.; Xue, Y.; Hu, Y.-P.; Zhou, Y.; Kang, J.-Y.; Wang, X.; Li, H.; Hua, M.-M.; et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef]
- Goh, W.S.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. PiRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef]
- Martinez, V.D.; Vucic, E.A.; Thu, K.L.; Hubaux, R.; Enfield, K.S.S.; Pikor, L.A.; Becker-Santos, D.D.; Brown, C.J.; Lam, S.; Lam, W.L. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci. Rep. 2015, 5, 10423. [Google Scholar] [CrossRef]
- Cheng, J.; Guo, J.-M.; Xiao, B.-X.; Miao, Y.; Jiang, Z.; Zhou, H.; Li, Q.-N. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta 2011, 412, 1621–1625. [Google Scholar] [CrossRef]
- Firmino, N.; Martinez, V.D.; Rowbotham, D.A.; Enfield, K.S.S.; Bennewith, K.L.; Lam, W.L. HPV status is associated with altered PIWI-interacting RNA expression pattern in head and neck cancer. Oral Oncol. 2016, 55, 43–48. [Google Scholar] [CrossRef]
- Singh, G.; Roy, J.; Rout, P.; Mallick, B. Genome-wide profiling of the PIWI-interacting RNA-mRNA regulatory networks in epithelial ovarian cancers. PLoS ONE 2018, 13, 1–24. [Google Scholar] [CrossRef]
- Chu, H.; Xia, L.; Qiu, X.; Gu, D.; Zhu, L.; Jin, J.; Hui, G.; Hua, Q.; Du, M.; Tong, N.; et al. Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer 2015, 121, 2044–2052. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.; Jacobs, D.I.; Hoffman, A.E.; Zheng, T.; Zhu, Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis 2015, 36, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, D.I.; Qin, Q.; Lerro, M.C.; Fu, A.; Dubrow, R.; Claus, E.B.; DeWan, A.T.; Wang, G.; Lin, H.; Zhu, Y. PIWI-interacting RNAs in gliomagenesis: Evidence from post-GWAS and functional analyses. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1073–1080. [Google Scholar] [CrossRef]
- Sudmant, P.H.; Rausch, T.; Gardner, E.J.; Handsaker, R.E.; Abyzov, A.; Huddleston, J.; Zhang, Y.; Ye, K.; Jun, G.; Hsi-Yang Fritz, M.; et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015, 526, 75–81. [Google Scholar] [CrossRef]
- Zhang, P.; Si, X.; Skogerbø, G.; Wang, J.; Cui, D.; Li, Y.; Sun, X.; Liu, L.; Sun, B.; Chen, R.; et al. piRBase: A web resource assisting piRNA functional study. Database 2014, 2014, bau110. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. Systematic Analysis of microRNA Targeting Impacted by Small Insertions and Deletions in Human Genome. PLoS ONE 2012, 7, e46176. [Google Scholar] [CrossRef][Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Calcagno, Q.; da Silva Mota, E.R.; Moreira, F.C.; de Sousa, S.B.M.; Burbano, R.R.; Assumpção, P.P. Role of PIWI-Interacting RNA (piRNA) as Epigenetic Regulation. In Handbook of Nutrition, Diet, and Epigenetics; Springer International Publisher: Berlin/Heidelberg, Germany, 2017; pp. 1–23. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Knaus, B.J.; Grünwald, N.J. vcfr: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 2017, 17, 44–53. [Google Scholar] [CrossRef]
- Obenchain, V.; Lawrence, M.; Carey, V.; Gogarten, S.; Shannon, P.; Morgan, M. VariantAnnotation: A Bioconductor package for exploration and annotation of genetic variants. Bioinformatics 2014, 30, 2076–2078. [Google Scholar] [CrossRef]
- Jobling, M.A.; Tyler-Smith, C. Human Y-chromosome variation in the genome-sequencing era. Nat. Rev. Genet. 2017, 18, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wang, Q.; Long, Q.; Ng, B.L.; Swerdlow, H.; Burton, J.; Tyler-Smith, C. Human Y Chromosome Base-Substitution Mutation Rate Measured by Direct Sequencing in a Deep-Rooting Pedigree. Curr. Biol. 2009, 19, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Hinch, A.G.; Altemose, N.; Noor, N.; Donnelly, P.; Myers, S.R. Recombination in the Human Pseudoautosomal Region PAR1. PLoS Genet. 2014, 10, e1004503. [Google Scholar] [CrossRef] [PubMed]
- Mohandas, T.K.; Speed, R.M.; Passage, M.B.; Yen, P.H.; Chandley, A.C.; Shapiro, L.J. Role of the pseudoautosomal region in sex-chromosome pairing during male meiosis: Meiotic studies in a man with a deletion of distal Xp. Am. J. Hum. Genet. 1992, 51, 526–533. [Google Scholar]
- Montgomery, S.B.; Goode, D.L.; Kvikstad, E.; Albers, C.A.; Zhang, Z.D.; Mu, X.J.; Ananda, G.; Howie, B.; Karczewski, K.J.; Smith, K.S.; et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013, 23, 749–761. [Google Scholar] [CrossRef]
- Clark, T.G.; Andrew, T.; Cooper, G.M.; Margulies, E.H.; Mullikin, J.C.; Balding, D.J. Functional constraint and small insertions and deletions in the ENCODE regions of the human genome. Genome Biol. 2007, 8, 1–14. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ishizu, H.; Saito, K.; Fukuhara, S.; Kamatani, M.K.; Bonnefond, L.; Matsumoto, N.; Nishizawa, T.; Nakanaga, K.; Aoki, J.; et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 2012, 491, 284–287. [Google Scholar] [CrossRef]
- Wang, W.; Yoshikawa, M.; Han, B.W.; Izumi, N.; Tomari, Y.; Weng, Z.; Zamore, P.D. The Initial Uridine of Primary piRNAs Does Not Create the Tenth Adenine that Is the Hallmark of Secondary piRNAs. Mol. Cell 2014, 56, 708–716. [Google Scholar] [CrossRef]
- Gebert, D.; Ketting, R.F.; Zischler, H.; Rosenkranz, D. piRNAs from Pig Testis Provide Evidence for a Conserved Role of the Piwi Pathway in Post-Transcriptional Gene Regulation in Mammals. PLoS ONE 2015, 10, e0124860. [Google Scholar] [CrossRef] [PubMed]
- Saunders, M.A.; Liang, H.; Li, W.-H. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tu, S.; Stubna, M.; Wu, W.-S.; Huang, W.-C.; Weng, Z.; Lee, H.-C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 2018, 359, 587–592. [Google Scholar] [CrossRef]
- Shen, E.-Z.; Chen, H.; Ozturk, A.R.; Tu, S.; Shirayama, M.; Tang, W.; Ding, Y.-H.; Dai, S.-Y.; Weng, Z.; Mello, C.C. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 2018, 172, 937–951.e18. [Google Scholar] [CrossRef]
- Rojas-Ríos, P.; Simonelig, M. piRNAs and PIWI proteins: Regulators of gene expression in development and stem cells. Development 2018, 145, dev161786. [Google Scholar] [CrossRef]
- Vourekas, A.; Alexiou, P.; Vrettos, N.; Maragkakis, M.; Mourelatos, Z. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 2016, 531, 390–394. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lima, J.R.S.; Azevedo-Pinheiro, J.; Andrade, R.B.; Khayat, A.S.; Assumpção, P.P.d.; Ribeiro-dos-Santos, Â.; Batista dos Santos, S.E.; Moreira, F.C. Identification and Characterization of Polymorphisms in piRNA Regions. Curr. Issues Mol. Biol. 2022, 44, 942-951. https://doi.org/10.3390/cimb44020062
Lima JRS, Azevedo-Pinheiro J, Andrade RB, Khayat AS, Assumpção PPd, Ribeiro-dos-Santos Â, Batista dos Santos SE, Moreira FC. Identification and Characterization of Polymorphisms in piRNA Regions. Current Issues in Molecular Biology. 2022; 44(2):942-951. https://doi.org/10.3390/cimb44020062
Chicago/Turabian StyleLima, José Roberto Sobrinho, Jhully Azevedo-Pinheiro, Roberta Borges Andrade, André Salim Khayat, Paulo Pimentel de Assumpção, Ândrea Ribeiro-dos-Santos, Sidney Emanuel Batista dos Santos, and Fabiano Cordeiro Moreira. 2022. "Identification and Characterization of Polymorphisms in piRNA Regions" Current Issues in Molecular Biology 44, no. 2: 942-951. https://doi.org/10.3390/cimb44020062
APA StyleLima, J. R. S., Azevedo-Pinheiro, J., Andrade, R. B., Khayat, A. S., Assumpção, P. P. d., Ribeiro-dos-Santos, Â., Batista dos Santos, S. E., & Moreira, F. C. (2022). Identification and Characterization of Polymorphisms in piRNA Regions. Current Issues in Molecular Biology, 44(2), 942-951. https://doi.org/10.3390/cimb44020062






